The Effect of DNA Methylation in the Development and Progression of Chronic Kidney Disease in the General Population: An Epigenome-Wide Association Study Using the Korean Genome and Epidemiology Study Database
Abstract
:1. Introduction
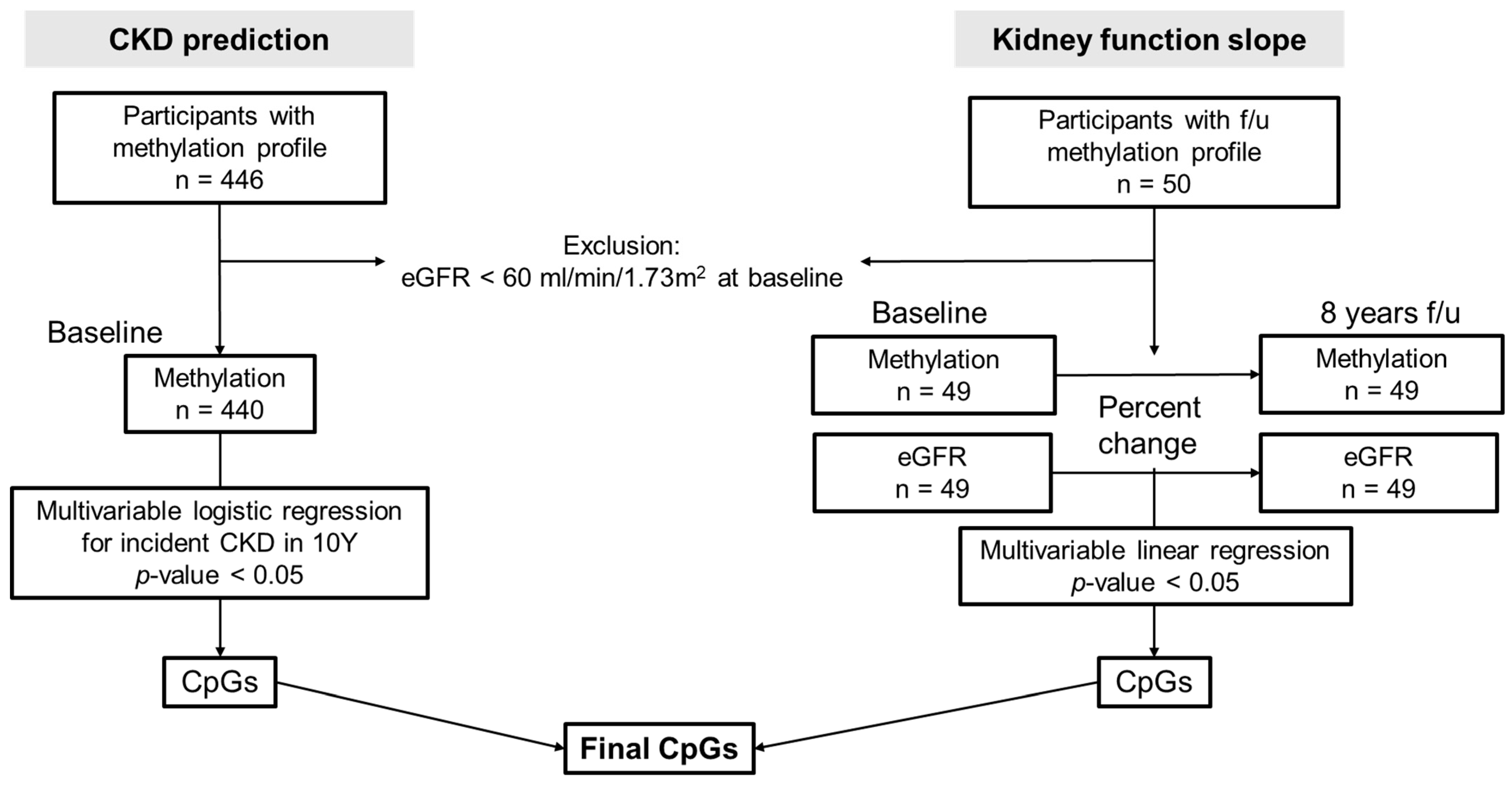
2. Materials and Methods
2.1. Study Population
2.2. DNA Methylation Profiling
2.3. Analysis Regarding Incidence and Progression of CKD
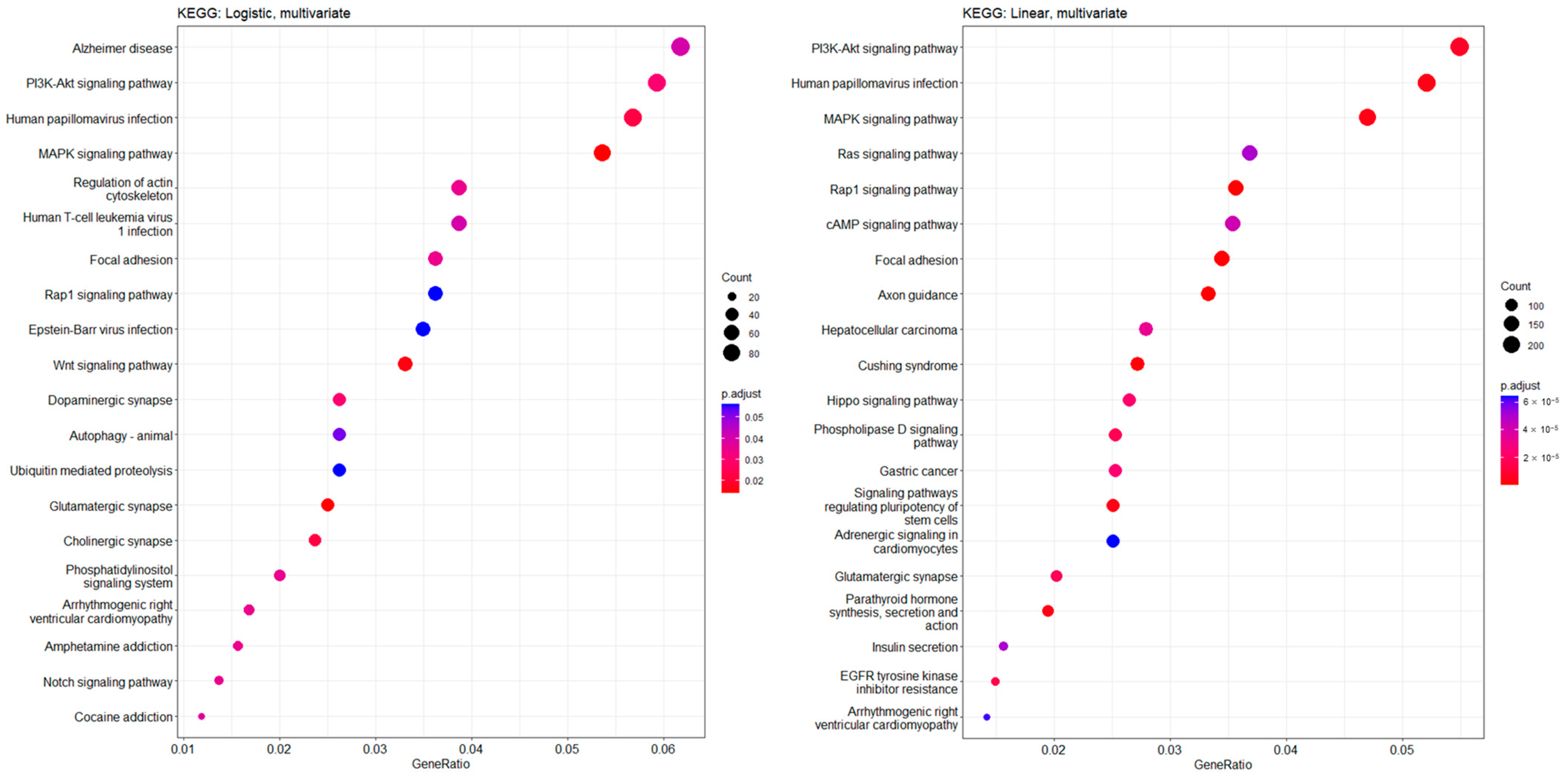
2.4. Statistical Analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
3. Results
3.1. Baseline Characteristics
3.2. Baseline Methylation Profiles Associated with CKD Development in 8 Years (CKD Prediction Analysis)
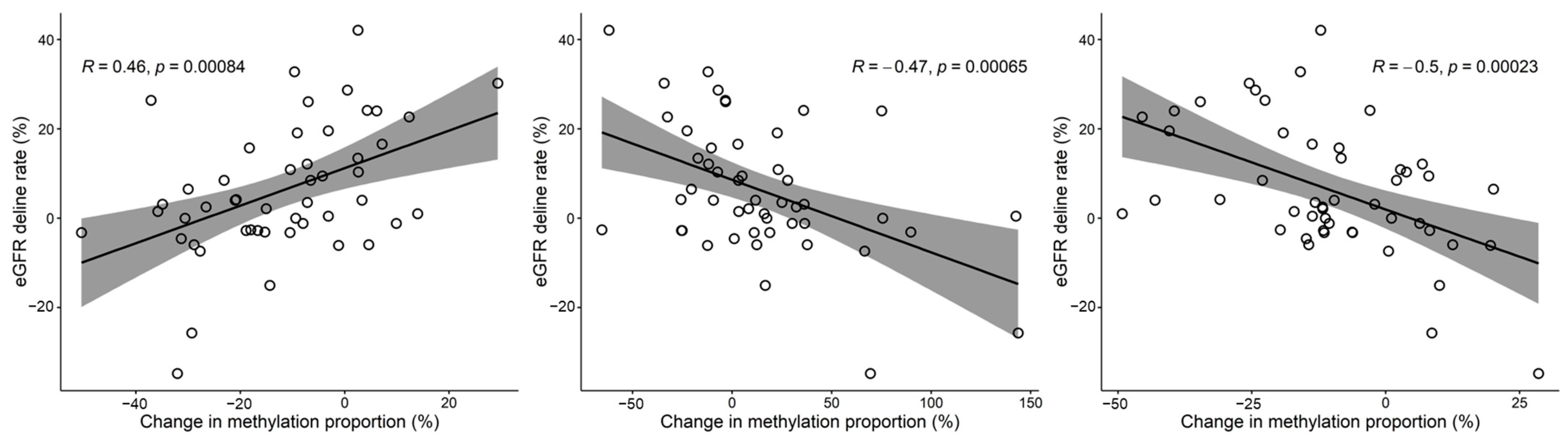
3.3. Relationship between Methylation and eGFR Changes over Time (Kidney Function Slope Analysis)
3.4. Functional Enrichment Features and Common Significant CpG Sites in CKD Prediction and Kidney Function Slope Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Levin, A.; Tonelli, M.; Bonventre, J.; Coresh, J.; Donner, J.A.; Fogo, A.B.; Fox, C.S.; Gansevoort, R.T.; Heerspink, H.J.L.; Jardine, M.; et al. Global kidney health 2017 and beyond: A roadmap for closing gaps in care, research, and policy. Lancet 2017, 390, 1888–1917. [Google Scholar] [CrossRef]
- Jha, V.; Garcia-Garcia, G.; Iseki, K.; Li, Z.; Naicker, S.; Plattner, B.; Saran, R.; Wang, A.Y.; Yang, C.W. Chronic kidney disease: Global dimension and perspectives. Lancet 2013, 382, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Stevens, P.E.; Levin, A. Kidney Disease: Improving Global Outcomes Chronic Kidney Disease Guideline Development Work Group Members. Evaluation and management of chronic kidney disease: Synopsis of the kidney disease: Improving global outcomes 2012 clinical practice guideline. Ann. Intern. Med. 2013, 158, 825–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Velde, M.; Matsushita, K.; Coresh, J.; Astor, B.C.; Woodward, M.; Levey, A.; de Jong, P.; Gansevoort, R.T.; Chronic Kidney Disease Prognosis Consortium; van der Velde, M.; et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with all-cause and cardiovascular mortality. A collaborative meta-analysis of high-risk population cohorts. Kidney Int. 2011, 79, 1341–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slee, A.D. Exploring metabolic dysfunction in chronic kidney disease. Nutr. Metab. 2012, 9, 36. [Google Scholar] [CrossRef] [Green Version]
