Trying to Kill a Killer; Impressive Killing of Patient Derived Glioblastoma Cultures Using NK-92 Natural Killer Cells Reveals Both Sensitive and Highly Resistant Glioblastoma Cells
Abstract
1. Introduction
2. Methods Section
2.1. Cell Culture Protocol of Glioblastoma Cell Lines
2.2. Culture of NK-92 Cells
2.3. Glioblastoma Killing Assays Using NK-92 Cells
2.4. ECIS Monitoring of NK-92 Mediated Killing of Glioblastoma Cells
2.5. ECIS Application and Contextualisation
2.6. Immunocytochemical Stain of NK-92-Glioblastoma Co-Cultures
2.7. Image Acquisition and Analysis Pipeline
2.8. Statistical Analysis of Data
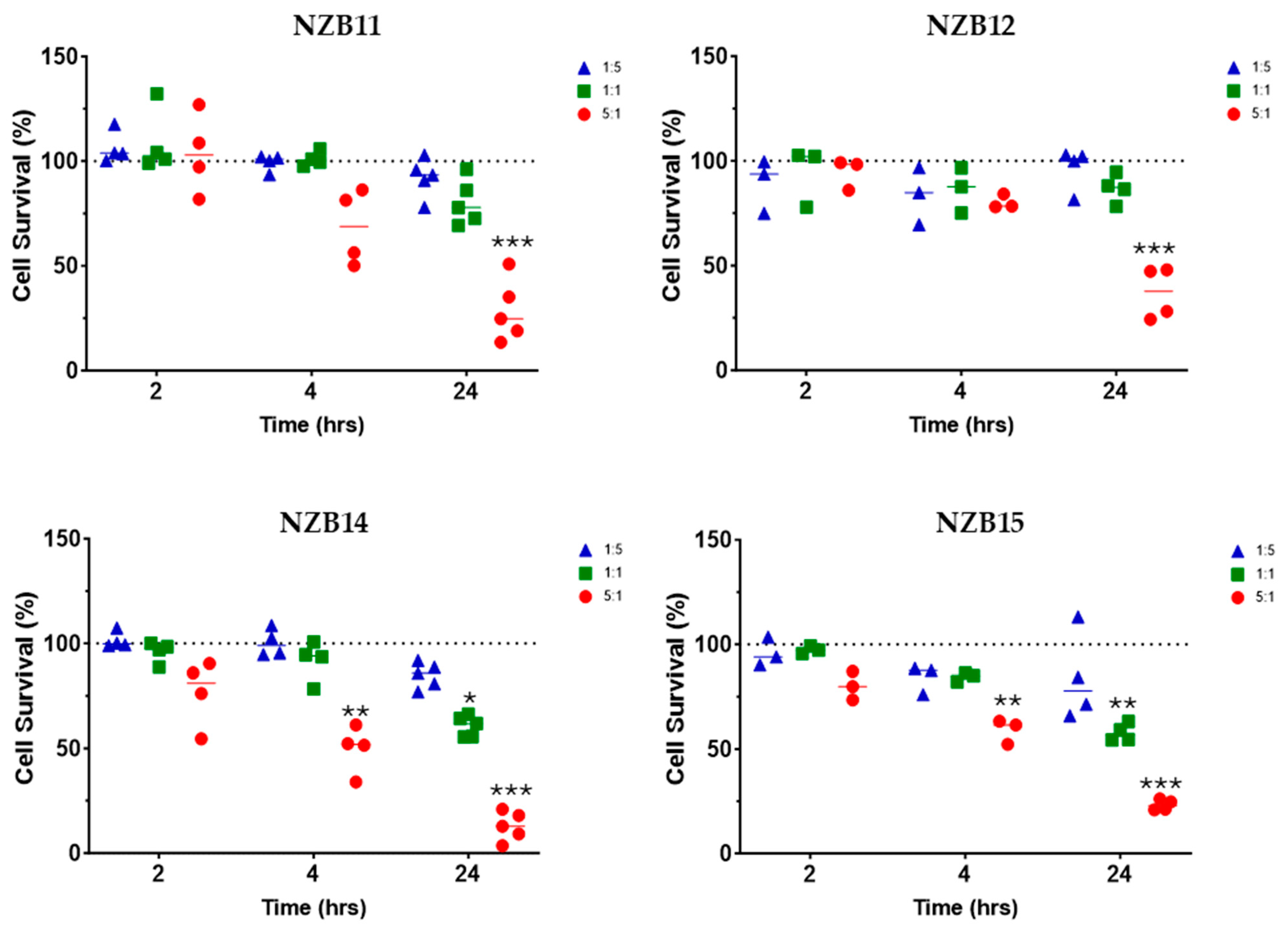
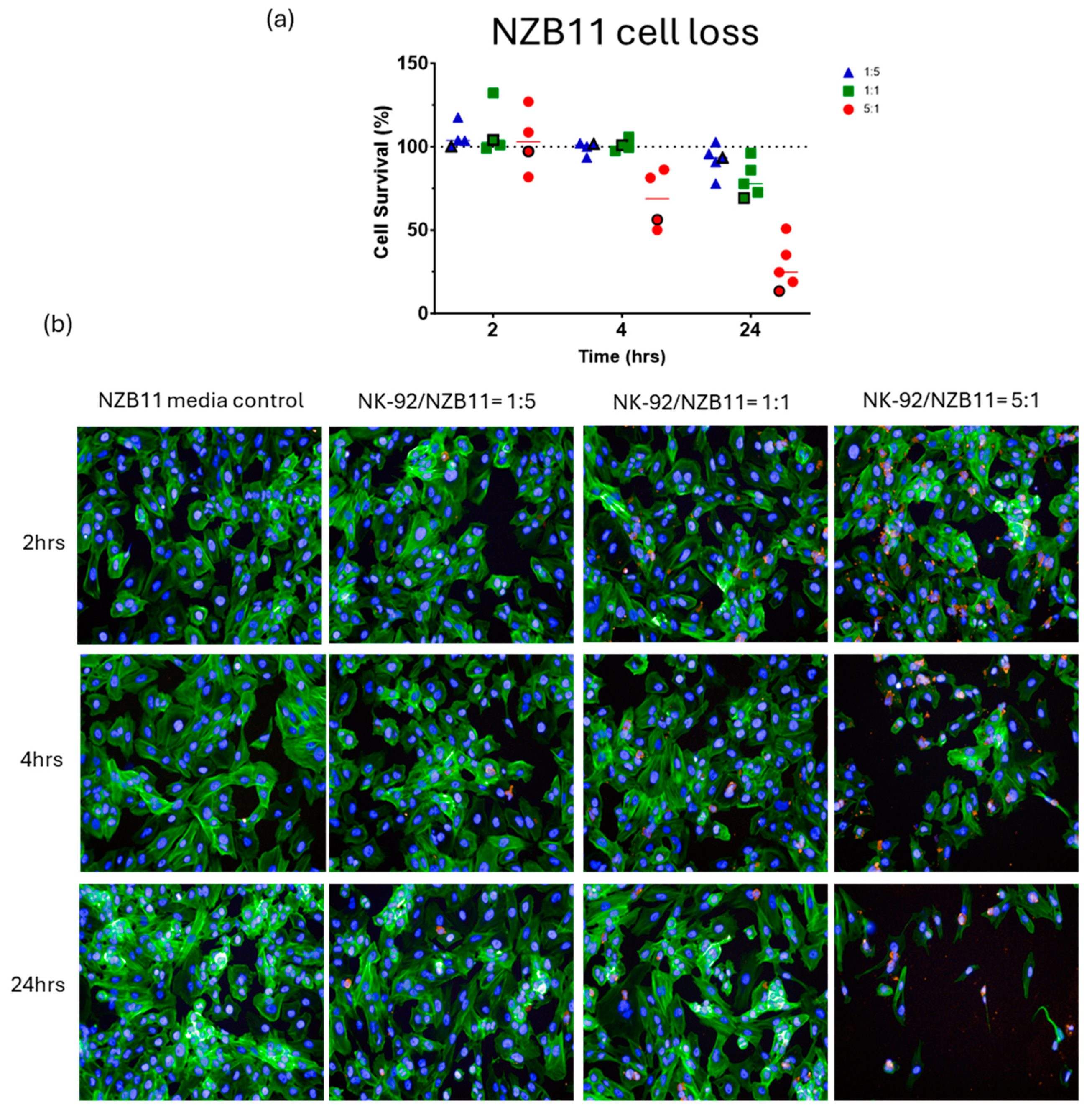
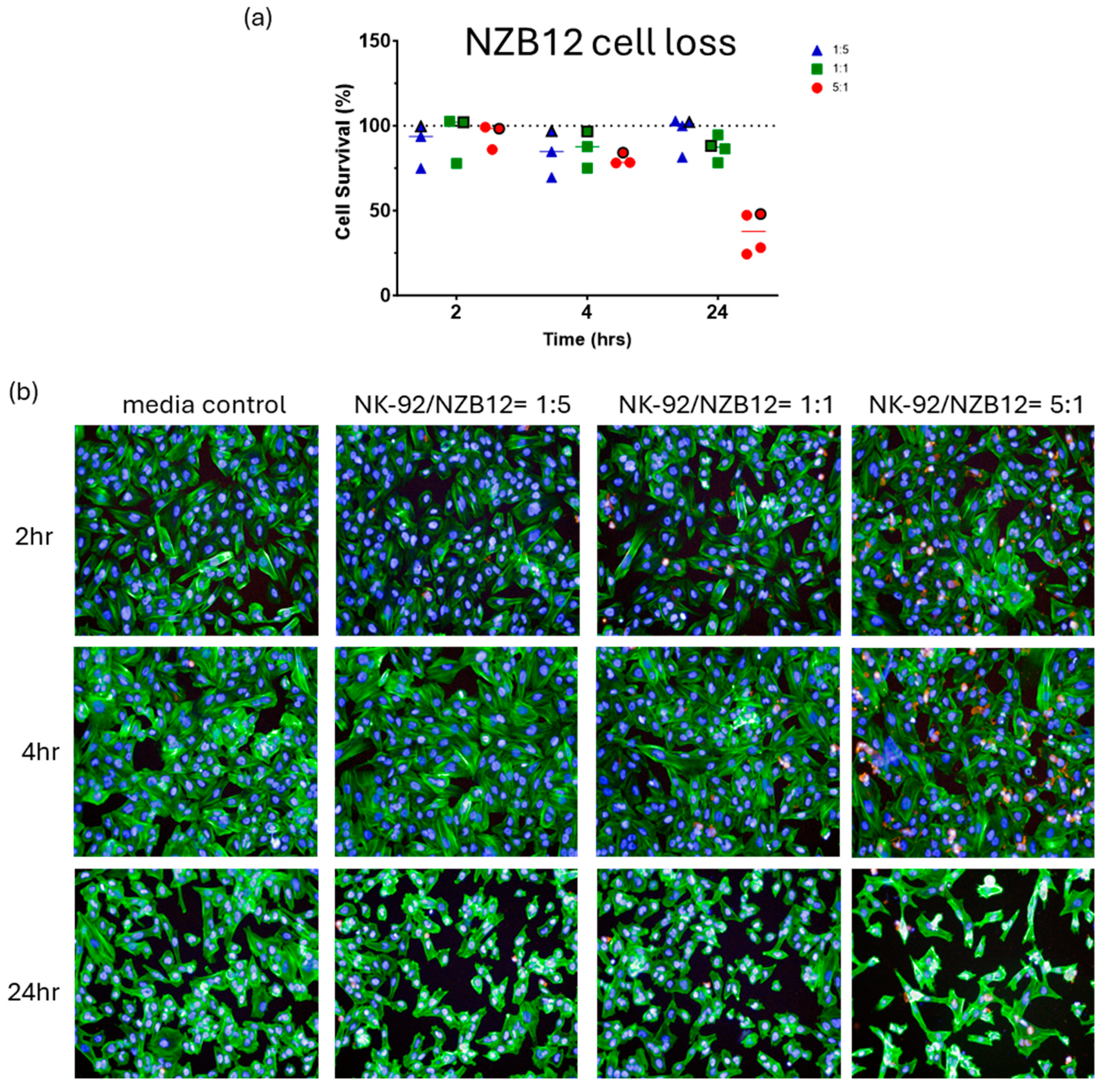
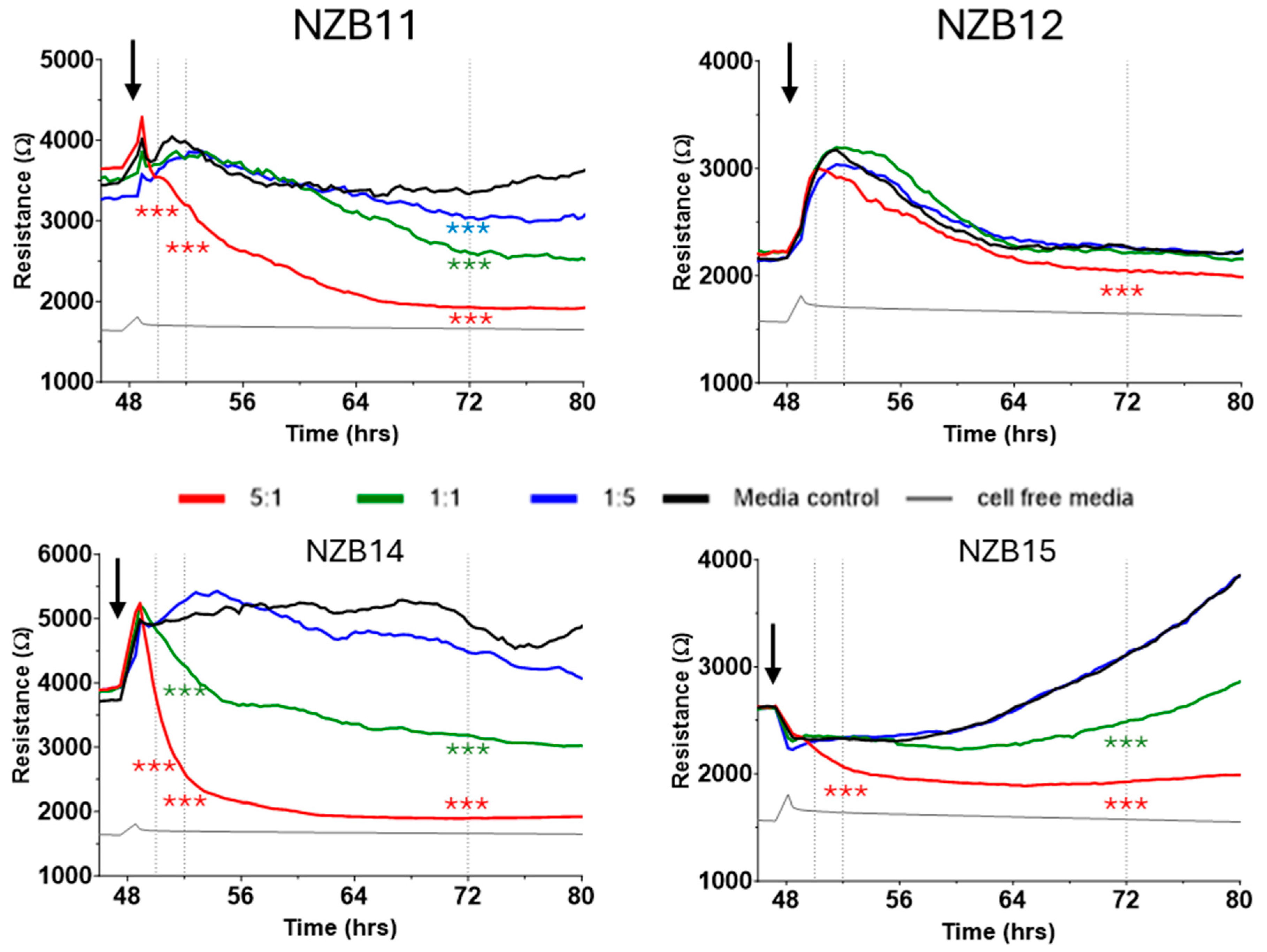
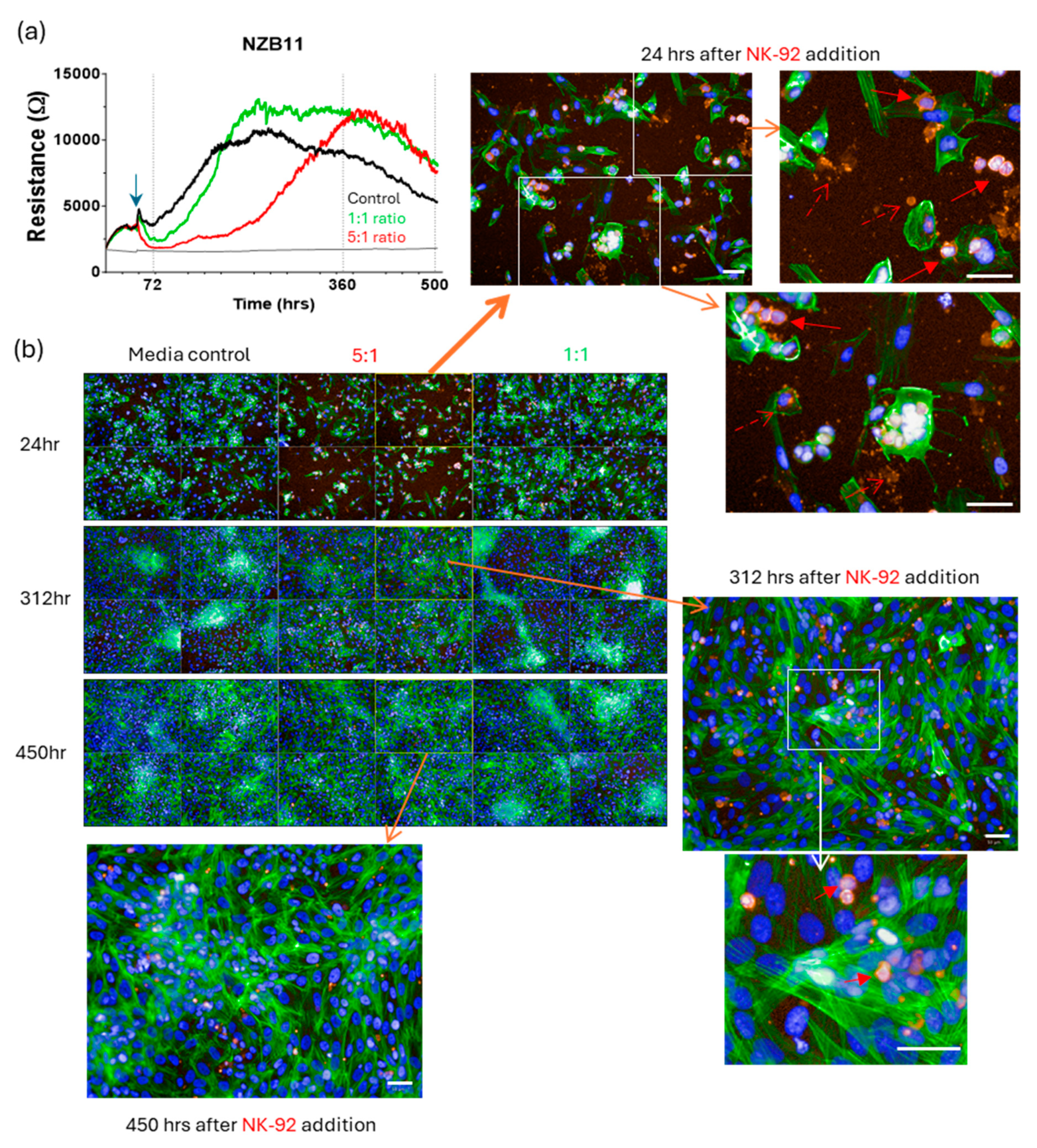
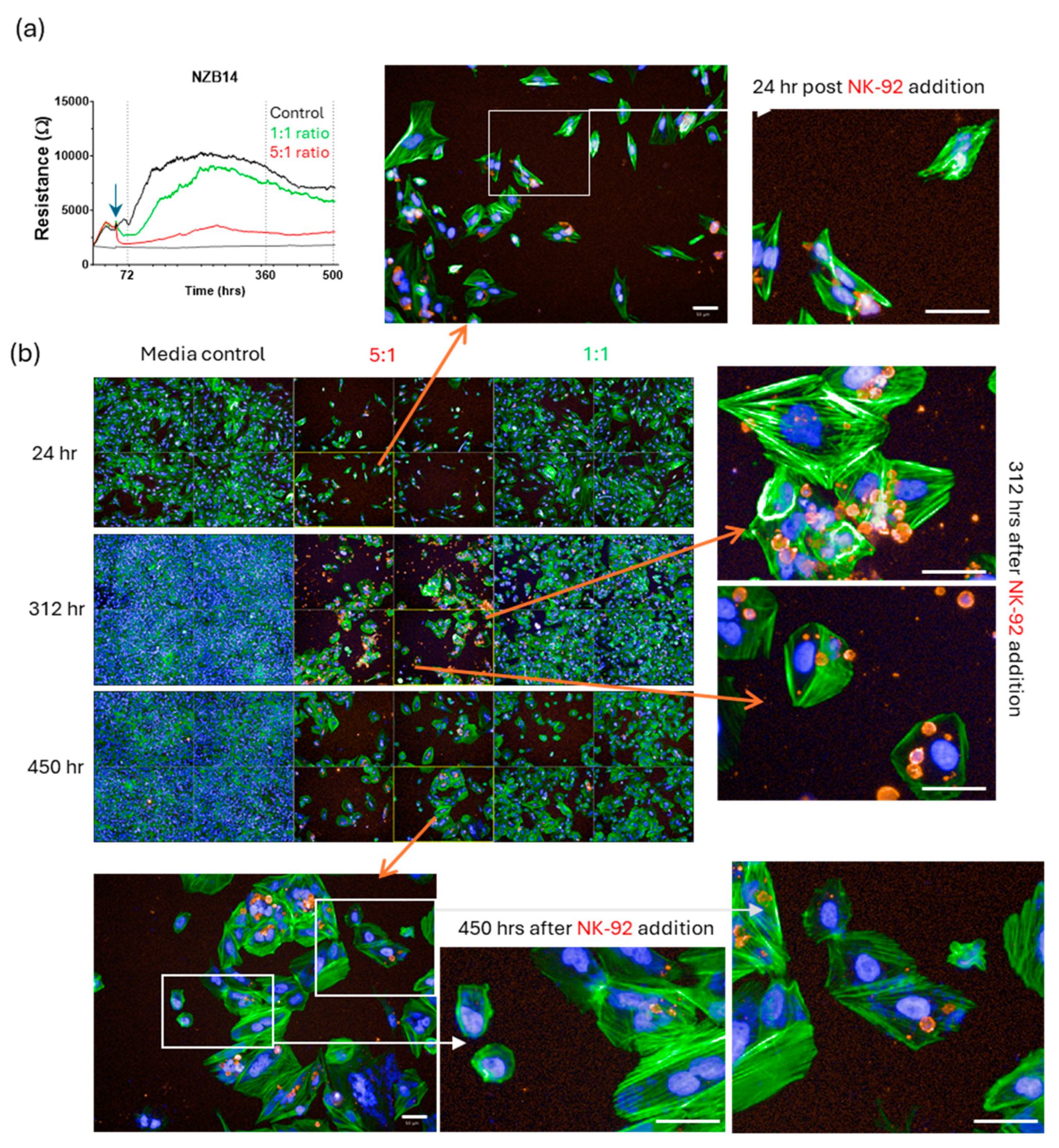
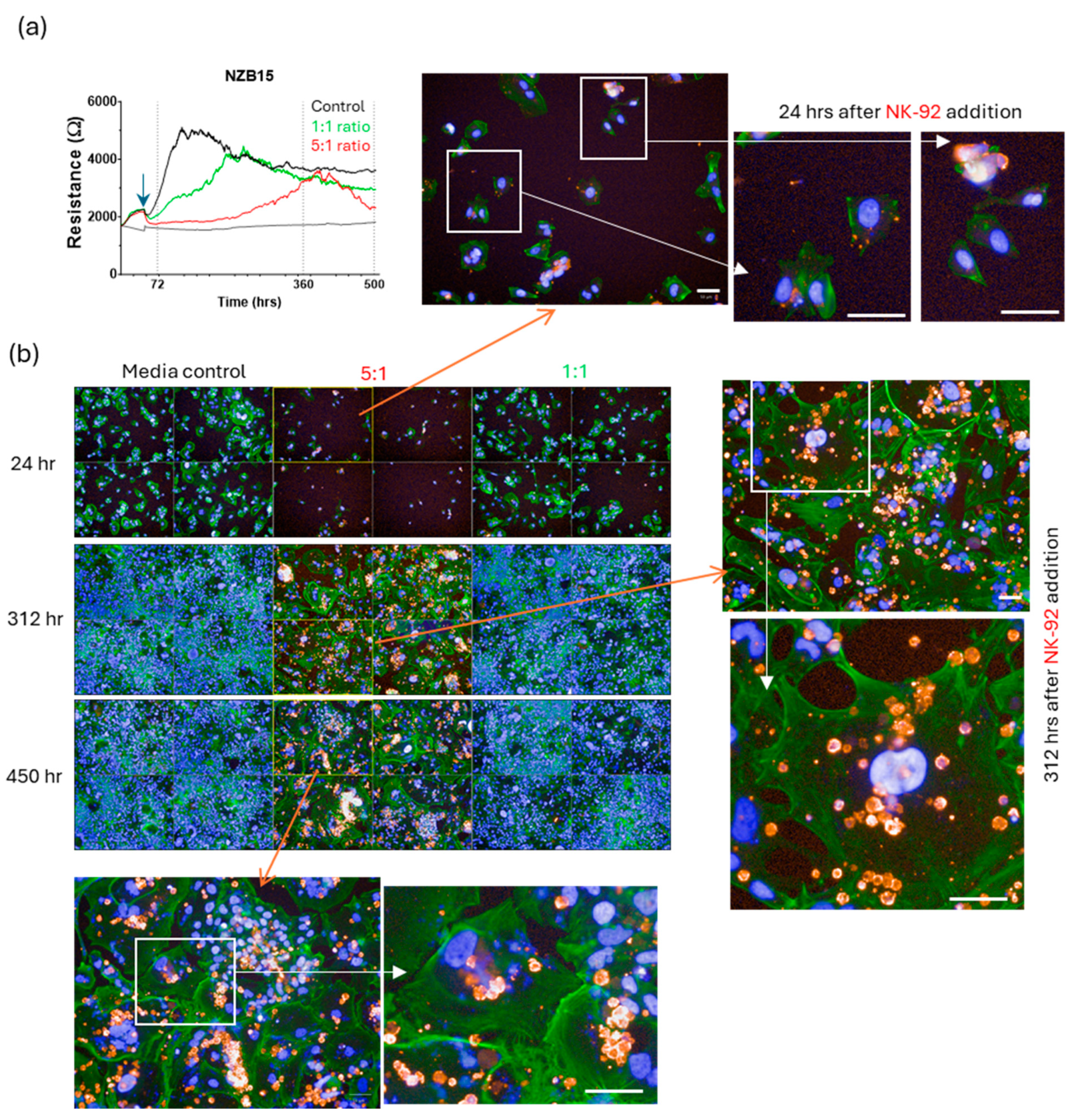
3. Results
Killing of Glioblastoma Cultures with NK-92 Cells
4. Discussion
5. Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, L.; Li, H.; Zheng, D.; Lin, S.; Ren, X. Glioblastoma patients’ survival and its relevant risk factors during the pre- and post-COVID-19 pandemic: Real-world cohort study in the USA and China. Int. J. Surg. 2024, 110, 2939–2949. [Google Scholar] [CrossRef]
- Goldman, D.A.; Hovinga, K.; Reiner, A.S.; Esquenazi, Y.; Tabar, V.; Panageas, K.S. The relationship between repeat resection and overall survival in patients with glioblastoma: A time-dependent analysis. J. Neurosurg. 2018, 129, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Mohammed, S.; Dinesan, M.; Ajayakumar, T. Survival and quality of life analysis in glioblastoma multiforme with adjuvant chemoradiotherapy: A retrospective study. Rep. Pract. Oncol. Radiother. 2022, 27, 1026–1036. [Google Scholar] [CrossRef]
