Revitalizing the Epigenome of Adult Jaw Periosteal Cells: Enhancing Diversity in iPSC-Derived Mesenchymal Stem Cells (iMSCs)
Abstract
1. Introduction
2. Methods
2.1. Cell Culture
2.2. Generation of Integration-Free iPSCs from JPCs Using srRNA
2.3. Differentiation of iPSCs into iMSCs
2.4. Flow Cytometric Analysis of JPCs, iPSCs, and iMSCs
2.5. Osteogenic Differentiation
2.6. Gene Expression Analysis of JPCs and iMSCs
2.7. Mouse Model for Teratoma Formation
2.8. DNA Methylation Profiling
2.9. Transcriptome Analysis
2.10. Statistical Analysis
3. Results
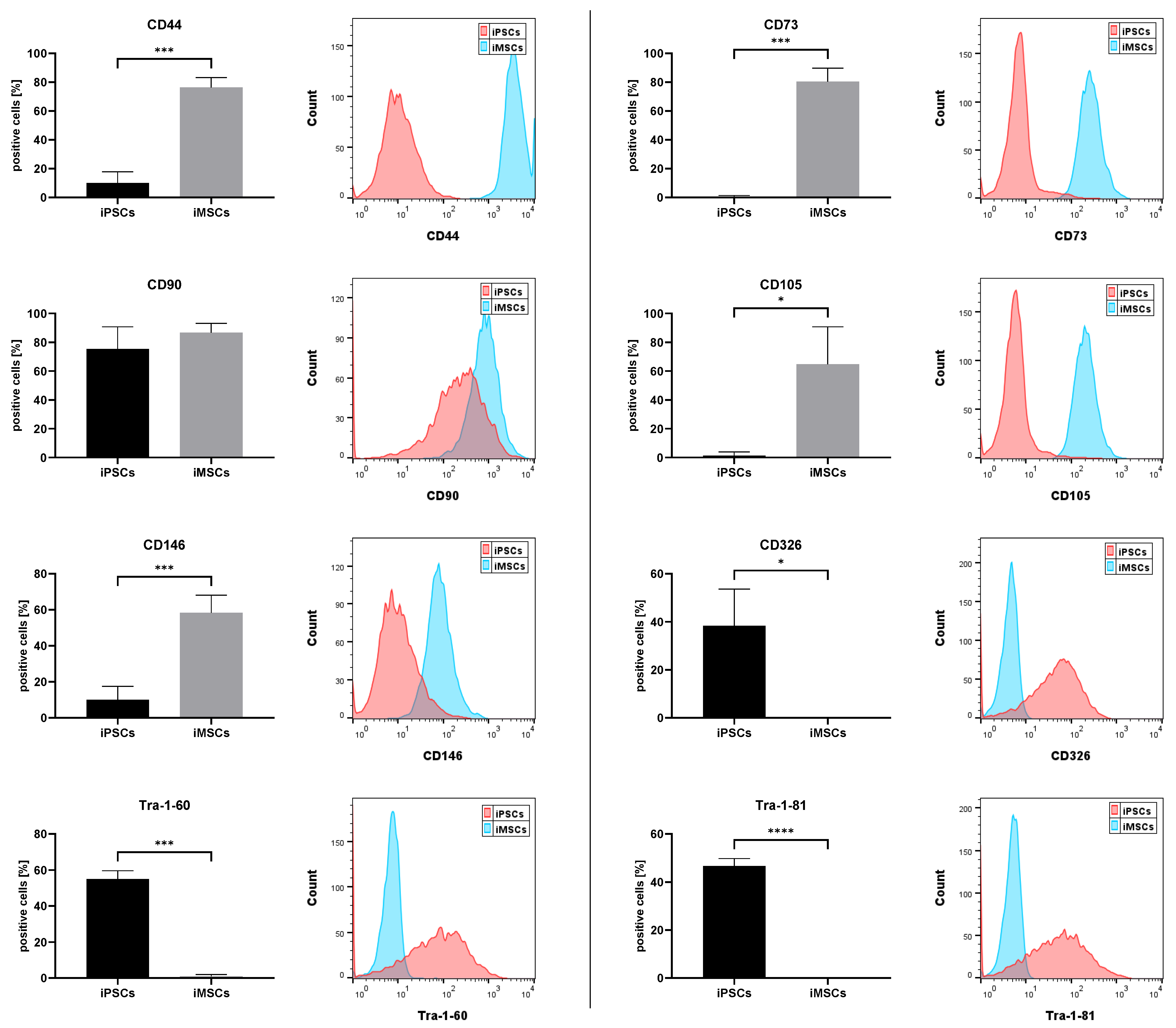
3.1. iMSC Characterization
3.1.1. MSC and iPSC Marker Expression
3.1.2. Osteogenic Differentiation
3.2. DNA Methylation
3.2.1. Differential Methylation Analysis
3.2.2. Enhancer Panel MSCs
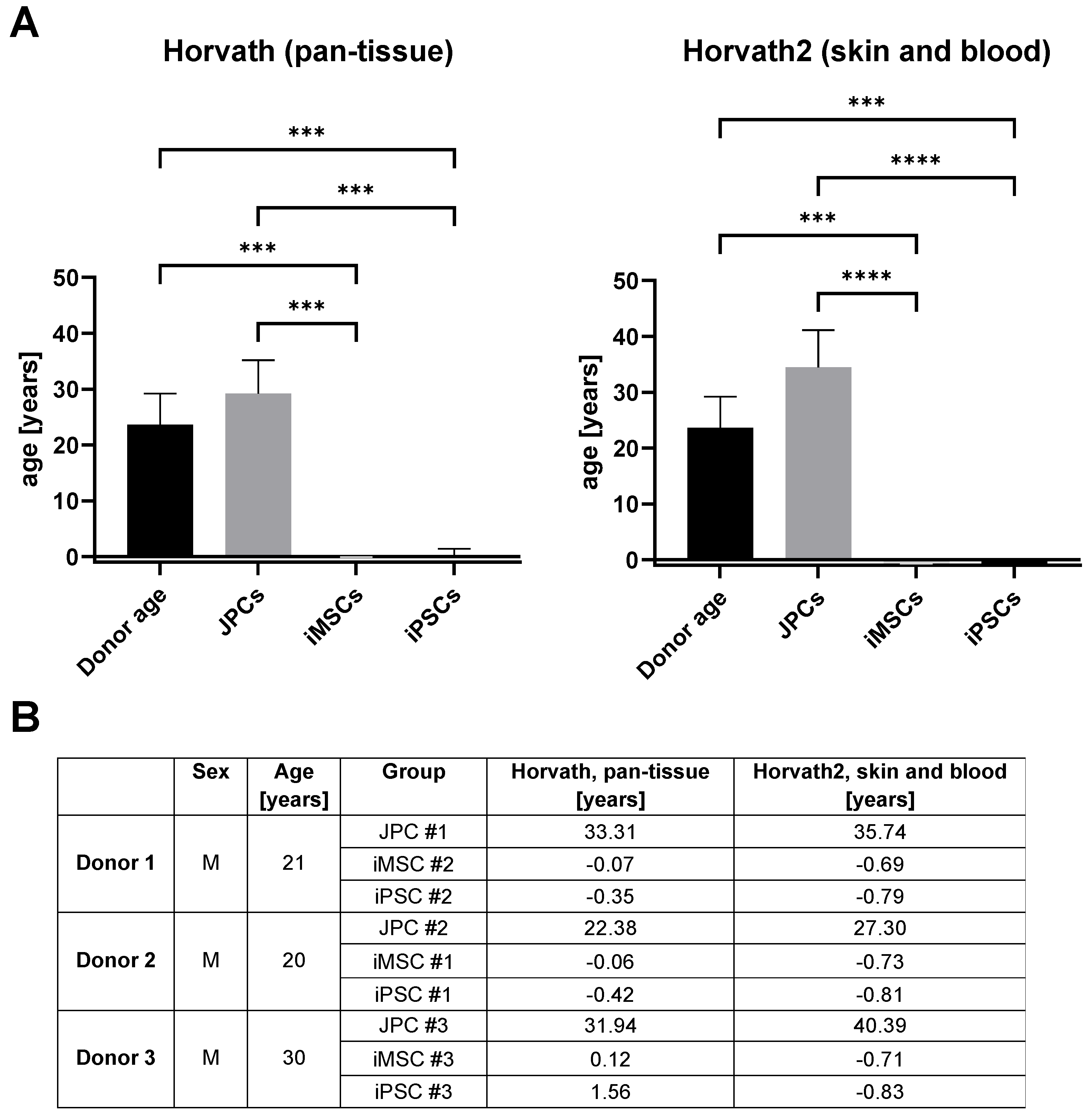
3.2.3. Age Prediction in iMSCs
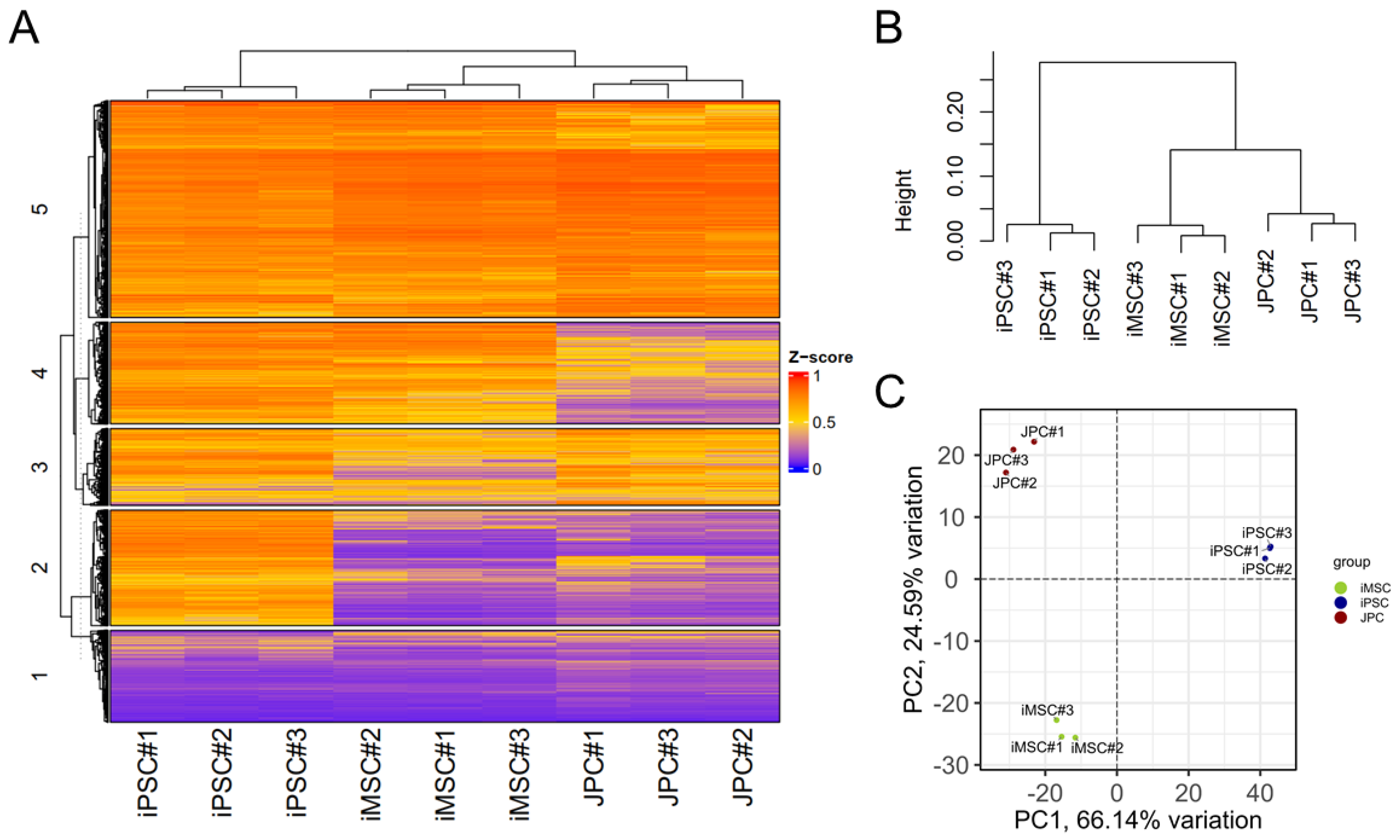
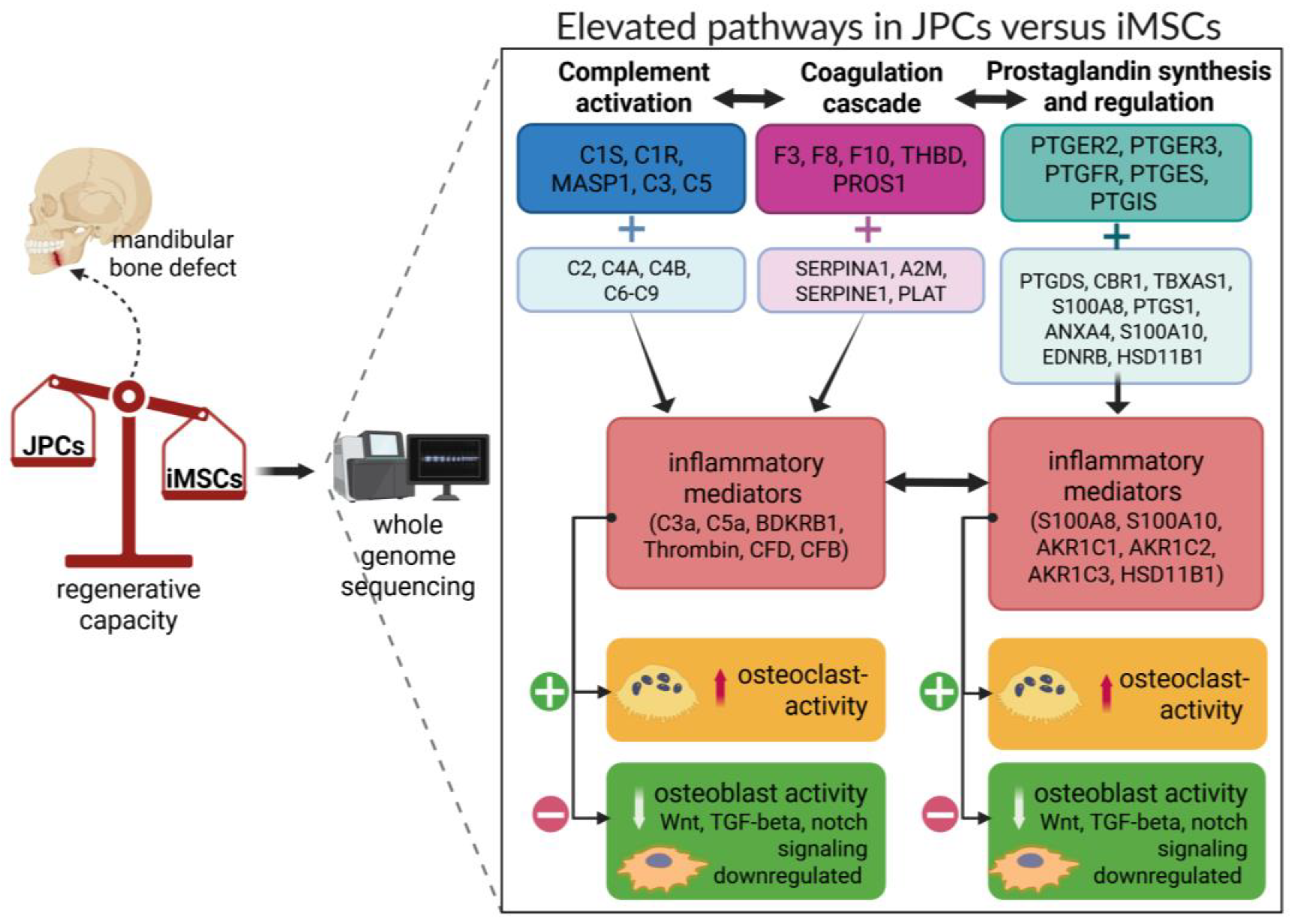
3.3. Transcriptome Analysis
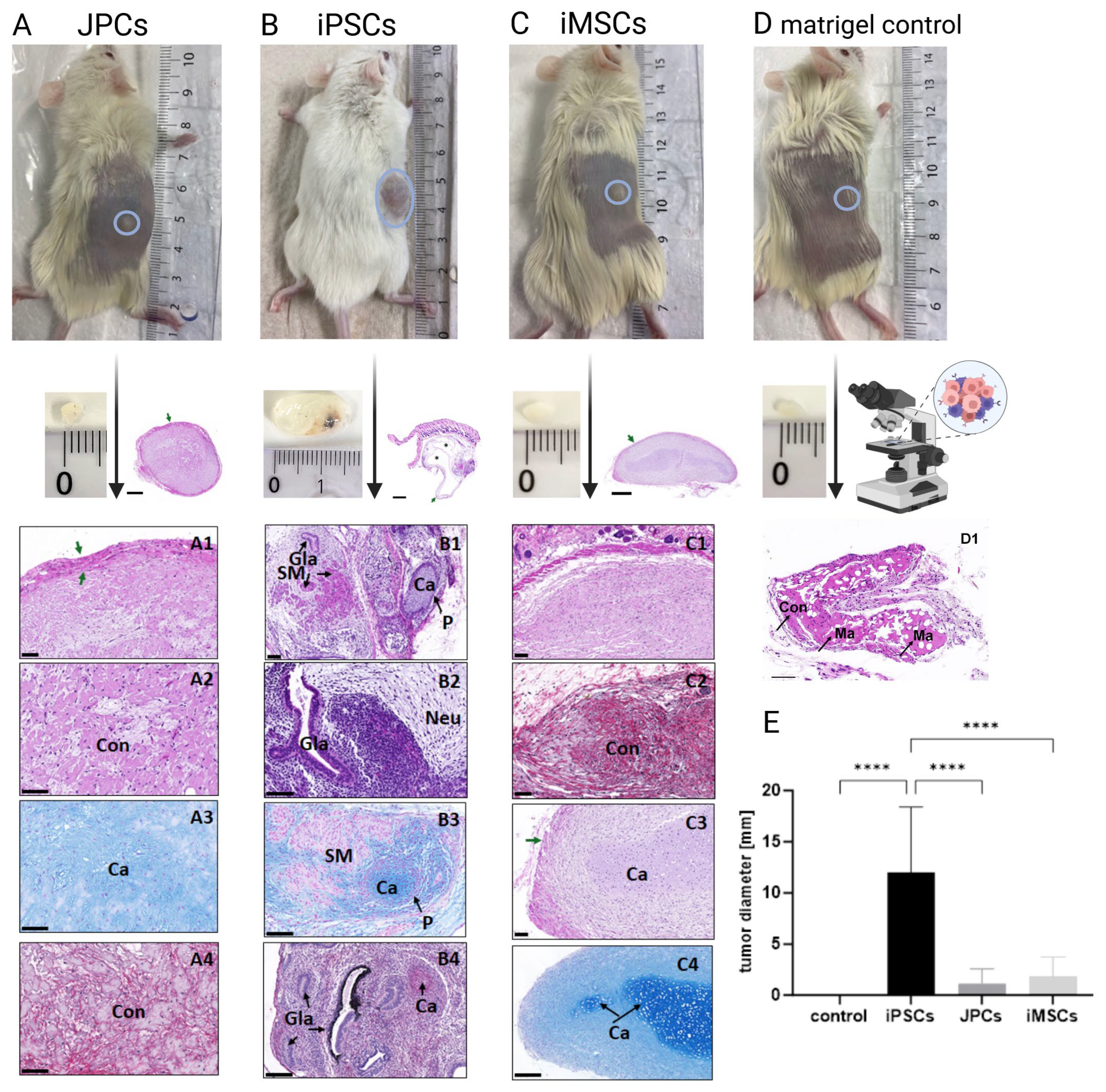
3.4. Teratoma Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BcM | B18R conditioned medium |
| DNAm | DNA methylation |
| ECM | extracellular matrix |
| fdr | false discovery rate |
| GO | gene ontology |
| GvHD | graft-versus-host disease |
| HE | hematoxylin–eosin |
| hPL | human platelet lysate |
| iPSCs | induced pluripotent stem cells |
| iMSCs | iPSC-derived mesenchymal stem cells |
| JPCs | jaw periosteal cells |
| MSCs | mesenchymal stem cells |
| PCA | principal component analysis |
| SASP | senescence-associated secretory phenotype |
| NaB | sodium butyrate |
| VTN | vitronectin |
References
- Divo, M.J.; Martinez, C.H.; Mannino, D.M. Ageing and the epidemiology of multimorbidity. Eur. Respir. J. 2014, 44, 1055–1068. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- Li, Y.; Charif, N.; Mainard, D.; Stoltz, J.F.; de Isla, N. The importance of mesenchymal stem cell donor’s age for cartilage engineering. Osteoarthr. Cartil. 2014, 22, S61. [Google Scholar] [CrossRef][Green Version]
- Alessio, N.; Acar, M.B.; Demirsoy, I.H.; Squillaro, T.; Siniscalco, D.; Di Bernardo, G.; Peluso, G.; Özcan, S.; Galderisi, U. Obesity is associated with senescence of mesenchymal stromal cells derived from bone marrow, subcutaneous and visceral fat of young mice. Aging 2020, 12, 12609–12621. [Google Scholar] [CrossRef]
- Cassidy, F.C.; Shortiss, C.; Murphy, C.G.; Kearns, S.R.; Curtin, W.; De Buitléir, C.; O’Brien, T.; Coleman, C.M. Impact of Type 2 Diabetes Mellitus on Human Bone Marrow Stromal Cell Number and Phenotypic Characteristics. Int. J. Mol. Sci. 2020, 21, 2476. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Yoshimura, H.; Muneta, T.; Nimura, A.; Yokoyama, A.; Koga, H.; Sekiya, I. Comparison of rat mesenchymal stem cells derived from bone marrow, synovium, periosteum, adipose tissue, and muscle. Cell Tissue Res. 2007, 327, 449–462. [Google Scholar] [CrossRef]
