Microglia Mitigate Neuronal Activation in a Zebrafish Model of Dravet Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Maintenance and Transgenic Lines
2.2. Morpholino Knockdown
2.3. Microglia Morphological Analysis
2.4. Morphological Clustering of Microglial Cells
2.5. RNA Isolation and Quantitative RT-PCR
2.6. Analysis of the Dynamics of Microglia Morphology Changes
2.7. Calcium Imaging
2.8. Local Field Potential (LFP) Recording
2.9. Locomotor Activity
2.10. Data Analysis
3. Results
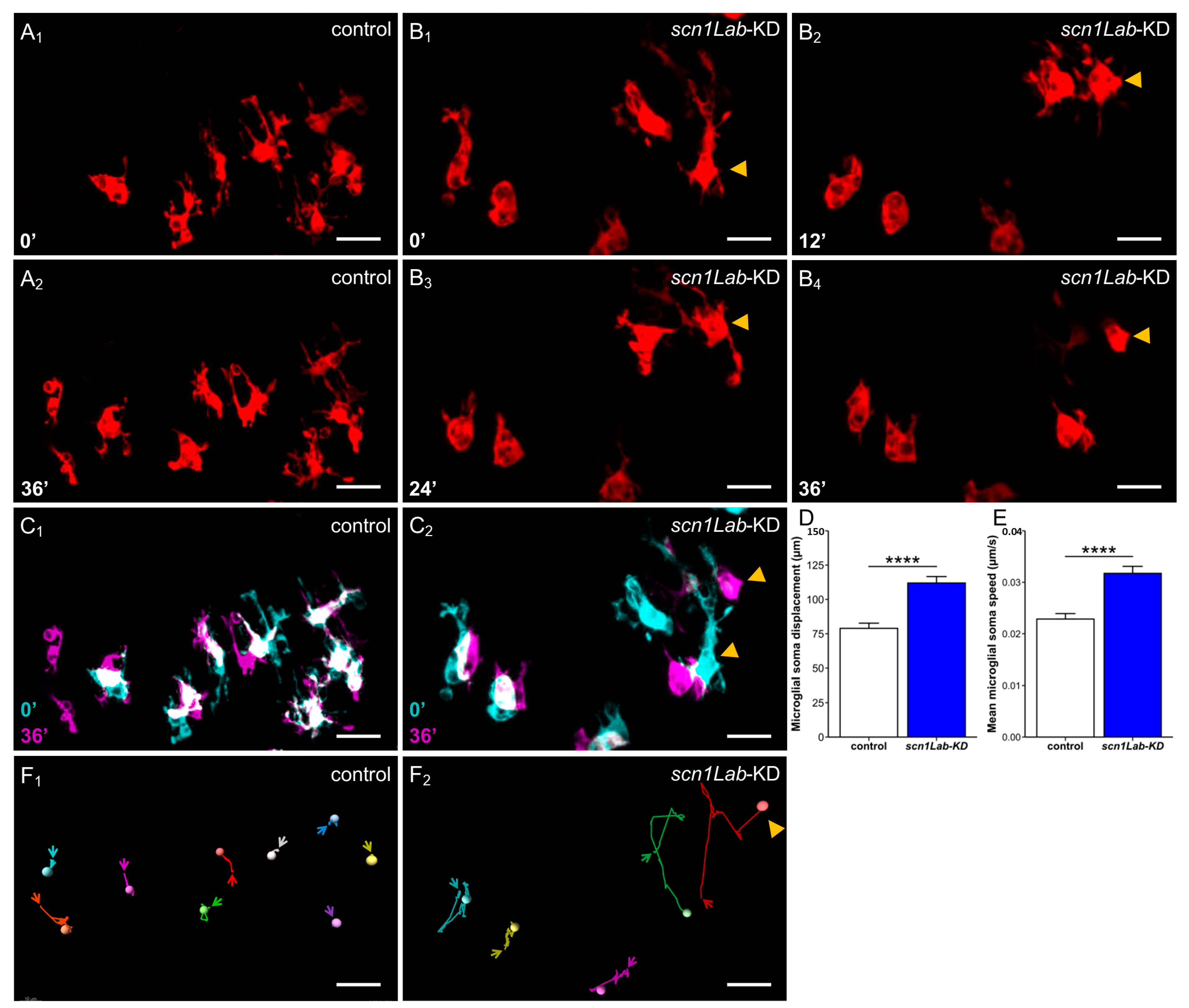
3.1. Microglia Morphology Changes in scn1Lab-KD Larvae
3.2. Increased Microglia-Mediated Brain Inflammation in scn1Lab-KD Larvae
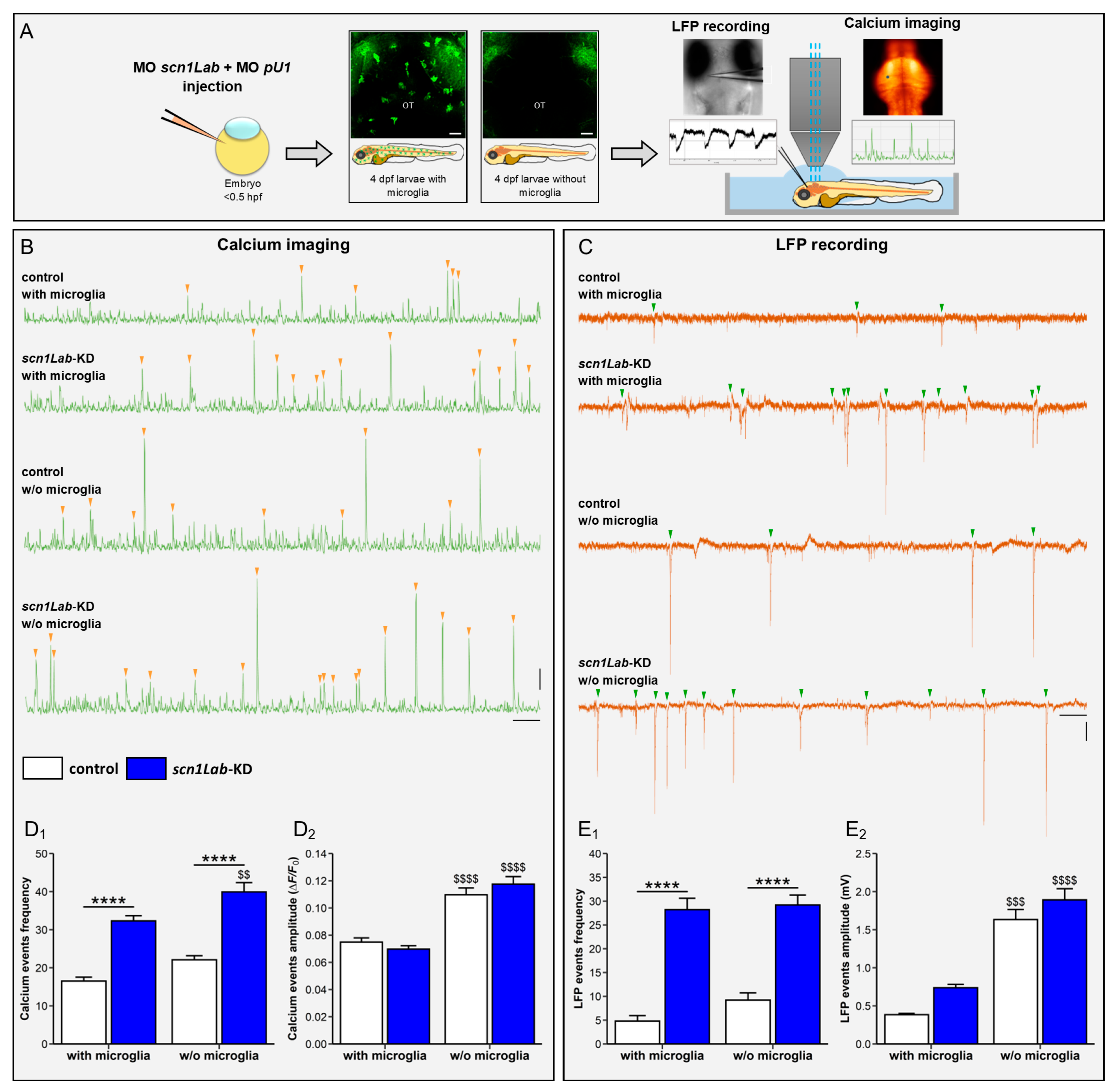
3.3. Microglia Ablation Increases Neuronal Activation in scn1Lab-KD Larvae
3.4. Microglia Ablation Exacerbates Seizure-like Swimming Behavior of scn1Lab-KD Larvae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE Official Report: A Practical Clinical Definition of Epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.S. Redefining Epilepsy. Curr. Opin. Neurol. 2015, 28, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Thurman, D.J.; Beghi, E.; Begley, C.E.; Berg, A.T.; Buchhalter, J.R.; Ding, D.; Hesdorffer, D.C.; Hauser, W.A.; Kazis, L.; Kobau, R.; et al. Standards for Epidemiologic Studies and Surveillance of Epilepsy. Epilepsia 2011, 52, 2–26. [Google Scholar] [CrossRef] [PubMed]
- Stafstrom, C.E.; Carmant, L. Seizures and Epilepsy: An Overview for Neuroscientists. Cold Spring Harb. Perspect. Biol. 2015, 7, a022426. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Epilepsy. Available online: https://www.who.int/news-room/fact-sheets/detail/epilepsy (accessed on 6 April 2024).