- Chandra, A.; Lopez-Rivera, V.; Ryba, B.; Chandran, A.S.; Brandel, M.G.; Dono, A.; Sheinberg, D.L.; Esquenazi, Y.L.; Aghi, M.K. Survival outcomes and prognostic factors of infratentorial glioblastoma in the elderly. Clin. Neurol. Neurosurg. 2024, 236, 108084. [Google Scholar] [CrossRef]
- Reinders, A.N.; Koshy, M.; Korpics, M. The Patterns of Failure and Prognostic Impact of Tumor Location in Patients Undergoing Reirradiation for Glioblastoma. Cureus 2024, 16, e68820. [Google Scholar] [CrossRef] [PubMed]
- Stadler, C.; Gramatzki, D.; Le Rhun, E.; Hottinger, A.F.; Hundsberger, T.; Roelcke, U.; Laubli, H.; Hofer, S.; Seystahl, K.; Wirsching, H.G.; et al. Glioblastoma in the oldest old: Clinical characteristics, therapy, and outcome in patients aged 80 years and older. Neurooncol. Pract. 2024, 11, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Sloan, A.E.; Winter, K.; Gilbert, M.R.; Aldape, K.; Choi, S.; Wen, P.Y.; Butowski, N.; Iwamoto, F.M.; Raval, R.R.; Voloschin, A.D.; et al. NRG-BN002: Phase I study of ipilimumab, nivolumab, and the combination in patients with newly diagnosed glioblastoma. Neuro. Oncol. 2024, 26, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A. Immune-checkpoint inhibitors for glioblastoma: What have we learned? Arq. De Neuro-Psiquiatr. 2022, 80, 266–269. [Google Scholar] [CrossRef]
- Adhikaree, J.; Moreno-Vicente, J.; Kaur, A.P.; Jackson, A.M.; Patel, P.M. Resistance Mechanisms and Barriers to Successful Immunotherapy for Treating Glioblastoma. Cells 2020, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.; Qi, J.; Thin, T.H.; Garcia-Barros, M.; Lee, B.; Hahn, M.; Mandeli, J.; Belani, P.; Nael, K.; Rashidipour, O.; et al. A Phase I Trial of VEGF-A Inhibition Combined with PD-L1 Blockade for Recurrent Glioblastoma. Cancer Res. Commun. 