- Nitsch, D.; Lawlor, D.A.; Patel, R.; Carson, C.; Ebrahim, S. The association of renal impairment with all-cause and cardiovascular disease mortality. Nephrol. Dial. Transplant. 2010, 25, 1191–1199. [Google Scholar] [CrossRef] [Green Version]
- Urbschat, A.; Obermuller, N.; Haferkamp, A. Biomarkers of kidney injury. Biomarkers 2011, 16 (Suppl. S1), S22–S30. [Google Scholar] [CrossRef] [PubMed]
- Breit, M.; Weinberger, K.M. Metabolic biomarkers for chronic kidney disease. Arch. Biochem. Biophys. 2016, 589, 62–80. [Google Scholar] [CrossRef]
- Hocher, B.; Adamski, J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 269–284. [Google Scholar] [CrossRef]
- Canadas-Garre, M.; Anderson, K.; McGoldrick, J.; Maxwell, A.P.; McKnight, A.J. Genomic approaches in the search for molecular biomarkers in chronic kidney disease. J. Transl. Med. 2018, 16, 292. [Google Scholar] [CrossRef] [Green Version]
- Wuttke, M.; Li, Y.; Li, M.; Sieber, K.B.; Feitosa, M.F.; Gorski, M.; Tin, A.; Wang, L.; Chu, A.Y.; Hoppmann, A.; et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2019, 51, 957–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teumer, A.; Li, Y.; Ghasemi, S.; Prins, B.P.; Wuttke, M.; Hermle, T.; Giri, A.; Sieber, K.B.; Qiu, C.; Kirsten, H.; et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat. Commun. 2019, 10, 4130. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin 2018, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011, 12, 529–541. [Google Scholar] [CrossRef] [Green Version]
- Wing, M.R.; Devaney, J.M.; Joffe, M.M.; Xie, D.; Feldman, H.I.; Dominic, E.A.; Guzman, N.J.; Ramezani, A.; Susztak, K.; Herman, J.G.; et al. DNA methylation profile associated with rapid decline in kidney function: Findings from the CRIC study. Nephrol. Dial. Transplant. 2014, 29, 864–872. [Google Scholar] [CrossRef] [Green Version]
- Sapienza, C.; Lee, J.; Powell, J.; Erinle, O.; Yafai, F.; Reichert, J.; Siraj, E.S.; Madaio, M. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 2011, 6, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Ingrosso, D.; Perna, A.F. DNA Methylation Dysfunction in Chronic Kidney Disease. Genes 2020, 11, 811. [Google Scholar] [CrossRef]
