Investigating the Impact of IL6 on Insulin Secretion: Evidence from INS-1 Cells, Human Pancreatic Islets, and Serum Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomic Analysis of Human Pancreatic Islets
2.2. Human Sampling and Data Collection
2.3. INS-1 Cell Culture Condition and Treatments
2.4. Human Islets Culture Condition and Treatments
2.5. siRNA Transfection
2.6. Insulin Release Assay
2.7. Western Blot Analysis
2.8. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.9. Cell Viability and Apoptosis Assay
2.10. Glucose Uptake
2.11. Reactive Oxygen Species (ROS) Measurements
2.12. Statistical Analysis
3. Results
3.1. RNA-Seq Expression Analysis of IL6 and IL6R in Human Pancreatic Islets
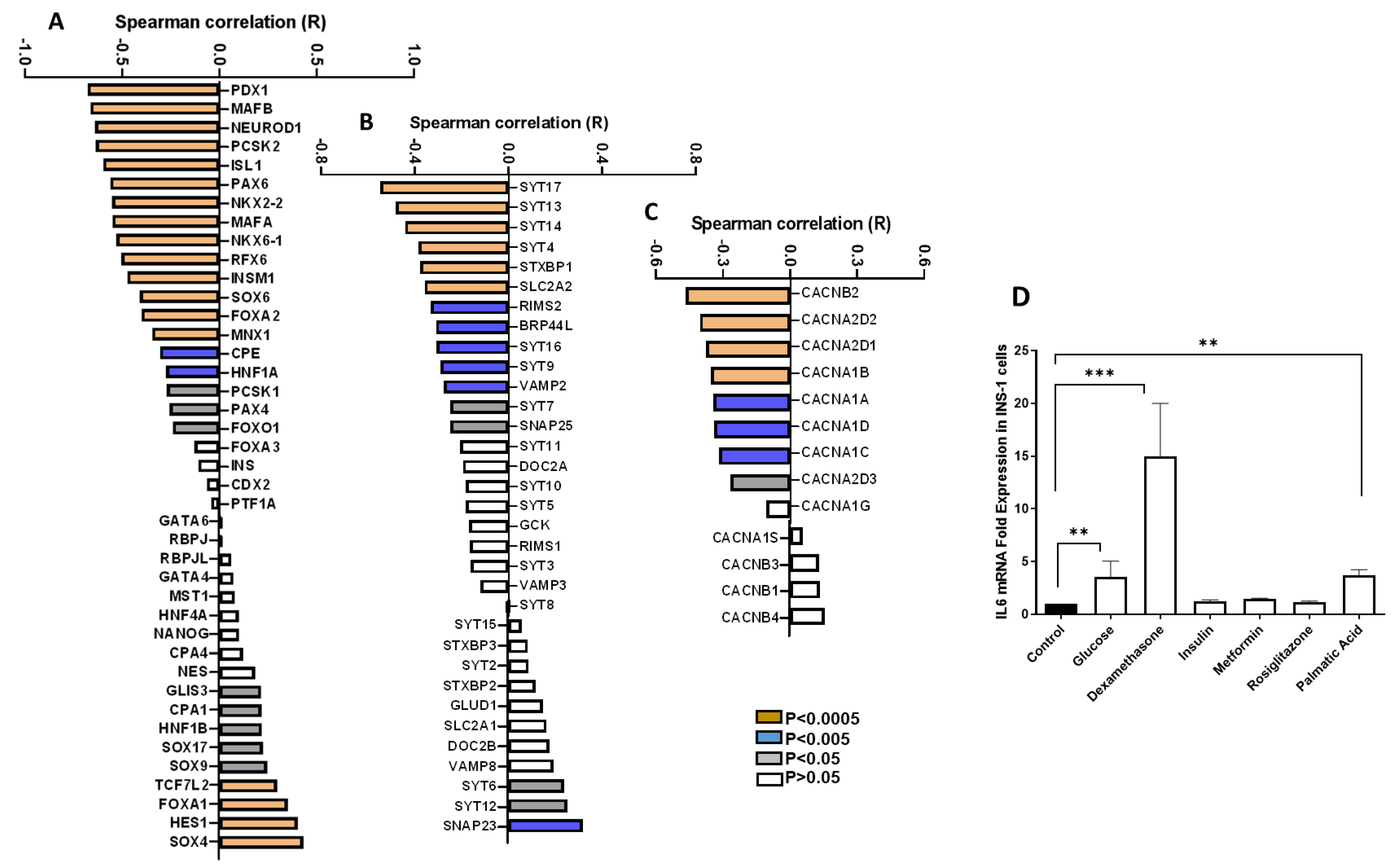
3.2. Correlation of IL6 Expression and Genes Involved in the Function of Pancreatic β-Cells
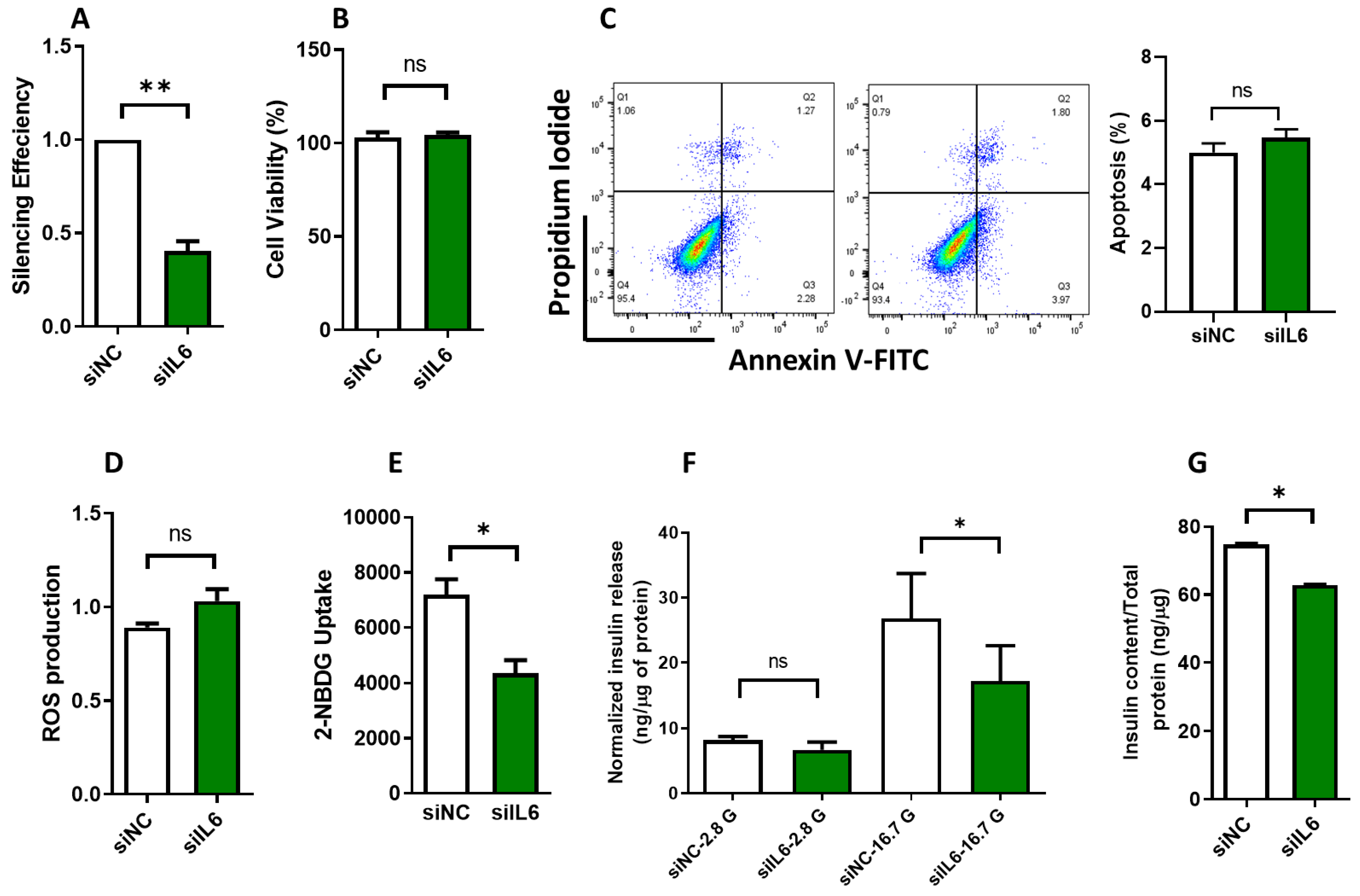
3.3. Effect of IL6 Silencing on Insulin Secretion and Glucose Uptake in INS-1 Cells
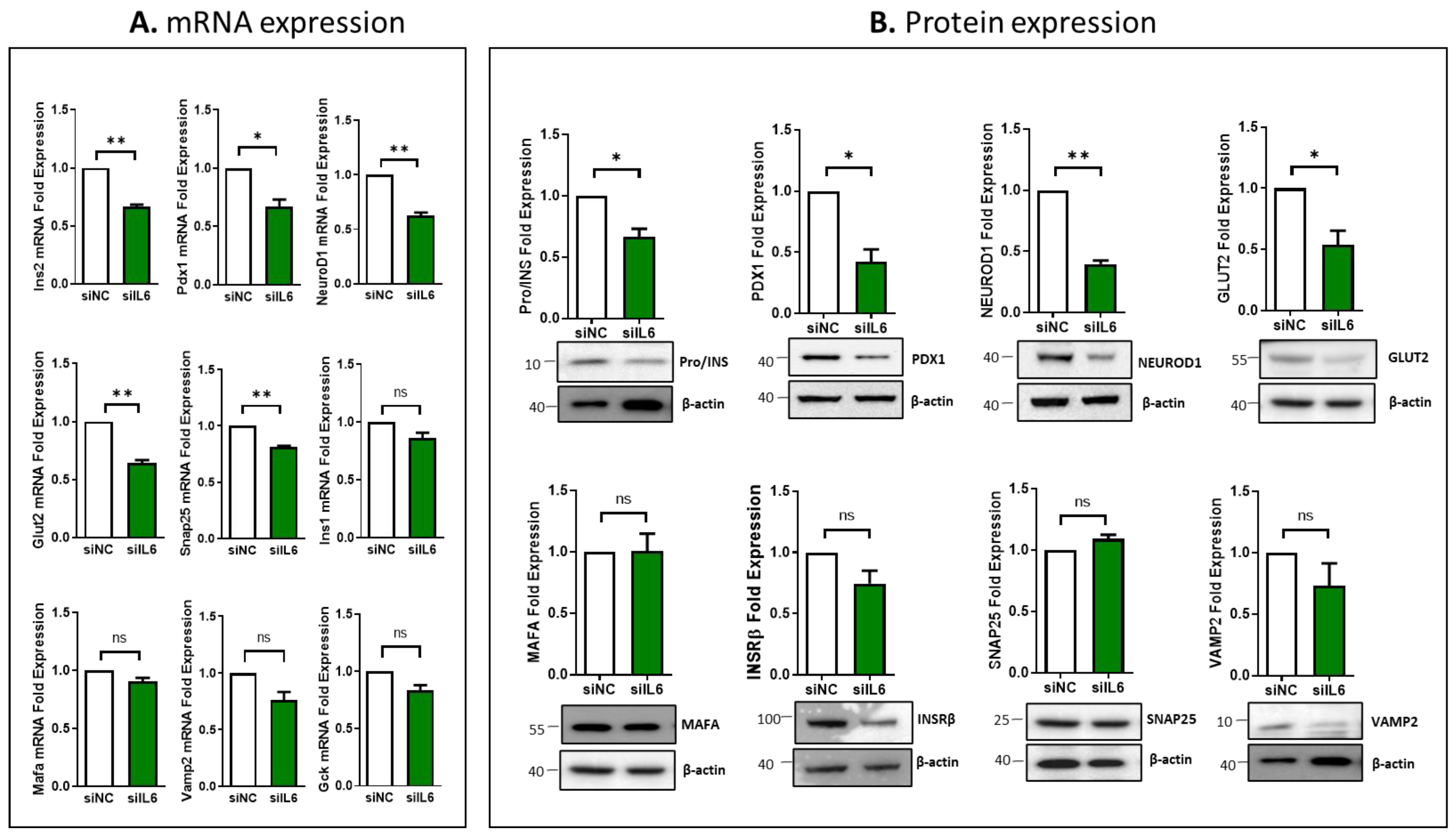
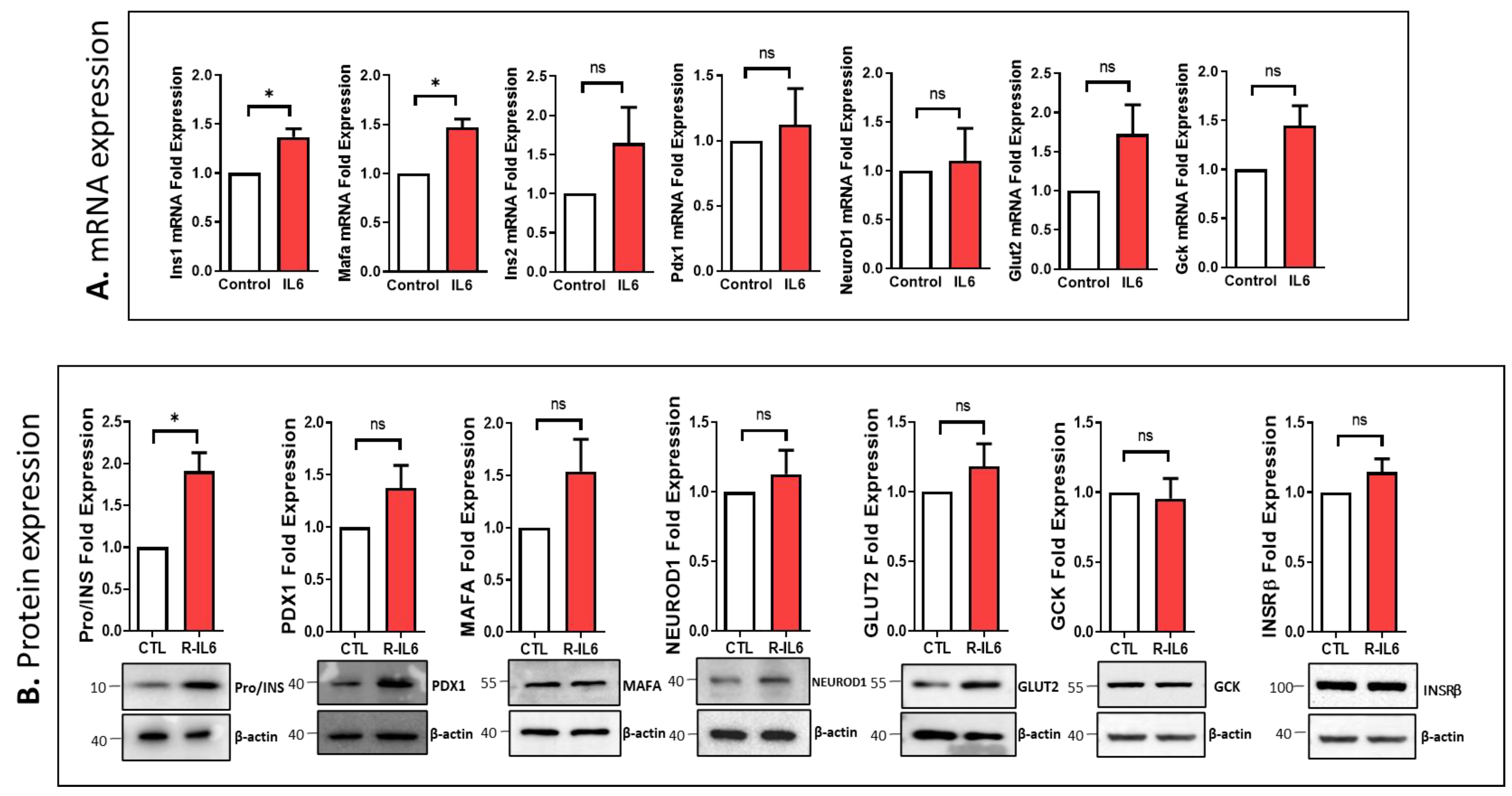
3.4. Impcat of IL6 Silencing on the Expression of Essential Genes Involved in β-Cell Function
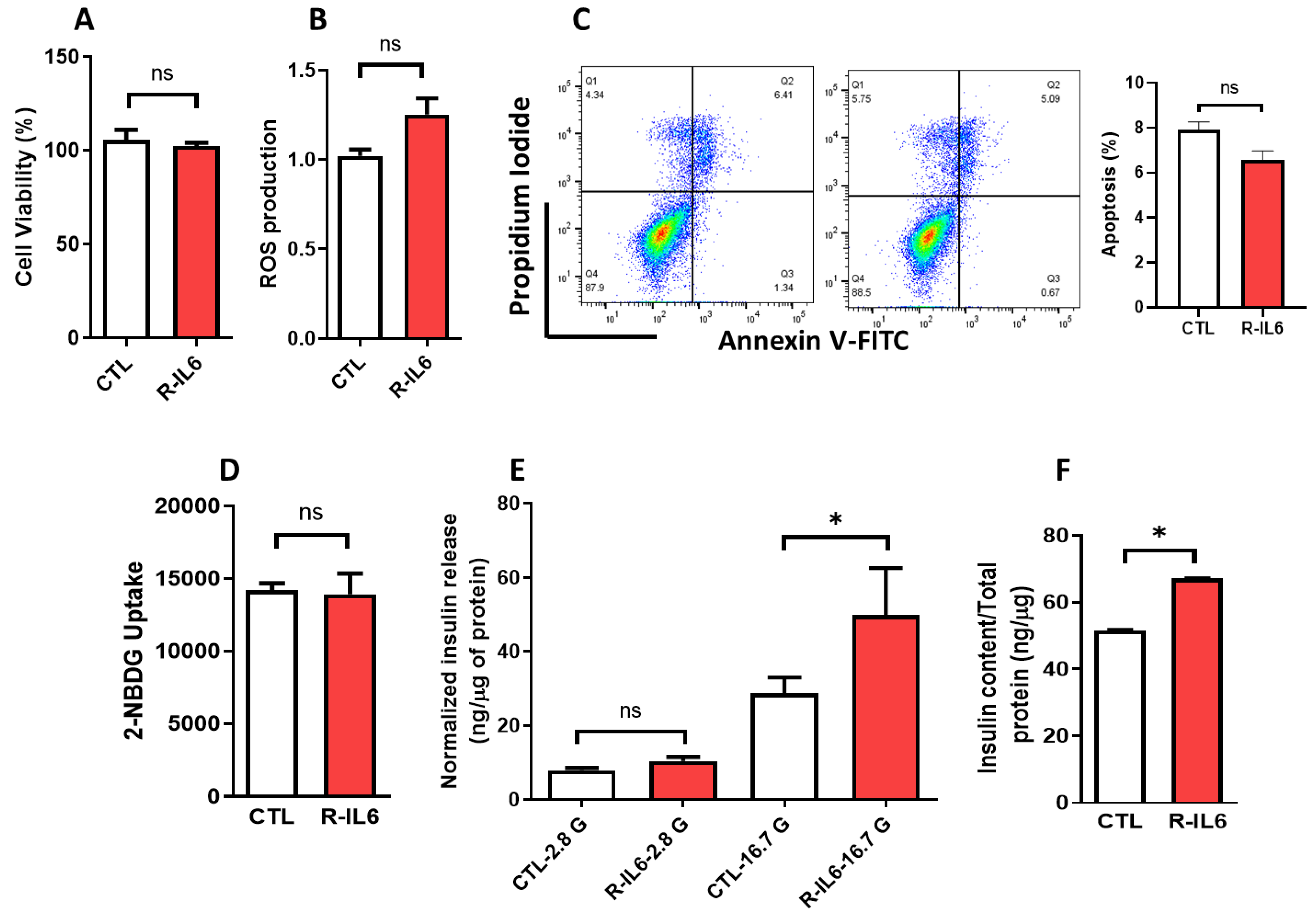
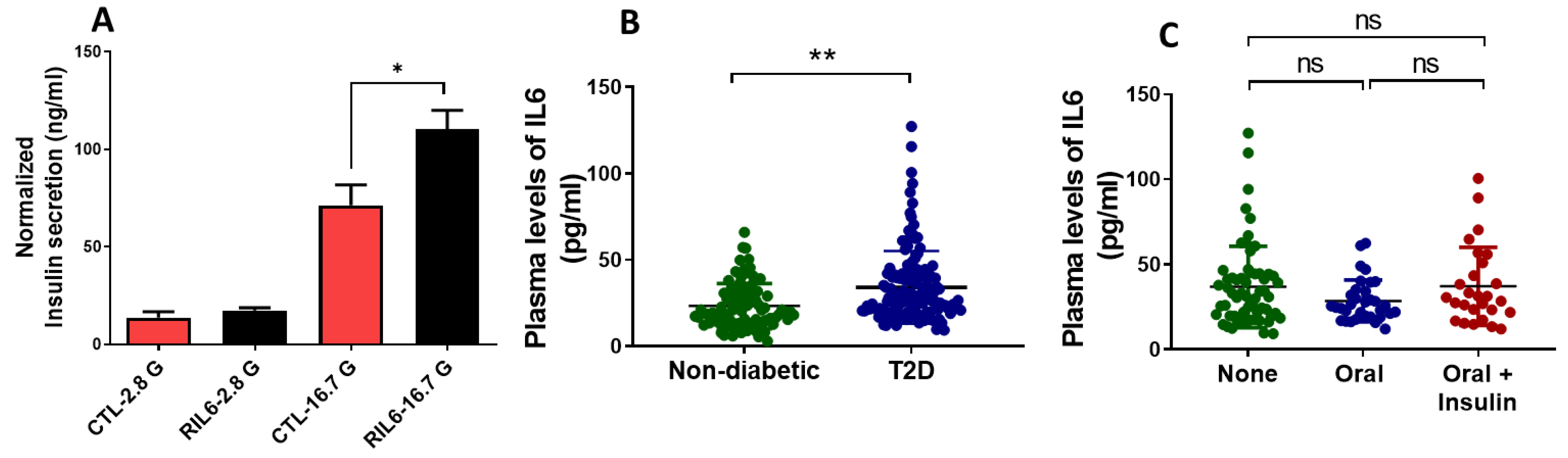
3.5. Effect of IL6 Treatment on the Function of Pancreatic β-Cells
3.6. Effect of IL6 Treatment in Human Islets and Measurements of Serum Levels in Patients with T2D
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef] [PubMed]
- Guest, C.B.; Park, M.J.; Johnson, D.R.; Freund, G.G. The implication of proinflammatory cytokines in type 2 diabetes. Front. Biosci. 2008, 13, 5187–5194. [Google Scholar] [CrossRef]
- King, G.L. The role of inflammatory cytokines in diabetes and its complications. J. Periodontol. 2008, 79, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Mirza, S.; Hossain, M.; Mathews, C.; Martinez, P.; Pino, P.; Gay, J.L.; Rentfro, A.; McCormick, J.B.; Fisher-Hoch, S.P. Type 2-diabetes is associated with elevated levels of TNF-alpha, IL-6 and adiponectin and low levels of leptin in a population of Mexican Americans: A cross-sectional study. Cytokine 2012, 57, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, A. An update on cytokines in the pathogenesis of insulin-dependent diabetes mellitus. Diabetes/Metab. Rev. 1998, 14, 129–151. [Google Scholar] [CrossRef]