2023, 3, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, W.M.; Scalzo, A.A. Natural killer cell activation receptors in innate immunity to infection. Microbes Infect. 2002, 4, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, N.; Mandelboim, O. The role of NK cells in innate immunity. Adv. Exp. Med. Biol. 2000, 479, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Mody, C.H.; Ogbomo, H.; Xiang, R.F.; Kyei, S.K.; Feehan, D.; Islam, A.; Li, S.S. Microbial killing by NK cells. J. Leukoc. Biol. 2019, 105, 1285–1296. [Google Scholar] [CrossRef]
- Littwitz-Salomon, E.; Dittmer, U.; Sutter, K. Insufficient natural killer cell responses against retroviruses: How to improve NK cell killing of retrovirus-infected cells. Retrovirology 2016, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Morgado, S.; Sanchez-Correa, B.; Casado, J.G.; Duran, E.; Gayoso, I.; Labella, F.; Solana, R.; Tarazona, R. NK cell recognition and killing of melanoma cells is controlled by multiple activating receptor-ligand interactions. J. Innate. Immun. 2011, 3, 365–373. [Google Scholar] [CrossRef]
- Gong, J.H.; Maki, G.; Klingemann, H.G. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia 1994, 8, 652–658. [Google Scholar]
- Klingemann, H.G.; Wong, E.; Maki, G. A cytotoxic NK-cell line (NK-92) for ex vivo purging of leukemia from blood. Biol. Blood Marrow Transplant. 1996, 2, 68–75. [Google Scholar]
- Suck, G.; Odendahl, M.; Nowakowska, P.; Seidl, C.; Wels, W.S.; Klingemann, H.G.; Tonn, T. NK-92: An ’off-the-shelf therapeutic’ for adoptive natural killer cell-based cancer immunotherapy. Cancer Immunol. Immunother. CII 2016, 65, 485–492. [Google Scholar] [CrossRef]
- Weitzman, J.; Betancur, M.; Boissel, L.; Rabinowitz, A.P.; Klein, A.; Klingemann, H. Variable contribution of monoclonal antibodies to ADCC in patients with chronic lymphocytic leukemia. Leuk Lymphoma 2009, 50, 1361–1368. [Google Scholar] [CrossRef]