- Hill, C.; Duffy, S.; Kettyle, L.M.; McGlynn, L.; Sandholm, N.; Salem, R.M.; Thompson, A.; Swan, E.J.; Kilner, J.; Rossing, P.; et al. Differential Methylation of Telomere-Related Genes Is Associated with Kidney Disease in Individuals with Type 1 Diabetes. Genes 2023, 14, 1029. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G.; KoGES Group. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, e20. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.P.; Koh, I.U.; Choi, N.H.; Kim, B.J.; Han, B.G.; Lee, S. Differential DNA methylation of MSI2 and its correlation with diabetic traits. PLoS ONE 2017, 12, e0177406. [Google Scholar] [CrossRef] [Green Version]
- Shim, S.M.; Cho, Y.K.; Hong, E.J.; Han, B.G.; Jeon, J.P. An epigenomic signature of postprandial hyperglycemia in peripheral blood leukocytes. J. Hum. Genet. 2016, 61, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, B.; Yi, M.; Qiu, H.; Yuan, X. A Prognostic Nomogram Model Based on mRNA Expression of DNA Methylation-Driven Genes for Gastric Cancer. Front. Oncol. 2020, 10, 584733. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Y.; Yu, H.; Liu, L.; Zhu, C.; Zuo, S.; Chen, Z. An aberrant DNA methylation signature for predicting hepatocellular carcinoma. Ann. Transl. Med. 2020, 8, 1667. [Google Scholar] [CrossRef]
- Zhang, E.; Hou, X.; Hou, B.; Zhang, M.; Song, Y. A risk prediction model of DNA methylation improves prognosis evaluation and indicates gene targets in prostate cancer. Epigenomics 2020, 12, 333–352. [Google Scholar] [CrossRef] [Green Version]
- Guastafierro, T.; Bacalini, M.G.; Marcoccia, A.; Gentilini, D.; Pisoni, S.; Di Blasio, A.M.; Corsi, A.; Franceschi, C.; Raimondo, D.; Spano, A.; et al. Genome-wide DNA methylation analysis in blood cells from patients with Werner syndrome. Clin. Epigenetics 2017, 9, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowdon, R.F.; Jang, H.S.; Wang, T. Evolution of Epigenetic Regulation in Vertebrate Genomes. Trends Genet. 2016, 32, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Lea, A.J.; Vockley, C.M.; Johnston, R.A.; Del Carpio, C.A.; Barreiro, L.B.; Reddy, T.E.; Tung, J. Genome-wide quantification of the effects of DNA methylation on human gene regulation. Elife 2018, 7, e37513. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef]
- Aran, D.; Sabato, S.; Hellman, A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013, 14, R21. [Google Scholar] [CrossRef] [Green Version]