- Kirkeby, A.; Main, H.; Carpenter, M. Pluripotent stem-cell-derived therapies in clinical trial: A 2025 update. Cell Stem Cell 2025, 32, 10–37. [Google Scholar] [CrossRef]
- Arakawa, M.; Sakamoto, Y.; Miyagawa, Y.; Nito, C.; Takahashi, S.; Nitahara-Kasahara, Y.; Suda, S.; Yamazaki, Y.; Sakai, M.; Kimura, K.; et al. iPSC-derived mesenchymal stem cells attenuate cerebral ischemia-reperfusion injury by inhibiting inflammatory signaling and oxidative stress. Mol. Ther.-Methods Clin. Dev. 2023, 30, 333–349. [Google Scholar] [CrossRef]
- Jungbluth, P.; Spitzhorn, L.S.; Grassmann, J.; Tanner, S.; Latz, D.; Rahman, M.S.; Bohndorf, M.; Wruck, W.; Sager, M.; Grotheer, V.; et al. Human iPSC-derived iMSCs improve bone regeneration in mini-pigs. Bone Res. 2019, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Poetsch, M.S.; Strano, A.; Guan, K. Human Induced Pluripotent Stem Cells: From Cell Origin, Genomic Stability, and Epigenetic Memory to Translational Medicine. Stem Cells 2022, 40, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, V.; Oltra, E. Methods to produce induced pluripotent stem cell-derived mesenchymal stem cells: Mesenchymal stem cells from induced pluripotent stem cells. World J. Stem Cells 2021, 13, 1094–1111. [Google Scholar] [CrossRef] [PubMed]
- Umrath, F.; Frick, S.L.; Wendt, V.; Naros, A.; Zimmerer, R.; Alexander, D. Inhibition of TGF-β signaling enhances osteogenic potential of iPSC-derived MSCs. Sci. Rep. 2025, 15, 7814. [Google Scholar] [CrossRef]
- Chang, S.-H.; Park, C.G. Comparing the Benefits and Drawbacks of Stem Cell Therapy Based on the Cell Origin or Manipulation Process: Addressing Immunogenicity. Immune Netw. 2023, 23, e44. [Google Scholar] [CrossRef]
- Kot, M.; Baj-Krzyworzeka, M.; Szatanek, R.; Musiał-Wysocka, A.; Suda-Szczurek, M.; Majka, M. The Importance of HLA Assessment in “Off-the-Shelf” Allogeneic Mesenchymal Stem Cells Based-Therapies. Int. J. Mol. Sci. 2019, 20, 5680. [Google Scholar] [CrossRef]
- Pearce, K.F.; Hildebrandt, M.; Greinix, H.; Scheding, S.; Koehl, U.; Worel, N.; Apperley, J.; Edinger, M.; Hauser, A.; Mischak-Weissinger, E.; et al. Regulation of advanced therapy medicinal products in Europe and the role of academia. Cytotherapy 2014, 16, 289–297. [Google Scholar] [CrossRef]
- Lapasset, L.; Milhavet, O.; Prieur, A.; Besnard, E.; Babled, A.; Aït-Hamou, N.; Leschik, J.; Pellestor, F.; Ramirez, J.-M.; De Vos, J. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011, 25, 2248–2253. [Google Scholar] [CrossRef]
- Frobel, J.; Hemeda, H.; Lenz, M.; Abagnale, G.; Joussen, S.; Denecke, B.; Saric, T.; Zenke, M.; Wagner, W. Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells. Stem Cell Rep. 2014, 3, 414–422. [Google Scholar] [CrossRef]