- Maki, G.; Tam, Y.K.; Berkahn, L.; Klingemann, H.G. Ex vivo purging with NK-92 prior to autografting for chronic myelogenous leukemia. Bone Marrow Transplant. 2003, 31, 1119–1125. [Google Scholar] [CrossRef]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genssler, S.; Schonfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2016, 108, 375. [Google Scholar] [CrossRef] [PubMed]
- Jochems, C.; Hodge, J.W.; Fantini, M.; Fujii, R.; Morillon, Y.M., 2nd; Greiner, J.W.; Padget, M.R.; Tritsch, S.R.; Tsang, K.Y.; Campbell, K.S.; et al. An NK cell line (haNK) expressing high levels of granzyme and engineered to express the high affinity CD16 allele. Oncotarget 2016, 7, 86359–86373. [Google Scholar] [CrossRef]
- Klingemann, H. The NK-92 cell line-30 years later: Its impact on natural killer cell research and treatment of cancer. Cytotherapy 2023, 25, 451–457. [Google Scholar] [CrossRef]
- Arai, S.; Meagher, R.; Swearingen, M.; Myint, H.; Rich, E.; Martinson, J.; Klingemann, H. Infusion of the allogeneic cell line NK-92 in patients with advanced renal cell cancer or melanoma: A phase I trial. Cytotherapy 2008, 10, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Robilliard, L.D.; Yu, J.; Anchan, A.; Finlay, G.; Angel, C.E.; Graham, E.S. Comprehensive Assessment of Secreted Immuno-Modulatory Cytokines by Serum-Differentiated and Stem-like Glioblastoma Cells Reveals Distinct Differences between Glioblastoma Phenotypes. Int. J. Mol. Sci. 2022, 23, 14164. [Google Scholar] [CrossRef] [PubMed]
- Robilliard, L.D.; Yu, J.; Jun, S.M.; Anchan, A.; Finlay, G.; Angel, C.E.; Graham, E.S. Can ECIS Biosensor Technology Be Used to Measure the Cellular Responses of Glioblastoma Stem Cells? Biosensors 2021, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Robilliard, L.D.; Yu, J.; Anchan, A.; Joseph, W.; Finlay, G.; Angel, C.E.; Scott