- Dayeh, T.; Volkov, P.; Salo, S.; Hall, E.; Nilsson, E.; Olsson, A.H.; Kirkpatrick, C.L.; Wollheim, C.B.; Eliasson, L.; Ronn, T.; et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014, 10, e1004160. [Google Scholar] [CrossRef] [PubMed]
- Bakulski, K.M.; Dolinoy, D.C.; Sartor, M.A.; Paulson, H.L.; Konen, J.R.; Lieberman, A.P.; Albin, R.L.; Hu, H.; Rozek, L.S. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J. Alzheimer’s Dis. 2012, 29, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Stenvinkel, P.; Karimi, M.; Johansson, S.; Axelsson, J.; Suliman, M.; Lindholm, B.; Heimburger, O.; Barany, P.; Alvestrand, A.; Nordfors, L.; et al. Impact of inflammation on epigenetic DNA methylation—A novel risk factor for cardiovascular disease? J. Intern. Med. 2007, 261, 488–499. [Google Scholar] [CrossRef]
- Smyth, L.J.; McKay, G.J.; Maxwell, A.P.; McKnight, A.J. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics 2014, 9, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Fuchshuber, A.; Jean, G.; Gribouval, O.; Gubler, M.C.; Broyer, M.; Beckmann, J.S.; Niaudet, P.; Antignac, C. Mapping a gene (SRN1) to chromosome 1q25-q31 in idiopathic nephrotic syndrome confirms a distinct entity of autosomal recessive nephrosis. Hum. Mol. Genet. 1995, 4, 2155–2158. [Google Scholar] [CrossRef]
- Caridi, G.; Perfumo, F.; Ghiggeri, G.M. NPHS2 (Podocin) mutations in nephrotic syndrome. Clinical spectrum and fine mechanisms. Pediatr. Res. 2005, 57, 54R–61R. [Google Scholar] [CrossRef] [PubMed]
- Roselli, S.; Heidet, L.; Sich, M.; Henger, A.; Kretzler, M.; Gubler, M.C.; Antignac, C. Early glomerular filtration defect and severe renal disease in podocin-deficient mice. Mol. Cell. Biol. 2004, 24, 550–560. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.Y.; Marlier, A.; Gribouval, O.; Gilbert, T.; Heidet, L.; Antignac, C.; Gubler, M.C. In vivo expression of podocyte slit diaphragm-associated proteins in nephrotic patients with NPHS2 mutation. Kidney Int. 2004, 66, 945–954. [Google Scholar] [CrossRef] [Green Version]
- Denison, M.S.; Nagy, S.R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 309–334. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Miao, H.; Wang, Y.N.; Chen, D.Q.; Wu, X.Q.; Chen, L.; Guo, Y.; Zou, L.; Vaziri, N.D.; Li, P.; et al. Intrarenal 1-methoxypyrene, an aryl hydrocarbon receptor agonist, mediates progressive tubulointerstitial fibrosis in mice. Acta Pharmacol. Sin. 2022, 43, 2929–2945. [Google Scholar] [CrossRef]