- Spitzhorn, L.-S.; Megges, M.; Wruck, W.; Rahman, M.S.; Otte, J.; Degistirici, Ö.; Meisel, R.; Sorg, R.V.; Oreffo, R.O.C.; Adjaye, J. Human iPSC-derived MSCs (iMSCs) from aged individuals acquire a rejuvenation signature. Stem Cell Res. Ther. 2019, 10, 100. [Google Scholar] [CrossRef]
- Neri, S.; Borzì, R.M. Molecular Mechanisms Contributing to Mesenchymal Stromal Cell Aging. Biomolecules 2020, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.; Boyette, L.B.; Tuan, R.S. Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone 2015, 70, 37–47. [Google Scholar] [CrossRef] [PubMed]
- D’Ippolito, G.; Schiller, P.C.; Ricordi, C.; Roos, B.A.; Howard, G.A. Age-related osteogenic potential of mesenchymal stromal stem cells from human vertebral bone marrow. J. Bone Min. Res. 1999, 14, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Abuna, R.P.; Stringhetta-Garcia, C.T.; Fiori, L.P.; Dornelles, R.C.; Rosa, A.L.; Beloti, M.M. Aging impairs osteoblast differentiation of mesenchymal stem cells grown on titanium by favoring adipogenesis. J. Appl. Oral Sci. 2016, 24, 376–382. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Horie, N.; Satoh, K.; Ishikawa, T.; Mori, T.; Maeda, H.; Fukuda, Y.; Ishizaka, S.; Hiu, T.; Morofuji, Y.; et al. Age of donor of human mesenchymal stem cells affects structural and functional recovery after cell therapy following ischaemic stroke. J. Cereb. Blood Flow Metab. 2018, 38, 1199–1212. [Google Scholar] [CrossRef]
- Liu, Q.; Song, S.; Song, L.; Bi, Y.; Zhu, K.; Qiao, X.; Wang, H.; Gao, C.; Cai, H.; Ji, G. Mesenchymal stem cells alleviate aging in vitro and in vivo. Ann. Transl. Med. 2022, 10, 1092. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Z.; Chen, V.P.; Wang, L.; Inman, C.L.; Zhou, Y.; Guo, C.; Tchkonia, T.; Rowe, D.W.; Kuchel, G.A.; et al. Transplanting cells from old but not young donors causes physical dysfunction in older recipients. Aging Cell 2020, 19, e13106. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Kabacik, S.; Lowe, D.; Fransen, L.; Leonard, M.; Ang, S.-L.; Whiteman, C.; Corsi, S.; Cohen, H.; Felton, S.; Bali, R.; et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nat. Aging 2022, 2, 484–493. [Google Scholar] [CrossRef]
- Umrath, F.; Weber, M.; Reinert, S.; Wendel, H.P.; Avci-Adali, M.; Alexander, D. iPSC-Derived MSCs Versus Originating Jaw Periosteal Cells: Comparison of Resulting Phenotype and Stem Cell Potential. Int. J. Mol. Sci. 2020, 21, 587. [Google Scholar] [CrossRef]
- Lowe, D.; Horvath, S.; Raj, K. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget 2016, 7, 8524–8531. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.Z.; Hernandez, D.G.; Tanaka, T.; Pilling, L.C.; Nalls, M.A.; Bandinelli, S.; Singleton, A.B.; Ferrucci, L. Change in Epigenome-Wide DNA Methylation Over 9 Years and Subsequent Mortality: Results From the InCHIANTI Study. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef]