- Kwan, P.; Schachter, S.C.; Brodie, M.J. Drug-Resistant Epilepsy. N. Engl. J. Med. 2011, 365, 2238–2240. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Sullivan, J.; McDaniel, S.S.; Meisler, M.H.; Walsh, E.M.; Li, S.X.; Kuzniewicz, M.W. Incidence of Dravet Syndrome in a US Population. Pediatrics 2015, 136, e1310–e1315. [Google Scholar] [CrossRef] [PubMed]
- Brunklaus, A.; Dorris, L.; Zuberi, S.M. Comorbidities and Predictors of Health-Related Quality of Life in Dravet Syndrome. Epilepsia 2011, 52, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Marxer, C.A.; Rüegg, S.; Rauch, M.S.; Panchaud, A.; Meier, C.R.; Spoendlin, J. A Review of the Evidence on the Risk of Congenital Malformations and Neurodevelopmental Disorders in Association with Antiseizure Medications during Pregnancy. Expert Opin. Drug Saf. 2021, 20, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Xue-Ping, W.; Hai-Jiao, W.; Li-Na, Z.; Xu, D.; Ling, L. Risk Factors for Drug-Resistant Epilepsy: A Systematic Review and Meta-Analysis. Medicine 2019, 98, e16402. [Google Scholar] [CrossRef]
- Vezzani, A.; Balosso, S.; Ravizza, T. Neuroinflammatory Pathways as Treatment Targets and Biomarkers in Epilepsy. Nat. Rev. Neurol. 2019, 15, 459–472. [Google Scholar] [CrossRef]
- Lukens, J.R.; Eyo, U.B. Microglia and Neurodevelopmental Disorders. Annu. Rev. Neurosci. 2022, 45, 425–445. [Google Scholar] [CrossRef]
- Woodburn, S.C.; Bollinger, J.L.; Wohleb, E.S. The Semantics of Microglia Activation: Neuroinflammation, Homeostasis, and Stress. J. Neuroinflamm. 2021, 18, 258. [Google Scholar] [CrossRef]
- Ravizza, T.; Gagliardi, B.; Noé, F.; Boer, K.; Aronica, E.; Vezzani, A. Innate and Adaptive Immunity during Epileptogenesis and Spontaneous Seizures: Evidence from Experimental Models and Human Temporal Lobe Epilepsy. Neurobiol. Dis. 2008, 29, 142–160. [Google Scholar] [CrossRef]
- Vezzani, A. Fetal Brain Inflammation May Prime Hyperexcitability and Behavioral Dysfunction Later in Life. Ann. Neurol. 2013, 74, 1–3. [Google Scholar] [CrossRef]
- Vezzani, A.; Ravizza, T.; Balosso, S.; Aronica, E. Glia as a Source of Cytokines: Implications for Neuronal Excitability and Survival. Epilepsia 2008, 49, 24–32. [Google Scholar] [CrossRef]
- Boer, K.; Jansen, F.; Nellist, M.; Redeker, S.; van den Ouweland, A.M.W.; Spliet, W.G.M.; van Nieuwenhuizen, O.; Troost, D.; Crino, P.B.; Aronica, E. Inflammatory Processes in Cortical Tubers and Subependymal Giant Cell Tumors of Tuberous Sclerosis Complex. Epilepsy Res. 2008, 78, 7–21. [Google Scholar] [CrossRef]
- Morin-Brureau, M.; Milior, G.; Royer, J.; Chali, F.; LeDuigou, C.; Savary, E.; Blugeon, C.; Jourdren, L.; Akbar, D.; Dupont, S.; et al. Microglial Phenotypes in the Human Epileptic Temporal Lobe. Brain 2018, 141, 3343–3360. [Google Scholar] [CrossRef]
- Eyo, U.B.; Peng, J.; Swiatkowski, P.; Mukherjee, A.; Bispo, A.; Wu, L.-J. Neuronal Hyperactivity Recruits Microglial Processes via Neuronal NMDA Receptors and Microglial P2Y12 Receptors after Status Epilepticus. J. Neurosci. 2014, 34, 10528–10540. [Google Scholar] [CrossRef]