- Lee, W.J.; Liu, S.H.; Chiang, C.K.; Lin, S.Y.; Liang, K.W.; Chen, C.H.; Tien, H.R.; Chen, P.H.; Wu, J.P.; Tsai, Y.C.; et al. Aryl Hydrocarbon Receptor Deficiency Attenuates Oxidative Stress-Related Mesangial Cell Activation and Macrophage Infiltration and Extracellular Matrix Accumulation in Diabetic Nephropathy. Antioxid. Redox Signal. 2016, 24, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.T.; Kim, S.S.; Jun, D.W.; Hwang, Y.H.; Park, W.H.; Pak, Y.K.; Lee, H.K. Serum arylhydrocarbon receptor transactivating activity is elevated in type 2 diabetic patients with diabetic nephropathy. J. Diabetes Investig. 2013, 4, 483–491. [Google Scholar] [CrossRef] [Green Version]
- Sokol, A.M.; Uszczynska-Ratajczak, B.; Collins, M.M.; Bazala, M.; Topf, U.; Lundegaard, P.R.; Sugunan, S.; Guenther, S.; Kuenne, C.; Graumann, J.; et al. Loss of the Mia40a oxidoreductase leads to hepato-pancreatic insufficiency in zebrafish. PLoS Genet. 2018, 14, e1007743. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Staples, O.; Thomas, L.W.; Briston, T.; Robson, M.; Poon, E.; Simoes, M.L.; El-Emir, E.; Buffa, F.M.; Ahmed, A.; et al. Human CHCHD4 mitochondrial proteins regulate cellular oxygen consumption rate and metabolism and provide a critical role in hypoxia signaling and tumor progression. J. Clin. Investig. 2012, 122, 600–611. [Google Scholar] [CrossRef]
- Thomas, L.W.; Staples, O.; Turmaine, M.; Ashcroft, M. CHCHD4 Regulates Intracellular Oxygenation and Perinuclear Distribution of Mitochondria. Front. Oncol. 2017, 7, 71. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Yang, J.; Zhu, X.; Zhong, J.; Elshaer, A.; Matsusaka, T.; Pastan, I.; Haase, V.H.; Yang, H.C.; Fogo, A.B. Stabilization of hypoxia-inducible factor ameliorates glomerular injury sensitization after tubulointerstitial injury. Kidney Int. 2021, 99, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Al-Habib, H.; Ashcroft, M. CHCHD4 (MIA40) and the mitochondrial disulfide relay system. Biochem. Soc. Trans. 2021, 49, 17–27. [Google Scholar] [CrossRef]
