The Effects of a Typhoon on the Dynamic of Microbial Community Structure and Water Quality of the Marine Bathing Beach
Abstract
:1. Introduction
2. Materials and Methods
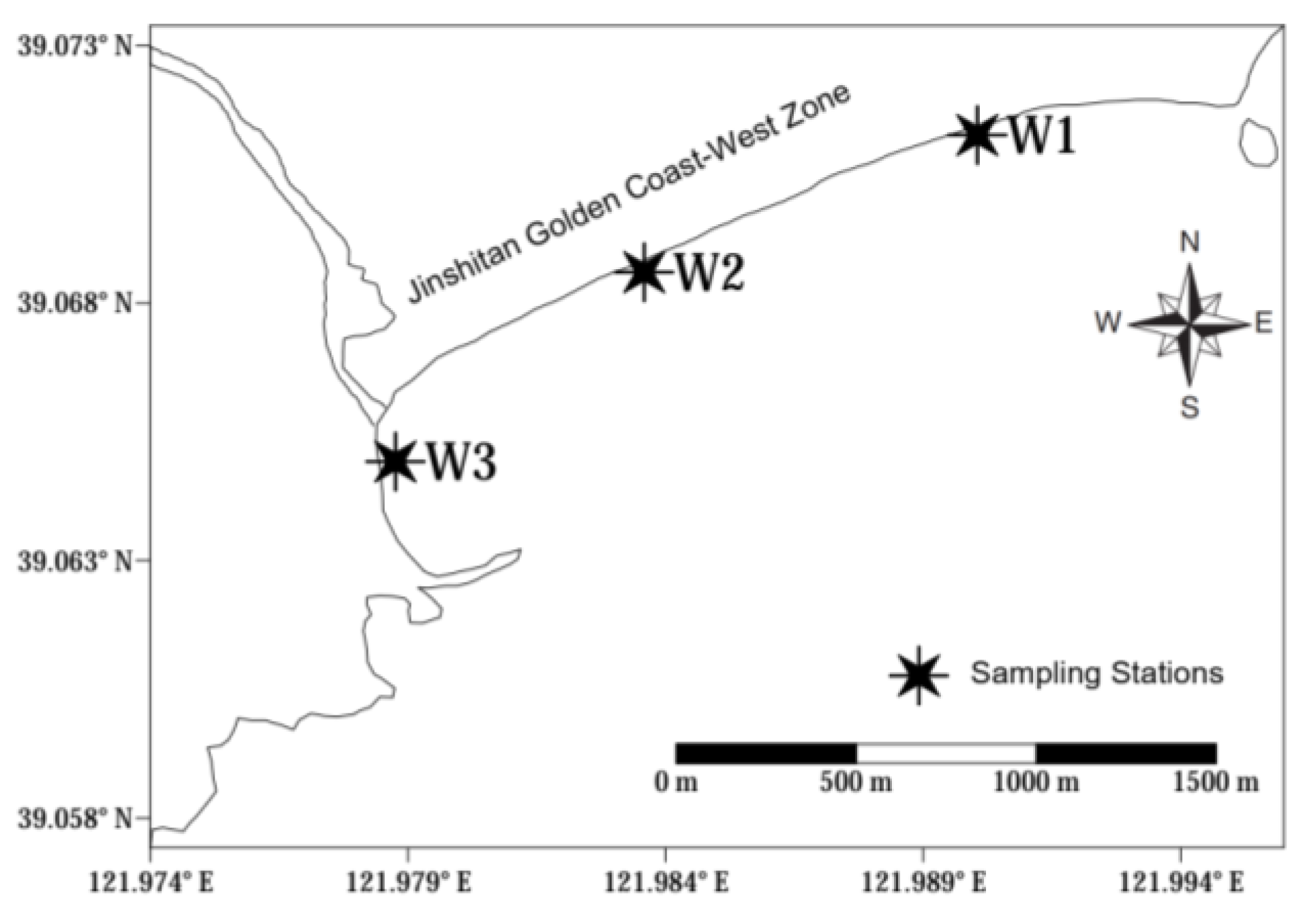
2.1. Sample Collection
2.2. Physical, Chemical, Meteorological, and Hydrological Parameters Analysis in Water and Beach Sand
2.3. Enterococci Cultivation
2.4. 16S rRNA Gene Quantification
2.5. 16S rRNA Gene Sequencing
2.6. Data Analysis
3. Results
3.1. The Physical-Chemical Factors of the Beach during the Typhoon Event