Graham, E. Comprehensive analysis of inhibitory checkpoint ligand expression by glioblastoma cells. Immunol. Cell Biol. 2021, 99, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Spurling, D.; Anchan, A.; Hucklesby, J.; Finlay, G.; Angel, C.E.; Graham, E.S. Melanoma Cells Produce Large Vesicular-Bodies That Cause Rapid Disruption of Brain Endothelial Barrier-Integrity and Disassembly of Junctional Proteins. Int. J. Mol. Sci. 2023, 24, 6082. [Google Scholar] [CrossRef]
- Hucklesby, J.J.W.; Anchan, A.; O’Carroll, S.J.; Unsworth, C.P.; Graham, E.S.; Angel, C.E. Comparison of Leading Biosensor Technologies to Detect Changes in Human Endothelial Barrier Properties in Response to Pro-Inflammatory TNFalpha and IL1beta in Real-Time. Biosensors 2021, 11, 159. [Google Scholar] [CrossRef]
- Anchan, A.; Martin, O.; Hucklesby, J.J.W.; Finlay, G.; Johnson, R.H.; Robilliard, L.D.; O’Carroll, S.J.; Angel, C.E.; Graham, E.S. Analysis of Melanoma Secretome for Factors That Directly Disrupt the Barrier Integrity of Brain Endothelial Cells. Int. J. Mol. Sci. 2020, 21, 8193. [Google Scholar] [CrossRef]
- Anchan, A.; Kalogirou-Baldwin, P.; Johnson, R.; Kho, D.T.; Joseph, W.; Hucklesby, J.; Finlay, G.J.; O’Carroll, S.J.; Angel, C.E.; Graham, E.S. Real-Time Measurement of Melanoma Cell-Mediated Human Brain Endothelial Barrier Disruption Using Electric Cell-Substrate Impedance Sensing Technology. Biosensors 2019, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Robilliard, L.D.; Kho, D.T.; Johnson, R.H.; Anchan, A.; O’Carroll, S.J.; Graham, E.S. The Importance of Multifrequency Impedance Sensing of Endothelial Barrier Formation Using ECIS Technology for the Generation of a Strong and Durable Paracellular Barrier. Biosensors 2018, 8, 64. [Google Scholar] [CrossRef]