- Liu, Y. Epithelial to mesenchymal transition in renal fibrogenesis: Pathologic significance, molecular mechanism, and therapeutic intervention. J. Am. Soc. Nephrol. 2004, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Winbanks, C.E.; Grimwood, L.; Gasser, A.; Darby, I.A.; Hewitson, T.D.; Becker, G.J. Role of the phosphatidylinositol 3-kinase and mTOR pathways in the regulation of renal fibroblast function and differentiation. Int. J. Biochem. Cell Biol. 2007, 39, 206–219. [Google Scholar] [CrossRef]
- Cheng, X.; Gao, W.; Dang, Y.; Liu, X.; Li, Y.; Peng, X.; Ye, X. Both ERK/MAPK and TGF-Beta/Smad signaling pathways play a role in the kidney fibrosis of diabetic mice accelerated by blood glucose fluctuation. J. Diabetes Res. 2013, 2013, 463740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.N.; Yang, S.H.; Kim, Y.C.; Hwang, J.H.; Park, J.Y.; Kim, D.K.; Kim, J.H.; Kim, D.W.; Hur, D.G.; Oh, Y.K.; et al. Periostin induces kidney fibrosis after acute kidney injury via the p38 MAPK pathway. Am. J. Physiol. Ren. Physiol. 2019, 316, F426–F437. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, P.H.; Xu, C.G.; Zhou, X.J.; Hu, W.; Zhang, J. Baicalein attenuates renal fibrosis by inhibiting inflammation via down-regulating NF-kappaB and MAPK signal pathways. J. Mol. Histol. 2015, 46, 283–290. [Google Scholar] [CrossRef]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.C.; Ried, J.S.; Zhang, W.; Yang, Y.; et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Marconett, C.N.; Duan, J.; Hyland, P.L.; Li, P.; Wang, Z.; Wheeler, W.; Zhou, B.; Campan, M.; Lee, D.S.; et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat. Commun. 2014, 5, 3365. [Google Scholar] [CrossRef] [Green Version]
- Schlosser, P.; Tin, A.; Matias-Garcia, P.R.; Thio, C.H.L.; Joehanes, R.; Liu, H.; Weihs, A.; Yu, Z.; Hoppmann, A.; Grundner-Culemann, F.; et al. Meta-analyses identify DNA methylation associated with kidney function and damage. Nat. Commun. 2021, 12, 7174. [Google Scholar] [CrossRef]
- Loyfer, N.; Magenheim, J.; Peretz, A.; Cann, G.; Bredno, J.; Klochendler, A.; Fox-Fisher, I.; Shabi-Porat, S.; Hecht, M.; Pelet, T.; et al. A DNA methylation atlas of normal human cell types. Nature 2023, 613, 355–364. [Google Scholar] [CrossRef]

| Baseline Variables | Values |
|---|---|
| Age, years [M ± SE] | 52.1 ± 8.4 |
| Female sex, [abs (%)] | 215 (48.9) |
| Body mass index, kg/m2 [M ± SE] | 24.6 ± 3.4 |
| Smoking tobacco, [abs (%)] | |
| Current | 121 (27.5) |
| Ex | 65 (14.8) |
| Never | 245 (55.7) |