- Avignone, E.; Lepleux, M.; Angibaud, J.; Nägerl, U.V. Altered Morphological Dynamics of Activated Microglia after Induction of Status Epilepticus. J. Neuroinflamm. 2015, 12, 202. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, N.; Navarro, F.P.; Ogier, M.; Belmeguenaï, A.; Lieutaud, T.; Georges, B.; Bodennec, J.; Guénot, M.; Streichenberger, N.; Ryvlin, P.; et al. Low Grade Inflammation in the Epileptic Hippocampus Contrasts with Explosive Inflammation Occurring in the Acute Phase Following Status Epilepticus in Rats: Translation to Patients with Epilepsy. bioRxiv 2021, bioRxiv:03.25.436701. [Google Scholar] [CrossRef]
- Vezzani, A.; Conti, M.; De Luigi, A.; Ravizza, T.; Moneta, D.; Marchesi, F.; De Simoni, M.G. Interleukin-1β Immunoreactivity and Microglia Are Enhanced in the Rat Hippocampus by Focal Kainate Application: Functional Evidence for Enhancement of Electrographic Seizures. J. Neurosci. 1999, 19, 5054–5065. [Google Scholar] [CrossRef] [PubMed]
- Andoh, M.; Ikegaya, Y.; Koyama, R. Synaptic Pruning by Microglia in Epilepsy. J. Clin. Med. 2019, 8, 2170. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.; Musto, A.E. The Role of Inflammation in the Development of Epilepsy. J. Neuroinflamm. 2018, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Badimon, A.; Strasburger, H.J.; Ayata, P.; Chen, X.; Nair, A.; Ikegami, A.; Hwang, P.; Chan, A.T.; Graves, S.M.; Uweru, J.O.; et al. Negative Feedback Control of Neuronal Activity by Microglia. Nature 2020, 586, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Du, X.; Liu, C.-S.; Wen, Z.-L.; Du, J.-L. Reciprocal Regulation between Resting Microglial Dynamics and Neuronal Activity In Vivo. Dev. Cell 2012, 23, 1189–1202. [Google Scholar] [CrossRef] [PubMed]
- Vinet, J.; van Weering, H.R.J.; Heinrich, A.; Kälin, R.E.; Wegner, A.; Brouwer, N.; Heppner, F.L.; van Rooijen, N.; Boddeke, H.W.G.M.; Biber, K. Neuroprotective Function for Ramified Microglia in Hippocampal Excitotoxicity. J. Neuroinflamm. 2012, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.J.; Manzanero, S.; Borges, K. Complex Alterations in Microglial M1/M2 Markers during the Development of Epilepsy in Two Mouse Models. Epilepsia 2015, 56, 895–905. [Google Scholar] [CrossRef]
- Dravet, C. Dravet Syndrome History. Dev. Med. Child Neurol. 2011, 53, 1–6. [Google Scholar] [CrossRef]
- Claes, L.; Del-Favero, J.; Ceulemans, B.; Lagae, L.; Broeckhoven, C.V.; De Jonghe, P. De Novo Mutations in the Sodium-Channel Gene SCN1A Cause Severe Myoclonic Epilepsy of Infancy. Am. J. Hum. Genet. 2001, 68, 1327–1332. [Google Scholar] [CrossRef]
- Brenet, A.; Hassan-Abdi, R.; Somkhit, J.; Yanicostas, C.; Soussi-Yanicostas, N. Defective Excitatory/Inhibitory Synaptic Balance and Increased Neuron Apoptosis in a Zebrafish Model of Dravet Syndrome. Cells 2019, 8, 1199. [Google Scholar] [CrossRef]
- Schoonheim, P.J.; Arrenberg, A.B.; Del Bene, F.; Baier, H. Optogenetic Localization and Genetic Perturbation of Saccade-Generating Neurons in Zebrafish. J. Neurosci. 2010, 30, 7111–7120. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, J.; Chen, T.W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderón, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef] [PubMed]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. Mpeg1 Promoter Transgenes Direct Macrophage-Lineage Expression in Zebrafish. Blood 2011, 117, e49. [Google Scholar] [CrossRef]