- Gu, A.Y.; Kho, D.T.; Johnson, R.H.; Graham, E.S.; O’Carroll, S.J. In Vitro Wounding Models Using the Electric Cell-Substrate Impedance Sensing (ECIS)-Ztheta Technology. Biosensors 2018, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Kho, D.T.; Johnson, R.H.; O’Carroll, S.J.; Angel, C.E.; Graham, E.S. Biosensor Technology Reveals the Disruption of the Endothelial Barrier Function and the Subsequent Death of Blood Brain Barrier Endothelial Cells to Sodium Azide and Its Gaseous Products. Biosensors 2017, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Kho, D.T.; Johnson, R.; Robilliard, L.; du Mez, E.; McIntosh, J.; O’Carroll, S.J.; Angel, C.E.; Graham, E.S. ECIS technology reveals that monocytes isolated by CD14+ve selection mediate greater loss of BBB integrity than untouched monocytes, which occurs to a greater extent with IL-1beta activated endothelium in comparison to TNFalpha. PLoS ONE 2017, 12, e0180267. [Google Scholar] [CrossRef]
- Wiltshire, R.; Nelson, V.; Kho, D.T.; Angel, C.E.; O’Carroll, S.J.; Graham, E.S. Regulation of human cerebro-microvascular endothelial baso-lateral adhesion and barrier function by S1P through dual involvement of S1P1 and S1P2 receptors. Sci. Rep. 2016, 6, 19814. [Google Scholar] [CrossRef] [PubMed]
- Tonn, T.; Schwabe, D.; Klingemann, H.G.; Becker, S.; Esser, R.; Koehl, U.; Suttorp, M.; Seifried, E.; Ottmann, O.G.; Bug, G. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy 2013, 15, 1563–1570. [Google Scholar] [CrossRef]
- Klingemann, H.; Boissel, L.; Toneguzzo, F. Natural Killer Cells for Immunotherapy—Advantages of the NK-92 Cell Line over Blood NK Cells. Front. Immunol. 2016, 7, 91. [Google Scholar] [CrossRef]
- Arndt, S.; Seebach, J.; Psathaki, K.; Galla, H.J.; Wegener, J. Bioelectrical impedance assay to monitor changes in cell shape during apoptosis. Biosens. Bioelectron. 2004, 19, 583–594. [Google Scholar] [CrossRef]