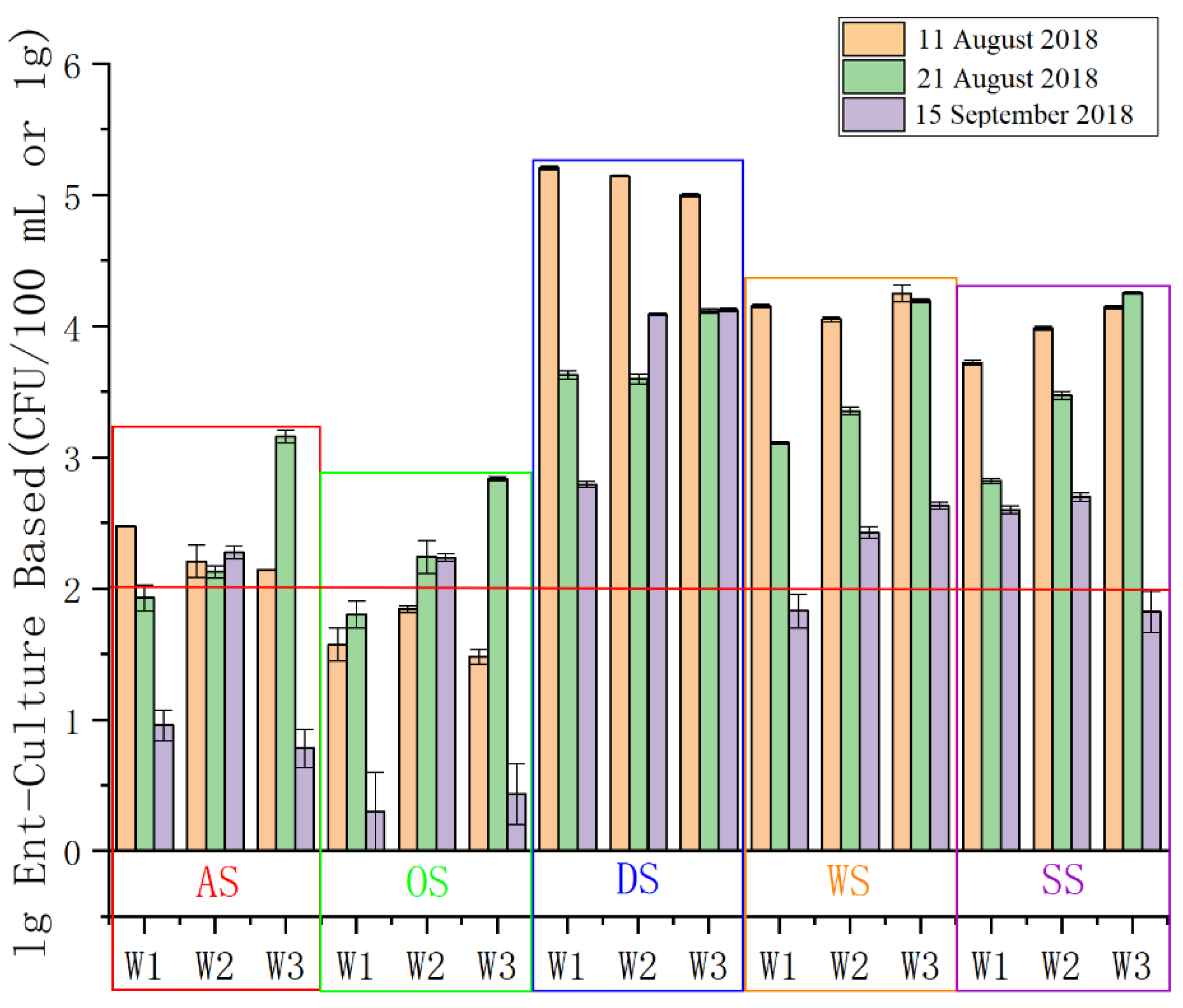
3.2. The Effect of the Typhoon Event on Water Quality According to Enterococci Concentration
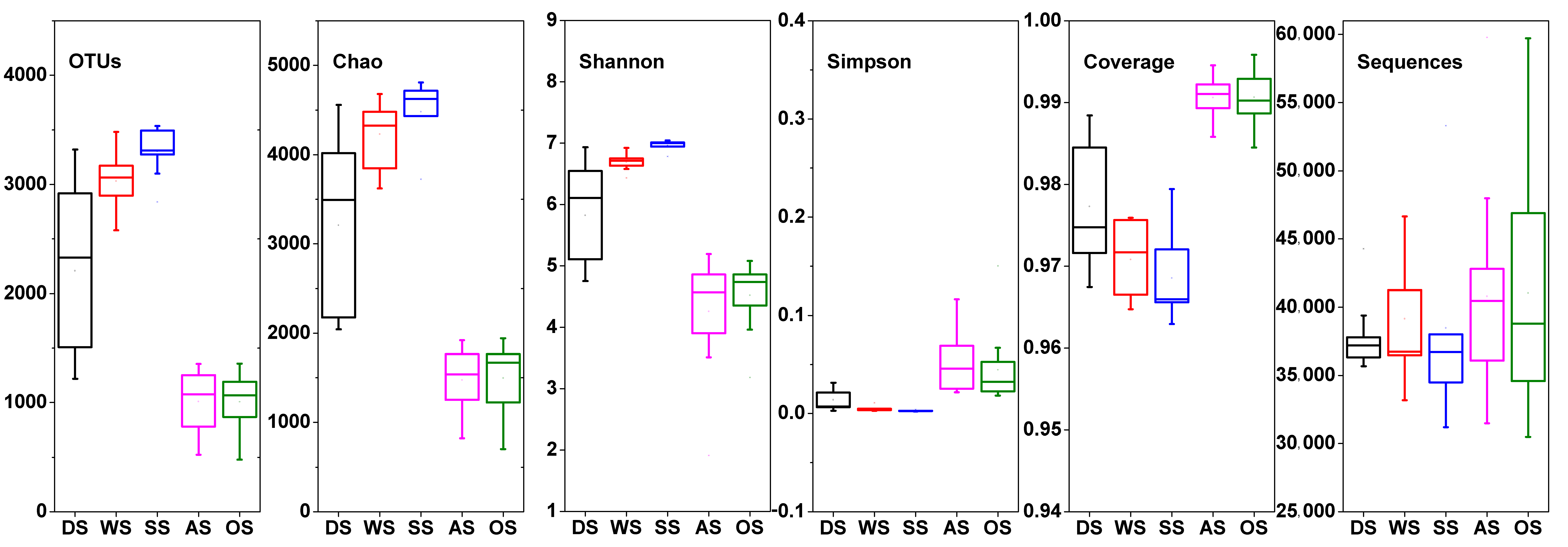
3.3. Abundance and Diversity of the Bacterial Community on the Beach
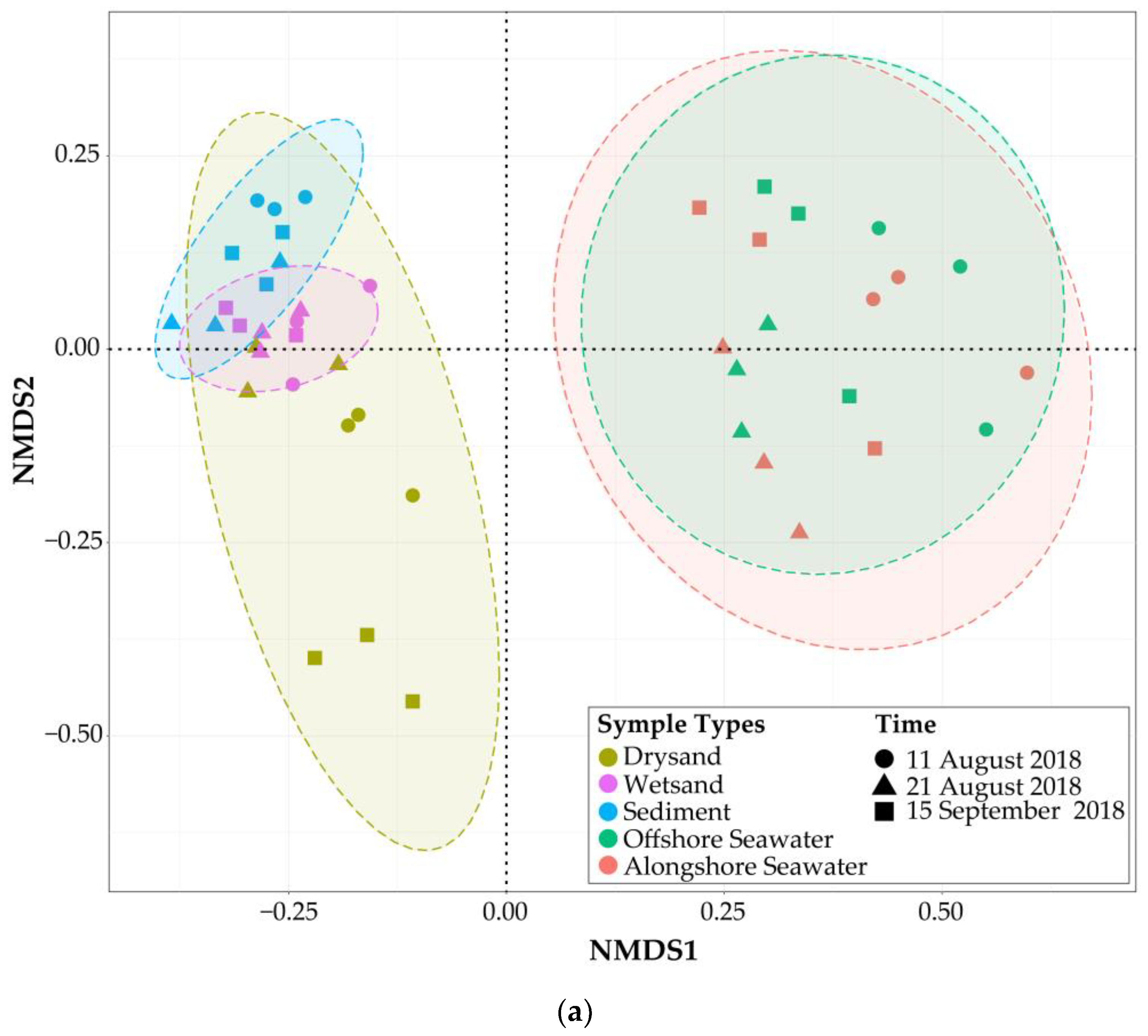
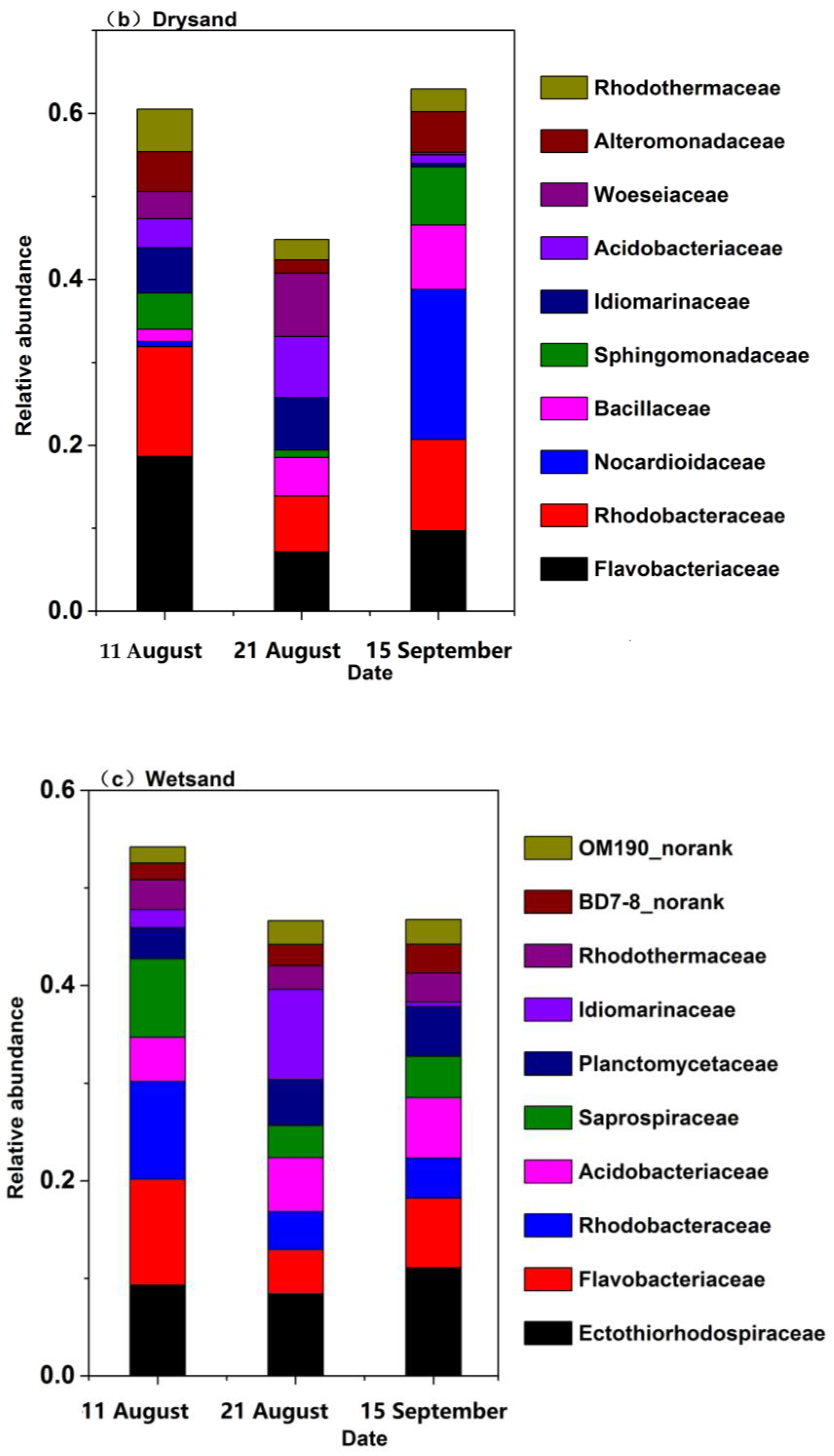
3.4. Bacterial Community Structure and Composition in Seawater and Sand Samples of the Bathing Beach
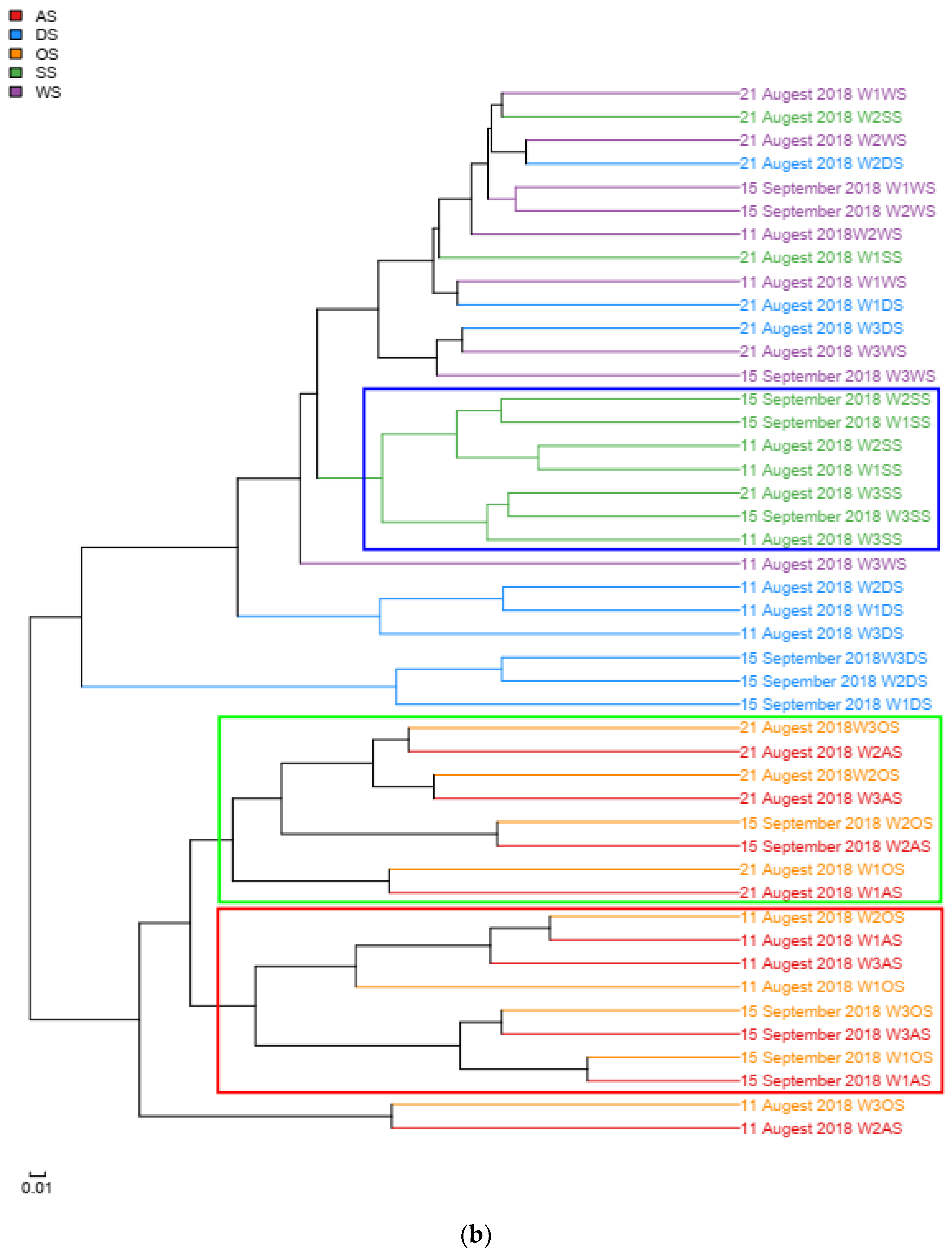