- Nguyen-Chi, M.; Laplace-Builhe, B.; Travnickova, J.; Luz-Crawford, P.; Tejedor, G.; Phan, Q.T.; Duroux-Richard, I.; Levraud, J.P.; Kissa, K.; Lutfalla, G.; et al. Identification of Polarized Macrophage Subsets in Zebrafish. eLife 2015, 4, e07288. [Google Scholar] [CrossRef] [PubMed]
- Hassan-Abdi, R.; Brenet, A.; Bennis, M.; Yanicostas, C.; Soussi-Yanicostas, N. Neurons Expressing Pathological Tau Protein Trigger Dramatic Changes in Microglial Morphology and Dynamics. Front. Neurosci. 2019, 13, 1199. [Google Scholar] [CrossRef] [PubMed]
- Somkhit, J.; Yanicostas, C.; Soussi-Yanicostas, N. Microglia Remodelling and Neuroinflammation Parallel Neuronal Hyperactivation Following Acute Organophosphate Poisoning. Int. J. Mol. Sci. 2022, 23, 8240. [Google Scholar] [CrossRef] [PubMed]
- Baraban, S.C.; Dinday, M.T.; Hortopan, G.A. Drug Screening in Scn1a Zebrafish Mutant Identifies Clemizole as a Potential Dravet Syndrome Treatment. Nat. Commun. 2013, 4, 2410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kecskés, A.; Copmans, D.; Langlois, M.; Crawford, A.D.; Ceulemans, B.; Lagae, L.; de Witte, P.A.M.; Esguerra, C.V. Pharmacological Characterization of an Antisense Knockdown Zebrafish Model of Dravet Syndrome: Inhibition of Epileptic Seizures by the Serotonin Agonist Fenfluramine. PLoS ONE 2015, 10, e0125898. [Google Scholar] [CrossRef]
- Travnickova, J.; Chau, V.T.; Julien, E.; Mateos-Langerak, J.; Gonzalez, C.; Lelièvre, E.; Lutfalla, G.; Tavian, M.; Kissa, K. Primitive Macrophages Control HSPC Mobilization and Definitive Haematopoiesis. Nat. Commun. 2015, 6, 6227. [Google Scholar] [CrossRef]
- Sieger, D.; Moritz, C.; Ziegenhals, T.; Prykhozhij, S.; Peri, F. Long-Range Ca2+ Waves Transmit Brain-Damage Signals to Microglia. Dev. Cell 2012, 22, 1138–1148. [Google Scholar] [CrossRef]
- Mildner, A. Ghosts in the Shell: Identification of Microglia in the Human Central Nervous System by P2y12 Receptor. Neural Regen. Res. 2017, 12, 570–571. [Google Scholar] [CrossRef] [PubMed]
- Peri, F.; Nüsslein-Volhard, C. Live Imaging of Neuronal Degradation by Microglia Reveals a Role for V0-ATPase A1 in Phagosomal Fusion In Vivo. Cell 2008, 133, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Satta, V.; Alonso, C.; Díez, P.; Martín-Suárez, S.; Rubio, M.; Encinas, J.M.; Fernández-Ruiz, J.; Sagredo, O. Neuropathological Characterization of a Dravet Syndrome Knock-In Mouse Model Useful for Investigating Cannabinoid Treatments. Front. Mol. Neurosci. 2021, 13, 602801. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yao, X.; Taylor, N.; Bai, Y.; Lovenberg, T.; Bhattacharya, A. RNA Sequencing Analysis Reveals Quiescent Microglia Isolation Methods from Postnatal Mouse Brains and Limitations of BV2 Cells. J. Neuroinflamm. 2018, 15, 153. [Google Scholar] [CrossRef]
- Rhodes, J.; Hagen, A.; Hsu, K.; Deng, M.; Liu, T.X.; Look, A.T.; Kanki, J.P. Interplay of Pu.1 and Gata1 Determines Myelo-Erythroid Progenitor Cell Fate in Zebrafish. Dev. Cell 2005, 8, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Baraban, S.C. Network Properties Revealed during Multi-Scale Calcium Imaging of Seizure Activity in Zebrafish. eNeuro 2019, 6, ENEURO.0041-19.2019. [Google Scholar] [CrossRef] [PubMed]