- Gyamfi, J.; Eom, M.; Koo, J.-S.; Choi, J. Multifaceted roles of interleukin-6 in adipocyte–breast cancer cell interaction. Transl. Oncol. 2018, 11, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Akbari, M.; Hassan-Zadeh, V. IL-6 signalling pathways and the development of type 2 diabetes. Inflammopharmacology 2018, 26, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, O.P.; Mandrup-Poulsen, T. Interleukin-6 and diabetes: The good, the bad, or the indifferent? Diabetes 2005, 54 (Suppl. S2), S114–S124. [Google Scholar] [CrossRef]
- Li, Y.-S.; Ren, H.-C.; Cao, J.-H. Roles of Interleukin-6-mediated immunometabolic reprogramming in COVID-19 and other viral infection-associated diseases. Int. Immunopharmacol. 2022, 110, 109005. [Google Scholar] [CrossRef]
- Mussa, B.M.; Srivastava, A.; Al-Habshi, A.; Mohammed, A.K.; Halwani, R.; Abusnana, S. Inflammatory biomarkers levels in T2DM Emirati patients with diabetic neuropathy. Diabetes Metab. Syndr. Obes. 2021, 14, 3389–3397. [Google Scholar] [CrossRef]
- Heinrich, P.C.; Behrmann, I.; Haan, S.; Hermanns, H.M.; Müller-Newen, G.; Schaper, F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003, 374, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, D.; Ishihara, K.; Hirano, T. IL-6 signal transduction and its physiological roles: The signal orchestration model. Rev. Physiol. Biochem. Pharmacol. 2004, 1–38. [Google Scholar] [CrossRef]
- Suzuki, T.; Imai, J.; Yamada, T.; Ishigaki, Y.; Kaneko, K.; Uno, K.; Hasegawa, Y.; Ishihara, H.; Oka, Y.; Katagiri, H. Interleukin-6 enhances glucose-stimulated insulin secretion from pancreatic β-cells: Potential involvement of the PLC-IP3–dependent pathway. Diabetes 2011, 60, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Krause, M.; Bittencourt, A.; Homem de Bittencourt, P.I.; McClenaghan, N.H.; Flatt, P.R.; Murphy, C.; Newsholme, P. Physiological concentrations of interleukin-6 directly promote insulin secretion, signal transduction, nitric oxide release, and redox status in a clonal pancreatic β-cell line and mouse islets. J. Endocrinol. 