- Wanner, Y.; Umrath, F.; Waidmann, M.; Reinert, S.; Alexander, D. Platelet Lysate: The Better Choice for Jaw Periosteal Cell Mineralization. Stem Cells Int. 2017, 2017, 8303959. [Google Scholar] [CrossRef]
- Weber, M.; Umrath, F.; Steinle, H.; Schmitt, L.-F.; Yu, L.T.; Schlensak, C.; Wendel, H.-P.; Reinert, S.; Alexander, D.; Avci-Adali, M. Influence of Human Jaw Periosteal Cells Seeded β-Tricalcium Phosphate Scaffolds on Blood Coagulation. Int. J. Mol. Sci. 2021, 22, 9942. [Google Scholar] [CrossRef]
- Umrath, F.; Steinle, H.; Weber, M.; Wendel, H.P.; Reinert, S.; Alexander, D.; Avci-Adali, M. Generation of iPSCs from Jaw Periosteal Cells Using Self-Replicating RNA. Int. J. Mol. Sci. 2019, 20, 1648. [Google Scholar] [CrossRef]
- Umrath, F.; Thomalla, C.; Poschel, S.; Schenke-Layland, K.; Reinert, S.; Alexander, D. Comparative Study of MSCA-1 and CD146 Isolated Periosteal Cell Subpopulations. Cell. Physiol. Biochem. 2018, 51, 1193–1206. [Google Scholar] [CrossRef]
- Sánchez-Porras, D.; Bermejo-Casares, F.; Carmona, R.; Weiss, T.; Campos, F.; Carriel, V. Tissue Fixation and Processing for the Histological Identification of Lipids. Methods Mol. Biol. 2023, 2566, 175–186. [Google Scholar]
- Sánchez-Porras, D.; Varas, J.; Godoy-Guzmán, C.; Bermejo-Casares, F.; San Martín, S.; Carriel, V. Histochemical and Immunohistochemical Methods for the Identification of Proteoglycans. Methods Mol. Biol. 2023, 2566, 85–98. [Google Scholar] [PubMed]
- Carriel, V.S.; Aneiros-Fernandez, J.; Arias-Santiago, S.; Garzón, I.J.; Alaminos, M.; Campos, A. A novel histochemical method for a simultaneous staining of melanin and collagen fibers. J. Histochem. Cytochem. 2011, 59, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Müller, F.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 2014, 11, 1138–1140. [Google Scholar] [CrossRef]
- Müller, F.; Scherer, M.; Assenov, Y.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. RnBeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biol. 2019, 20, 55. [Google Scholar] [CrossRef]
- Pidsley, R.; Wong, C.C.; Volta, M.; Lunnon, K.; Mill, J.; Schalkwyk, L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genom. 2013, 14, 293. [Google Scholar] [CrossRef]
- Pelegí-Sisó, D.; de Prado, P.; Ronkainen, J.; Bustamante, M.; González, J.R. methylclock: A Bioconductor package to estimate DNA methylation age. Bioinformatics 2021, 37, 1759–1760. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data, Version 0.11.8; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential analysis of count data–the DESeq2 package. Genome Biol. 2014, 15, 10–1186. [Google Scholar]