3.5. The Bacterial Community Structure Transition Due to the Typhoon Event
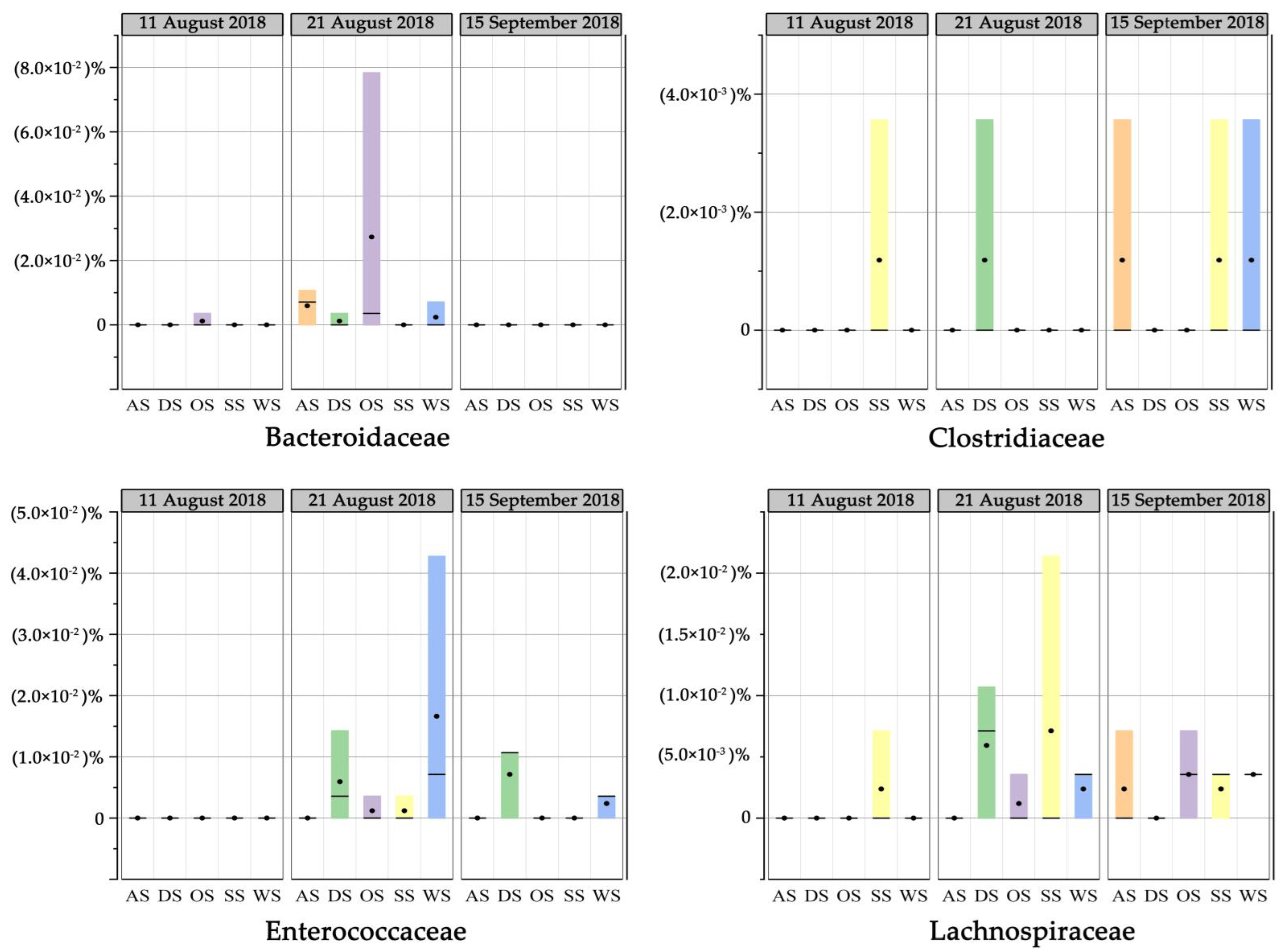
3.6. Fecal Contamination Based on DNA Sequencing during the Typhoon Event
3.7. The Main Environmental Factor Affecting Bacterial Community Structure
4. Discussions
4.1. Effects of the Typhoon on Water Quality
4.2. Typhoon-Induced Redistribution of Bacterial Community
4.3. The Recovery of Bacterial Community Structure after the Typhoon
5. Conclusions
- (1)