2012, 214, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Ohtani, K.; Kato, Y.; Mori, M. Interleukin-6 increases insulin secretion and preproinsulin mRNA expression via Ca^ 2^+-dependent mechanism. J. Endocrinol. 2000, 166, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Ellingsgaard, H.; Ehses, J.A.; Hammar, E.B.; Van Lommel, L.; Quintens, R.; Martens, G.; Kerr-Conte, J.; Pattou, F.; Berney, T.; Pipeleers, D. Interleukin-6 regulates pancreatic α-cell mass expansion. Proc. Natl. Acad. Sci. USA 2008, 105, 13163–13168. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-E.; Choi, K.-M.; Yoon, I.-H.; Shin, J.-Y.; Kim, J.-S.; Park, W.-Y.; Han, D.-J.; Kim, S.-C.; Ahn, C.; Kim, J.-Y. IL-6 protects pancreatic islet beta cells from pro-inflammatory cytokines-induced cell death and functional impairment in vitro and in vivo. Transpl. Immunol. 2004, 13, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Linnemann, A.K.; Blumer, J.; Marasco, M.R.; Battiola, T.J.; Umhoefer, H.M.; Han, J.Y.; Lamming, D.W.; Davis, D.B. Interleukin 6 protects pancreatic β cells from apoptosis by stimulation of autophagy. FASEB J. 2017, 31, 4140. [Google Scholar] [CrossRef] [PubMed]
- Wallenius, V.; Wallenius, K.; Ahrén, B.; Rudling, M.; Carlsten, H.; Dickson, S.L.; Ohlsson, C.; Jansson, J.-O. Interleukin-6-deficient mice develop mature-onset obesity. Nat. Med. 2002, 8, 75–79. [Google Scholar] [CrossRef]
- Andreozzi, F.; Laratta, E.; Cardellini, M.; Marini, M.A.; Lauro, R.; Hribal, M.L.; Perticone, F.; Sesti, G. Plasma interleukin-6 levels are independently associated with insulin secretion in a cohort of Italian-Caucasian nondiabetic subjects. Diabetes 2006, 55, 2021–2024. [Google Scholar] [CrossRef]