- Akhmedov, M.; Martinelli, A.; Geiger, R.; Kwee, I. Omics Playground: A comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom. Bioinform. 2019, 2, lqz019. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Qian, J. EnhancerAtlas 2.0: An updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020, 48, D58–D64. [Google Scholar] [CrossRef] [PubMed]
- Wruck, W.; Graffmann, N.; Spitzhorn, L.-S.; Adjaye, J. Human Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Acquire Rejuvenation and Reduced Heterogeneity. Front. Cell Dev. Biol. 2021, 9, 717772. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.M.; Chen, A.; Koh, P.W.; Deng, T.Z.; Sinha, R.; Tsai, J.M.; Barkal, A.A.; Shen, K.Y.; Jain, R.; Morganti, R.M.; et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 2016, 166, 451–467. [Google Scholar] [CrossRef]
- Kelly, K.; Bloor, A.J.C.; Griffin, J.E.; Radia, R.; Yeung, D.T.; Rasko, J.E.J. Two-year safety outcomes of iPS cell-derived mesenchymal stromal cells in acute steroid-resistant graft-versus-host disease. Nat. Med. 2024, 30, 1556–1558. [Google Scholar] [CrossRef]
- Wu, H.; Sun, Y.E. Epigenetic Regulation of Stem Cell Differentiation. Pediatr. Res. 2006, 59, 21–25. [Google Scholar] [CrossRef]
- Schellenberg, A.; Lin, Q.; Schüler, H.; Koch, C.M.; Joussen, S.; Denecke, B.; Walenda, G.; Pallua, N.; Suschek, C.V.; Zenke, M.; et al. Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging 2011, 3, 873–888. [Google Scholar] [CrossRef]
- Reinisch, A.; Etchart, N.; Thomas, D.; Hofmann, N.A.; Fruehwirth, M.; Sinha, S.; Chan, C.K.; Senarath-Yapa, K.; Seo, E.-Y.; Wearda, T.; et al. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood 2015, 125, 249–260. [Google Scholar] [CrossRef]
- de Almeida, D.C.; Ferreira, M.R.; Franzen, J.; Weidner, C.I.; Frobel, J.; Zenke, M.; Costa, I.G.; Wagner, W. Epigenetic Classification of Human Mesenchymal Stromal Cells. Stem Cell Rep. 2016, 6, 168–175. [Google Scholar] [CrossRef]
- Danalache, M.; Gaa, L.K.; Burgun, C.; Umrath, F.; Naros, A.; Alexander, D. Mesenchymal Stem Cell Plasticity: What Role Do Culture Conditions and Substrates Play in Shaping Biomechanical Signatures? Bioengineering 2024, 11, 1282. [Google Scholar] [CrossRef]
- Ocampo, A.; Reddy, P.; Martinez-Redondo, P.; Platero-Luengo, A.; Hatanaka, F.; Hishida, T.; Li, M.; Lam, D.; Kurita, M.; Beyret, E.; et al. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 2016, 167, 1719–1733.e12. [Google Scholar] [CrossRef] [PubMed]
- Browder, K.C.; Reddy, P.; Yamamoto, M.; Haghani, A.; Guillen, I.G.; Sahu, S.; Wang, C.; Luque, Y.; Prieto, J.; Shi, L.; et al. In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nat. Aging 2022, 2, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Brommer, B.; Tian, X.; Krishnan, A.; Meer, M.; Wang, C.; Vera, D.L.; Zeng, Q.; Yu, D.; Bonkowski, M.S.; et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature 2020, 588, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.; Parry, A.; Santos, F.; Okkenhaug, H.; Todd, C.D.; Hernando-Herraez, I.; Stubbs, T.M.; Milagre, I.; Reik, W. Multi-omic rejuvenation of human cells by maturation phase transient reprogramming. eLife 2022, 11, e71624. [Google Scholar] [CrossRef]