- Curtis, T.M.; Tabb, J.; Romeo, L.; Schwager, S.J.; Widder, M.W.; van der Schalie, W.H. Improved cell sensitivity and longevity in a rapid impedance-based toxicity sensor. J. Appl. Toxicol. JAT 2009, 29, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Opp, D.; Wafula, B.; Lim, J.; Huang, E.; Lo, J.C.; Lo, C.M. Use of electric cell-substrate impedance sensing to assess in vitro cytotoxicity. Biosens. Bioelectron. 2009, 24, 2625–2629. [Google Scholar] [CrossRef]
- Qiu, Y.; Liao, R.; Zhang, X. Intervention of cardiomyocyte death based on real-time monitoring of cell adhesion through impedance sensing. Biosens. Bioelectron. 2009, 25, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Stolwijk, J.A.; Matrougui, K.; Renken, C.W.; Trebak, M. Impedance analysis of GPCR-mediated changes in endothelial barrier function: Overview and fundamental considerations for stable and reproducible measurements. Pflügers Arch.-Eur. J. Physiol. 2014, 467, 2193–2218. [Google Scholar] [CrossRef]
- Sertel, O.; Catalyurek, U.V.; Shimada, H.; Gurcan, M.N. Computer-aided prognosis of neuroblastoma: Detection of mitosis and karyorrhexis cells in digitized histological images. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 1433–1436. [Google Scholar]
- Fradera, J. Erythroid karyorrhexis in myelodysplasia: Bone marrow aspirate. Blood 2008, 112, 479. [Google Scholar]
- Zamzami, N.; Kroemer, G. Condensed matter in cell death. Nature 1999, 401, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.V.; Rao, P.V.; Cantor, A.B.; Altshuler, G.; Shuster, J.J.; Castleberry, R.P. Modified histologic grading of neuroblastomas by replacement of mitotic rate with mitosis karyorrhexis index. A clinicopathologic study of 223 cases from the Pediatric Oncology Group. Cancer 1996, 77, 1582–1588. [Google Scholar] [CrossRef]