- Darko, S.N.; Yar, D.D.; Owusu-Dabo, E.; Awuah, A.A.-A.; Dapaah, W.; Addofoh, N.; Salifu, S.P.; Awua-Boateng, N.Y.; Adomako-Boateng, F. Variations in levels of IL-6 and TNF-α in type 2 diabetes mellitus between rural and urban Ashanti Region of Ghana. BMC Endocr. Disord. 2015, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.; Carter, S.; Solomon, T.P. Probing the effect of physiological concentrations of IL-6 on insulin secretion by INS-1 832/3 insulinoma cells under diabetic-like conditions. Int. J. Mol. Sci. 2018, 19, 1924. [Google Scholar] [CrossRef] [PubMed]
- Fadista, J.; Vikman, P.; Laakso, E.O.; Mollet, I.G.; Esguerra, J.L.; Taneera, J.; Storm, P.; Osmark, P.; Ladenvall, C.; Prasad, R.B. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 13924–13929. [Google Scholar] [CrossRef] [PubMed]
- Hamoudi, R.; Saheb Sharif-Askari, N.; Saheb Sharif-Askari, F.; Abusnana, S.; Aljaibeji, H.; Taneera, J.; Sulaiman, N. Prediabetes and diabetes prevalence and risk factors comparison between ethnic groups in the United Arab Emirates. Sci. Rep. 2019, 9, 17437. [Google Scholar] [CrossRef] [PubMed]
- Taneera, J.; Dhaiban, S.; Hachim, M.; Mohammed, A.K.; Mukhopadhyay, D.; Bajbouj, K.; Hamoudi, R.; Salehi, A.; Hamad, M. Reduced expression of Chl1 gene impairs insulin secretion by down-regulating the expression of key molecules of β-cell function. Exp. Clin. Endocrinol. Diabetes 2021, 129, 864–872. [Google Scholar] [CrossRef]
- Taneera, J.; Mohammed, I.; Mohammed, A.K.; Hachim, M.; Dhaiban, S.; Malek, A.; Dunér, P.; Elemam, N.M.; Sulaiman, N.; Hamad, M. Orphan G-protein coupled receptor 183 (GPR183) potentiates insulin secretion and prevents glucotoxicity-induced β-cell dysfunction. Mol. Cell Endocrinol. 2020, 499, 110592. [Google Scholar] [CrossRef] [PubMed]
- El-Huneidi, W.; Anjum, S.; Mohammed, A.K.; Unnikannan, H.; Saeed, R.; Bajbouj, K.; Abu-Gharbieh, E.; Taneera, J. Copine 3 “CPNE3” is a novel regulator for insulin secretion and glucose uptake in pancreatic β-cells. Sci. Rep. 2021, 11, 20692. [Google Scholar] [CrossRef] [PubMed]
- Asplund, O.; Storm, P.; Chandra, V.; Hatem, G.; Ottosson-Laakso, E.; Mansour-Aly, D.; Krus, U.; Ibrahim, H.; Ahlqvist, E.; Tuomi, T. Islet Gene View—A tool to facilitate islet research. Life Sci. Alliance 2022, 5, e202201376. [Google Scholar] [CrossRef]