- Chondronasiou, D.; Gill, D.; Mosteiro, L.; Urdinguio, R.G.; Berenguer-Llergo, A.; Aguilera, M.; Durand, S.; Aprahamian, F.; Nirmalathasan, N.; Abad, M.; et al. Multi-omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell 2022, 21, e13578. [Google Scholar] [CrossRef]
- Mödinger, Y.; Löffler, B.; Huber-Lang, M.; Ignatius, A. Complement involvement in bone homeostasis and bone disorders. Semin. Immunol. 2018, 37, 53–65. [Google Scholar] [CrossRef]
- Ghosh, M.; Rana, S. The anaphylatoxin C5a: Structure, function, signaling, physiology, disease, and therapeutics. Int. Immunopharmacol. 2023, 118, 110081. [Google Scholar] [CrossRef]
- Ishida, M.; Kawao, N.; Mizukami, Y.; Takafuji, Y.; Kaji, H. Serpinb1a suppresses osteoclast formation. Biochem. Biophys. Rep. 2021, 26, 101004. [Google Scholar] [CrossRef]
- Meghadri, S.H.; Martinez-Delgado, B.; Ostermann, L.; Gomez-Mariano, G.; Perez-Luz, S.; Tumpara, S.; Wrenger, S.; DeLuca, D.S.; Maus, U.A.; Welte, T.; et al. Loss of Serpina1 in Mice Leads to Altered Gene Expression in Inflammatory and Metabolic Pathways. Int. J. Mol. Sci. 2022, 23, 10425. [Google Scholar] [CrossRef]
- Mirsaidi, A.; Tiaden, A.N.; Richards, P.J. Prostaglandin E(2) inhibits matrix mineralization by human bone marrow stromal cell-derived osteoblasts via Epac-dependent cAMP signaling. Sci. Rep. 2017, 7, 2243. [Google Scholar] [CrossRef]
- Cmoch, A.; Strzelecka-Kiliszek, A.; Palczewska, M.; Groves, P.; Pikula, S. Matrix vesicles isolated from mineralization-competent Saos-2 cells are selectively enriched with annexins and S100 proteins. Biochem. Biophys. Res. Commun. 2011, 412, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Okura, G.C.; Bharadwaj, A.G.; Waisman, D.M. Recent Advances in Molecular and Cellular Functions of S100A10. Biomolecules 2023, 13, 1450. [Google Scholar] [CrossRef] [PubMed]
- Di Ceglie, I.; Kruisbergen, N.N.L.; van den Bosch, M.H.J.; van Lent, P. Fc-gamma receptors and S100A8/A9 cause bone erosion during rheumatoid arthritis. Do they act as partners in crime? Rheumatology 2019, 58, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lian, X.; Liu, X.; Du, Y.; Zhu, Y.; Hu, M.; Zhang, P.; Liu, Y.; Zhou, Y. Aldo-keto reductase family 1 member C1 regulates the osteogenic differentiation of human ASCs by targeting the progesterone receptor. Stem Cell Res. Ther. 2021, 12, 383. [Google Scholar] [CrossRef]