- Vezzani, A.; Moneta, D.; Richichi, C.; Perego, C.; De Simoni, M.G. Functional Role of Proinflammatory and Anti-Inflammatory Cytokines in Seizures. Adv. Exp. Med. Biol. 2004, 548, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Maupu, C.; Enderlin, J.; Igert, A.; Oger, M.; Auvin, S.; Hassan-Abdi, R.; Soussi-Yanicostas, N.; Brazzolotto, X.; Nachon, F.; Dal Bo, G.; et al. Diisopropylfluorophosphate-Induced Status Epilepticus Drives Complex Glial Cell Phenotypes in Adult Male Mice. Neurobiol. Dis. 2021, 152, 105276. [Google Scholar] [CrossRef] [PubMed]
- Altmann, A.; Ryten, M.; Di Nunzio, M.; Ravizza, T.; Tolomeo, D.; Reynolds, R.H.; Somani, A.; Bacigaluppi, M.; Iori, V.; Micotti, E.; et al. A Systems-Level Analysis Highlights Microglial Activation as a Modifying Factor in Common Epilepsies. Neuropathol. Appl. Neurobiol. 2022, 48, e12758. [Google Scholar] [CrossRef]
- Nakajima, K.; Kohsaka, S. Microglia: Activation and Their Significance in the Central Nervous System. J. Biochem. 2001, 130, 169–175. [Google Scholar] [CrossRef]
- Leal, B.; Chaves, J.; Carvalho, C.; Rangel, R.; Santos, A.; Bettencourt, A.; Lopes, J.; Ramalheira, J.; Silva, B.M.; da Silva, A.M.; et al. Brain Expression of Inflammatory Mediators in Mesial Temporal Lobe Epilepsy Patients. J. Neuroimmunol. 2017, 313, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Salgueiro-Pereira, A.R.; Duprat, F.; Pousinha, P.A.; Loucif, A.; Douchamps, V.; Regondi, C.; Ayrault, M.; Eugie, M.; Stunault, M.I.; Escayg, A.; et al. A Two-Hit Story: Seizures and Genetic Mutation Interaction Sets Phenotype Severity in SCN1A Epilepsies. Neurobiol. Dis. 2019, 125, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Mirrione, M.M.; Konomos, D.K.; Gravanis, I.; Dewey, S.L.; Aguzzi, A.; Heppner, F.L.; Tsirka, S.E. Microglial Ablation and Lipopolysaccharide Preconditioning Affects Pilocarpine-Induced Seizures in Mice. Neurobiol. Dis. 2010, 39, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Di Nunzio, M.; Di Sapia, R.; Sorrentino, D.; Kebede, V.; Cerovic, M.; Gullotta, G.S.; Bacigaluppi, M.; Audinat, E.; Marchi, N.; Ravizza, T.; et al. Microglia Proliferation Plays Distinct Roles in Acquired Epilepsy Depending on Disease Stages. Epilepsia 2021, 62, 1931–1945. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brenet, A.; Somkhit, J.; Csaba, Z.; Ciura, S.; Kabashi, E.; Yanicostas, C.; Soussi-Yanicostas, N. Microglia Mitigate Neuronal Activation in a Zebrafish Model of Dravet Syndrome. Cells 2024, 13, 684. https://doi.org/10.3390/cells13080684
Brenet A, Somkhit J, Csaba Z, Ciura S, Kabashi E, Yanicostas C, Soussi-Yanicostas N. Microglia Mitigate Neuronal Activation in a Zebrafish Model of Dravet Syndrome. Cells. 2024; 13(8):684. https://doi.org/10.3390/cells13080684
Chicago/Turabian StyleBrenet, Alexandre, Julie Somkhit, Zsolt Csaba, Sorana Ciura, Edor Kabashi, Constantin Yanicostas, and Nadia Soussi-Yanicostas. 2024. "Microglia Mitigate Neuronal Activation in a Zebrafish Model of Dravet Syndrome" Cells 13, no. 8: 684. https://doi.org/10.3390/cells13080684
APA StyleBrenet, A., Somkhit, J., Csaba, Z., Ciura, S., Kabashi, E., Yanicostas, C., & Soussi-Yanicostas, N. (2024). Microglia Mitigate Neuronal Activation in a Zebrafish Model of Dravet Syndrome. Cells, 13(8), 684. https://doi.org/10.3390/cells13080684