- Poitout, V.; Hagman, D.; Stein, R.; Artner, I.; Robertson, R.P.; Harmon, J.S. Regulation of the insulin gene by glucose and fatty acids. J. Nutr. 2006, 136, 873–876. [Google Scholar] [CrossRef]
- Prentki, M.; Joly, E.; El-Assaad, W.; Roduit, R. Malonyl-CoA signaling, lipid partitioning, and glucolipotoxicity: Role in β-cell adaptation and failure in the etiology of diabetes. Diabetes 2002, 51, S405–S413. [Google Scholar] [CrossRef]
- Louet, J.-F.; LeMay, C.; Mauvais-Jarvis, F. Antidiabetic actions of estrogen: Insight from human and genetic mouse models. Curr. Atheroscler. Rep. 2004, 6, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Nakamura, T.; Iino, T.; Hagi, T.; Kita, K.; Asanuma, K.; Sudo, A. Expression of interleukin-6 and the interleukin-6 receptor predicts the clinical outcomes of patients with soft tissue sarcomas. Cancers 2020, 12, 585. [Google Scholar] [CrossRef] [PubMed]
- Bacos, K.; Perfilyev, A.; Karagiannopoulos, A.; Cowan, E.; Ofori, J.K.; Bertonnier-Brouty, L.; Rönn, T.; Lindqvist, A.; Luan, C.; Ruhrmann, S. Type 2 diabetes candidate genes, including PAX5, cause impaired insulin secretion in human pancreatic islets. J. Clin. Investig. 2023, 133, e163612. [Google Scholar] [CrossRef]
- Böni-Schnetzler, M.; Meier, D.T. Islet inflammation in type 2 diabetes. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 501–513. [Google Scholar]
- Kurauti, M.A.; Costa-Júnior, J.M.; Ferreira, S.M.; Santos, G.J.; Sponton, C.H.; Carneiro, E.M.; Telles, G.D.; Chacon-Mikahil, M.P.; Cavaglieri, C.R.; Rezende, L.F. Interleukin-6 increases the expression and activity of insulin-degrading enzyme. Sci. Rep. 2017, 7, 46750. [Google Scholar] [CrossRef]
- Rehman, K.; Akash, M.S.H.; Liaqat, A.; Kamal, S.; Qadir, M.I.; Rasul, A. Role of interleukin-6 in development of insulin resistance and type 2 diabetes mellitus. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 229–236. [Google Scholar] [CrossRef]