- Kaftalli, J.; Donato, K.; Bonetti, G.; Dhuli, K.; Macchia, A.; Maltese, P.E.; Louise Herbst, K.; Michelini, S.; Chiurazzi, P.; Hill, M.; et al. Aldo-keto reductase 1C2 (AKR1C2) as the second gene associated to non-syndromic primary lipedema: Investigating activating mutation or overexpression as causative factors. Eur. Rev. Med. Pharmacol. Sci. 2023, 27 (Suppl. S6), 127–136. [Google Scholar]
- Hu, G.; Yu, Y.; Sharma, D.; Pruett-Miller, S.M.; Ren, Y.; Zhang, G.-F.; Karner, C.M. Glutathione limits RUNX2 oxidation and degradation to regulate bone formation. JCI Insight 2023, 8, e166888. [Google Scholar] [CrossRef]
- Maldonado, V.V.; Patel, N.H.; Smith, E.E.; Barnes, C.L.; Gustafson, M.P.; Rao, R.R.; Samsonraj, R.M. Clinical utility of mesenchymal stem/stromal cells in regenerative medicine and cellular therapy. J. Biol. Eng. 2023, 17, 44. [Google Scholar] [CrossRef]
- Fernández-Garza, L.E.; Barrera-Barrera, S.A.; Barrera-Saldaña, H.A. Mesenchymal Stem Cell Therapies Approved by Regulatory Agencies around the World. Pharmaceuticals 2023, 16, 1334. [Google Scholar] [CrossRef]




| Human Antigen | Isotype | Conjugate | Company |
|---|---|---|---|
| SSEA4 | human recombinant antibody (REA) | PE | Miltenyi, Bergisch Gladbach, Germany |
| TRA-1-60 | PE | ||
| TRA-1-81 | PE | ||
| REA-Isotype | PE | ||
| CD73 | mouse IgG1 | PE | BD Biosciences, Franklin Lakes, NJ, USA |
| CD90 | PE | ||
| CD105 | APC | BioLegend, San Diego, CA, USA | |
| IgG1-Isotype | APC | ||
| IgG1-Isotype | PE | R&D Systems, Minneapolis, MN, USA |
| Genes | Average Methylation | p-Value | GO Terms | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| JPCs | iMSCs | iPSCs | JPCs vs. iMSCs | JPCs vs. iPSCs | iMSCs vs. iPSCs | Cell Adhesion | Bone Development | ECM | SSD | SSM | |
| CCDC80 | 0.32 ± 0.2 | 0.49 ± 0.33 | 0.69 ± 0.15 | 0.0003 | <0.0001 | 0.001 | X | ||||
| CD34 | 0.43 ± 0.28 | 0.54 ± 0.29 | 0.55 ± 0.27 | <0.0001 | 0.0034 | 0.9671 | X | ||||
| CDH13 | 0.56 ± 0.2 | 0.63 ± 0.27 | 0.67 ± 0.2 | <0.0001 | <0.0001 | 0.0079 | X | ||||
| CFDP1 | 0.45 ± 0.27 | 0.49 ± 0.31 | 0.56 ± 0.28 | 0.1963 | 0.0022 | 0.1178 | X | ||||
| COL1A1 | 0.23 ± 0.12 | 0.57 ± 0.33 | 0.59 ± 0.29 | <0.0001 | <0.0001 | 0.5364 | X | X | X | X | |
| COL4A2 | 0.76 ± 0.19 | 0.69 ± 0.25 | 0.74 ± 0.16 | <0.0001 | 0.3314 | 0.0006 | X | ||||
| DDR2 | 0.5 ± 0.31 | 0.61 ± 0.29 | 0.73 ± 0.09 | 0.0027 | <0.0001 | 0.0091 | X | X | |||
| DLC1 | 0.53 ± 0.3 | 0.5 ± 0.3 | 0.63 ± 0.26 | 0.2942 | <0.0001 | <0.0001 | X | ||||
| EFNA5 | 0.61 ± 0.28 | 0.53 ± 0.29 | 0.57 ± 0.27 | <0.0001 | 0.2943 | 0.1527 | X | ||||
| EMILIN1 | 0.42 ± 0.32 | 0.44 ± 0.34 | 0.61 ± 0.21 | 0.4037 | 0.0121 | 0.0332 | X | X | |||
| EPHA2 | 0.59 ± 0.32 | 0.48 ± 0.35 | 0.53 ± 0.32 | 0.0002 | 0.0226 | 0.0669 | X | ||||
| FES | 0.53 ± 0.28 | 0.68 ± 0.21 | 0.6 ± 0.25 | <0.0001 | 0.0276 | <0.0001 | X | ||||
| FMOD | 0.52 ± 0.3 | 0.7 ± 0.13 | 0.64 ± 0.19 | 0.0044 | 0.0374 | 0.1394 | X | ||||