| No response | 9 (2.1) |
| Alcohol consumption, [abs (%)] | |
| Current | 218 (49.6) |
| Ex | 30 (6.8) |
| Never | 186 (42.3) |
| No response | 6 (1.4) |
| Hypertension, [abs (%)] | 63 (14.3) |
| Diabetes mellitus, [abs (%)] | 43 (9.8) |
| Dyslipidemia, [abs (%)] | 7 (1.6) |
| Myocardial infarction, [abs (%)] | 1 (0.2) |
| Congestive heart failure, [abs (%)] | 1 (0.2) |
| Cerebrovascular disease, [abs (%)] | 4 (0.9) |
| Systolic blood pressure, mmHg [M ± SE] | 121.9 ± 17.8 |
| Diastolic blood pressure, mmHg [M ± SE] | 81.2 ± 11.2 |
| eGFR CKD-EPI, mL/min/1.73 m2 [M ± SE] | 91.8 ± 12.8 |
| Serum blood urea nitrogen, mg/dL [M ± SE] | 14.4 ± 3.4 |
| Serum creatinine, mg/dL [M ± SE] | 0.85 ± 0.17 |
| Serum total cholesterol, mg/dL [M ± SE] | 193.3 ± 35.0 |
| Blood hemoglobin, g/dL [M ± SE] | 13.7 ± 1.6 |
| Serum albumin, g/dL [M ± SE] | 4.3 ± 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-E.; Jo, M.-J.; Cho, E.; Ahn, S.-Y.; Kwon, Y.-J.; Gim, J.-A.; Ko, G.-J. The Effect of DNA Methylation in the Development and Progression of Chronic Kidney Disease in the General Population: An Epigenome-Wide Association Study Using the Korean Genome and Epidemiology Study Database. Genes 2023, 14, 1489. https://doi.org/10.3390/genes14071489
Kim J-E, Jo M-J, Cho E, Ahn S-Y, Kwon Y-J, Gim J-A, Ko G-J. The Effect of DNA Methylation in the Development and Progression of Chronic Kidney Disease in the General Population: An Epigenome-Wide Association Study Using the Korean Genome and Epidemiology Study Database. Genes. 2023; 14(7):1489. https://doi.org/10.3390/genes14071489
Chicago/Turabian StyleKim, Ji-Eun, Min-Jee Jo, Eunjung Cho, Shin-Young Ahn, Young-Joo Kwon, Jeong-An Gim, and Gang-Jee Ko. 2023. "The Effect of DNA Methylation in the Development and Progression of Chronic Kidney Disease in the General Population: An Epigenome-Wide Association Study Using the Korean Genome and Epidemiology Study Database" Genes 14, no. 7: 1489. https://doi.org/10.3390/genes14071489
APA StyleKim, J. -E., Jo, M. -J., Cho, E., Ahn, S. -Y., Kwon, Y. -J., Gim, J. -A., & Ko, G. -J. (2023). The Effect of DNA Methylation in the Development and Progression of Chronic Kidney Disease in the General Population: An Epigenome-Wide Association Study Using the Korean Genome and Epidemiology Study Database. Genes, 14(7), 1489. https://doi.org/10.3390/genes14071489