- Fasshauer, M.; Klein, J.; Lossner, U.; Paschke, R. Interleukin (IL)-6 mRNA expression is stimulated by insulin, isoproterenol, tumour necrosis factor alpha, growth hormone, and IL-6 in 3T3-L1 adipocytes. Horm. Metab. Res. 2003, 35, 147–152. [Google Scholar] [CrossRef]
- Ershler, W.B.; Keller, E.T. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu. Rev. Med. 2000, 51, 245–270. [Google Scholar] [CrossRef] [PubMed]
- Maggio, M.; Guralnik, J.M.; Longo, D.L.; Ferrucci, L. Interleukin-6 in aging and chronic disease: A magnificent pathway. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2006, 61, 575–584. [Google Scholar] [CrossRef]
- Wyczalkowska-Tomasik, A.; Czarkowska-Paczek, B.; Zielenkiewicz, M.; Paczek, L. Inflammatory markers change with age, but do not fall beyond reported normal ranges. Arch. Immunol. Et Ther. Exp. 2016, 64, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Panagi, L.; Poole, L.; Hackett, R.A.; Steptoe, A. Sex differences in interleukin-6 stress responses in people with Type 2 diabetes. Psychophysiology 2019, 56, e13334. [Google Scholar] [CrossRef]
- El-Mikkawy, D.M.; EL-Sadek, M.A.; EL-Badawy, M.A.; Samaha, D. Circulating level of interleukin-6 in relation to body mass indices and lipid profile in Egyptian adults with overweight and obesity. Egypt. Rheumatol. Rehabil. 2020, 47, 7. [Google Scholar] [CrossRef]
- Nordmann, T.M.; Dror, E.; Schulze, F.; Traub, S.; Berishvili, E.; Barbieux, C.; Böni-Schnetzler, M.; Donath, M.Y. The role of inflammation in β-cell dedifferentiation. Sci. Rep. 2017, 7, 6285. [Google Scholar] [CrossRef] [PubMed]
- Marasco, M.R.; Conteh, A.M.; Reissaus, C.A.; Cupit, J.E.; Appleman, E.M.; Mirmira, R.G.; Linnemann, A.K. Interleukin-6 reduces β-cell oxidative stress by linking autophagy with the antioxidant response. Diabetes 2018, 67, 1576–1588. [Google Scholar] [CrossRef] [PubMed]
- Bowker, N.; Shah, R.L.; Sharp, S.J.; Stewart, I.D.; Wheeler, E.; Ferreira, M.A.; Baras, A.; Wareham, N.J.; Langenberg, C.; Lotta, L.A. Meta-analysis investigating the role of interleukin-6 mediated inflammation in type 2 diabetes. EBioMedicine 2020, 61. [Google Scholar] [CrossRef] [PubMed]