| FOXF2 | 0.57 ± 0.3 | 0.19 ± 0.1 | 0.12 ± 0.05 | <0.0001 | <0.0001 | 0.0011 | X | ||||
| GNAS | 0.59 ± 0.22 | 0.4 ± 0.26 | 0.38 ± 0.25 | <0.0001 | <0.0001 | <0.0001 | X | X | X | X | |
| ISLR | 0.45 ± 0.24 | 0.76 ± 0.13 | 0.72 ± 0.11 | <0.0001 | <0.0001 | 0.1003 | X | ||||
| ITGBL1 | 0.58 ± 0.27 | 0.71 ± 0.14 | 0.72 ± 0.13 | <0.0001 | 0.0005 | 0.8764 | X | ||||
| LPP | 0.67 ± 0.24 | 0.65 ± 0.24 | 0.69 ± 0.18 | 0.0731 | 0.2886 | 0.0056 | X | ||||
| LRRC17 | 0.42 ± 0.25 | 0.51 ± 0.31 | 0.63 ± 0.18 | 0.0803 | 0.0021 | 0.168 | X | X | |||
| MGP | 0.4 ± 0.28 | 0.73 ± 0.09 | 0.72 ± 0.12 | 0.0047 | 0.0056 | 0.9947 | X | X | |||
| MYH9 | 0.61 ± 0.26 | 0.62 ± 0.28 | 0.69 ± 0.21 | 0.6502 | 0.0001 | 0.0016 | X | ||||
| PCDHA1 | 0.47 ± 0.17 | 0.68 ± 0.16 | 0.66 ± 0.18 | <0.0001 | <0.0001 | 0.6186 | X | ||||
| PCDHB15 | 0.33 ± 0.14 | 0.62 ± 0.16 | 0.56 ± 0.2 | <0.0001 | <0.0001 | 0.0675 | X | ||||
| PCDHGA2 | 0.46 ± 0.21 | 0.68 ± 0.2 | 0.68 ± 0.2 | <0.0001 | <0.0001 | 0.9797 | X | ||||
| PCDHGA4 | 0.44 ± 0.2 | 0.71 ± 0.21 | 0.65 ± 0.24 | <0.0001 | <0.0001 | <0.0001 | X | ||||
| SH3PXD2B | 0.57 ± 0.29 | 0.68 ± 0.28 | 0.68 ± 0.24 | <0.0001 | <0.0001 | 0.9922 | X | X | X | ||
| SORBS3 | 0.46 ± 0.31 | 0.52 ± 0.31 | 0.45 ± 0.29 | 0.005 | 0.7766 | 0.0007 | X | ||||
| SRCIN1 | 0.57 ± 0.23 | 0.41 ± 0.29 | 0.39 ± 0.3 | <0.0001 | <0.0001 | 0.2304 | X | ||||
| TGFB3 | 0.37 ± 0.29 | 0.54 ± 0.3 | 0.52 ± 0.29 | 0.0056 | 0.0102 | 0.6925 | X | X | X | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umrath, F.; Wendt, V.; Gasparoni, G.; Narknava, Y.; Walter, J.; Lethaus, B.; Weber, J.; Carriel, V.; Avci-Adali, M.; Alexander, D. Revitalizing the Epigenome of Adult Jaw Periosteal Cells: Enhancing Diversity in iPSC-Derived Mesenchymal Stem Cells (iMSCs). Cells 2025, 14, 627. https://doi.org/10.3390/cells14090627
Umrath F, Wendt V, Gasparoni G, Narknava Y, Walter J, Lethaus B, Weber J, Carriel V, Avci-Adali M, Alexander D. Revitalizing the Epigenome of Adult Jaw Periosteal Cells: Enhancing Diversity in iPSC-Derived Mesenchymal Stem Cells (iMSCs). Cells. 2025; 14(9):627. https://doi.org/10.3390/cells14090627
Chicago/Turabian StyleUmrath, Felix, Valerie Wendt, Gilles Gasparoni, Yasser Narknava, Jörn Walter, Bernd Lethaus, Josefin Weber, Victor Carriel, Meltem Avci-Adali, and Dorothea Alexander. 2025. "Revitalizing the Epigenome of Adult Jaw Periosteal Cells: Enhancing Diversity in iPSC-Derived Mesenchymal Stem Cells (iMSCs)" Cells 14, no. 9: 627. https://doi.org/10.3390/cells14090627
APA StyleUmrath, F., Wendt, V., Gasparoni, G., Narknava, Y., Walter, J., Lethaus, B., Weber, J., Carriel, V., Avci-Adali, M., & Alexander, D. (2025). Revitalizing the Epigenome of Adult Jaw Periosteal Cells: Enhancing Diversity in iPSC-Derived Mesenchymal Stem Cells (iMSCs). Cells, 14(9), 627. https://doi.org/10.3390/cells14090627