| Staining Target | Staining Dye/Antibody | CAT# | Dilution |
|---|---|---|---|
| NK-92 | Mouse anti-CD45 antibody (Primary Antibody) | ab30470-100ug | 1:400 |
| Anti mouse Alexa Fluor™ 594 (Secondary antibody) | A11005 | 1:400 | |
| Nuclei | Hoechst 33342 (nuclei/DNA) | H3570 | 1:10,000 |
| Glioblastoma cells | ActinGreen™ 488 ReadyProbes™ (AlexaFluor™ 488 phalloidin) | R37110 | 1 drop per mL of buffer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Kim, H.J.; Reinecke, J.; Hucklesby, J.; Read, T.; Anchan, A.; Angel, C.E.; Graham, E.S. Trying to Kill a Killer; Impressive Killing of Patient Derived Glioblastoma Cultures Using NK-92 Natural Killer Cells Reveals Both Sensitive and Highly Resistant Glioblastoma Cells. Cells 2025, 14, 53. https://doi.org/10.3390/cells14010053
Yu J, Kim HJ, Reinecke J, Hucklesby J, Read T, Anchan A, Angel CE, Graham ES. Trying to Kill a Killer; Impressive Killing of Patient Derived Glioblastoma Cultures Using NK-92 Natural Killer Cells Reveals Both Sensitive and Highly Resistant Glioblastoma Cells. Cells. 2025; 14(1):53. https://doi.org/10.3390/cells14010053
Chicago/Turabian StyleYu, Jane, Hyeon Joo Kim, Jordyn Reinecke, James Hucklesby, Tennille Read, Akshata Anchan, Catherine E. Angel, and Euan Scott Graham. 2025. "Trying to Kill a Killer; Impressive Killing of Patient Derived Glioblastoma Cultures Using NK-92 Natural Killer Cells Reveals Both Sensitive and Highly Resistant Glioblastoma Cells" Cells 14, no. 1: 53. https://doi.org/10.3390/cells14010053
APA StyleYu, J., Kim, H. J., Reinecke, J., Hucklesby, J., Read, T., Anchan, A., Angel, C. E., & Graham, E. S. (2025). Trying to Kill a Killer; Impressive Killing of Patient Derived Glioblastoma Cultures Using NK-92 Natural Killer Cells Reveals Both Sensitive and Highly Resistant Glioblastoma Cells. Cells, 14(1), 53. https://doi.org/10.3390/cells14010053