- Southern, C.; Schulster, D.; Green, I. Inhibition of insulin secretion from rat islets of Langerhans by interleukin-6. An effect distinct from that of interleukin-1. Biochem. J. 1990, 272, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.R.; Paula, C.C.; Taniguchi, S.; Ortis, F. Pre-treatment with IL-6 potentiates β-cell death induced by pro-inflammatory cytokines. BMC Mol. Cell Biol. 2023, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Wueest, S.; Konrad, D. The controversial role of IL-6 in adipose tissue on obesity-induced dysregulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E607–E613. [Google Scholar] [CrossRef] [PubMed]
- Senn, J.J.; Klover, P.J.; Nowak, I.A.; Mooney, R.A. Interleukin-6 induces cellular insulin resistance in hepatocytes. Diabetes 2002, 51, 3391–3399. [Google Scholar] [CrossRef]
- Choy, E.H.; De Benedetti, F.; Takeuchi, T.; Hashizume, M.; John, M.R.; Kishimoto, T. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 2020, 16, 335–345. [Google Scholar] [CrossRef]

| Gene | Forward Sequence (5′-3′) | Reverse Sequence (5′-3′) |
|---|---|---|
| Snap25 | GGCGTTTGCTGAATGACAAC | CAGAGCCTGACACCCTAAGA |
| Vamp2 | TGGTGGACATCATGAGGGTG | GCTTGGCTGCACTTGTTTCA |
| Mafa | GAGGAGGAGCGCAAGATCGG | AGCAAAAGTTTCGTGCTGTCAA |
| Neurod1 | CCC TAACTGATTGCACCAGC | TGCAGGGTAGTGCATGGTAA |
| Hprt1 | TTGTGTCATCAGCGAAAGTGG | CACAGGACTAGAACGTCTGCT |
| Variables (N) | Controls (99) | T2DM (134) | p-Value |
|---|---|---|---|
| Gender (Male/Female) | 35/64 | 66/68 | - |
| Smokers, n (%) | 6 (6.9) | 17 (13.6) | 0.12 |
| Age (Years) | 36.4 ± 9.7 | 51.7 ± 13.7 | <0.0001 |
| BMI (kg/m2) | 27.6 ± 4.8 | 31.3 ± 5.6 | <0.0001 |
| Waist circumference | 88.7 ± 13.4 | 104.0 ± 15.2 | <0.0001 |
| SBP (mmHg) | 118.0 ± 15.5 | 131.2 ± 21.2 | <0.0001 |
| DBP (mmHg) | 77.5 ± 10.1 | 82.8 ± 10.4 | <0.0001 |
| Fasting Glucose (mmol/L) | 5.2 ± 0.6 | 9.7 ± 4.1 | <0.0001 |
| HbA1c (%) | 5.3 ± 0.4 | 7.7 ± 2.1 | <0.0001 |
| Total Cholesterol (mmol/L) | 4.9 ± 0.9 | 4.6 ± 1.1 | 0.03 |
| HDL-Cholesterol (mmol/L) | 3.20 ± 0.83 | 2.85 ± 1.0 | 0.005 |
| LDL-Cholesterol (mmol/L) | 1.33 ± 0.34 | 1.16 ± 0.39 | 0.001 |
| Triglycerides (mmol/L) | 1.19 ± 0.71 | 1.77 ± 0.92 | <0.0001 |
| IL-6 | 24.4 ± 16.9 | 34.1 ± 21.1 | <0.0001 |
| Model 1 | Model 2 | |||
|---|---|---|---|---|
| Characteristics | r | p-Value | r | p-Value |
| Age | 0.26 | <0.0001 | - | - |
| BMI (kg/m2) | 0.10 | 0.20 | 0.03 | 0.72 |
| Waist circumference | 0.25 | <0.0001 | 0.07 | 0.35 |
| SBP (mmHg) | 0.08 | 0.23 | 0.003 | 0.97 |
| DBP (mmHg) | 0.008 | 0.90 | −0.06 | 0.44 |
| Fasting Glucose (mmol/L) # | 0.22 | 0.001 | 0.18 | 0.02 |
| HbA1c (%) # | 0.30 | 0.002 | 0.25 | 0.001 |
| Total Cholesterol (mmol/L) | −0.009 | 0.89 | 0.08 | 0.29 |
| Triglycerides (mmol/L) # | 0.36 | <0.0001 | 0.35 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taneera, J.; Khalique, A.; Mohammed, A.K.; Mussa, B.M.; Sulaiman, N.; Abu-Gharbieh, E.; El-Huneidi, W.; Saber-Ayad, M.M. Investigating the Impact of IL6 on Insulin Secretion: Evidence from INS-1 Cells, Human Pancreatic Islets, and Serum Analysis. Cells 2024, 13, 685. https://doi.org/10.3390/cells13080685
Taneera J, Khalique A, Mohammed AK, Mussa BM, Sulaiman N, Abu-Gharbieh E, El-Huneidi W, Saber-Ayad MM. Investigating the Impact of IL6 on Insulin Secretion: Evidence from INS-1 Cells, Human Pancreatic Islets, and Serum Analysis. Cells. 2024; 13(8):685. https://doi.org/10.3390/cells13080685
Chicago/Turabian StyleTaneera, Jalal, Anila Khalique, Abdul Khader Mohammed, Bashair M. Mussa, Nabil Sulaiman, Eman Abu-Gharbieh, Waseem El-Huneidi, and Maha M. Saber-Ayad. 2024. "Investigating the Impact of IL6 on Insulin Secretion: Evidence from INS-1 Cells, Human Pancreatic Islets, and Serum Analysis" Cells 13, no. 8: 685. https://doi.org/10.3390/cells13080685
APA StyleTaneera, J., Khalique, A., Mohammed, A. K., Mussa, B. M., Sulaiman, N., Abu-Gharbieh, E., El-Huneidi, W., & Saber-Ayad, M. M. (2024). Investigating the Impact of IL6 on Insulin Secretion: Evidence from INS-1 Cells, Human Pancreatic Islets, and Serum Analysis. Cells, 13(8), 685. https://doi.org/10.3390/cells13080685