- The concentration of enterococci in sand samples decreased while it increased in seawater, and the water quality significantly deteriorated after the typhoon and heavy storm. Human and other fecal taxa increased immediately after the typhoon caused by runoff or riverine input. The transportation of fecal bacteria was observed from sand or runoff to seawater on the bathing beach.
- (2)
- There were significant differences in microbial community structure between seawater and beach sand samples even after the typhoon. Immediately after the typhoon, the bacterial community diversity commonly increased. One month later, the bacterial diversity in seawater and dry sand decreased, while there was a significant difference between before and one month after the typhoon in the dry sand. However, in wet or submerged sand, the diversity nearly recovered.
- (3)
- The bacterial community in Jinshitan bathing beach changed after the typhoon. Cyanobacteria decreased in seawater, while Acidobacteriota and Planctomycetota increased. Bacteroidetes in seawater and sand decreased. However, Alphaproteobacteria and Gammaproteobacteria fluctuated during the typhoon event.
- (4)
- The water quality parameters of seawater had a high degree of interpretation for their community structure, among which pH, ORP, and water temperature were the most significant factors. Bacteria abundance was another vital factor. NH4+ was the most driven factor of the bacterial community in the sand. Apart from the impact of the typhoon, the geographical location was another important factor in the changed bacterial community.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- United Nations. The Ocean Conference. Available online: https://www.un.org/sustainabledevelopment/wp-content/uploads/2017/05/Oceanfact-sheet-package.pdf (accessed on 30 March 2017).
- WHO. Chapter 4: Faecal pollution and water quality. In Guidelines for Safe Recreational Water Environments. Volume 1, Coastal and Fresh Water; World Health Organization: Geneva, Switzerland, 2003; pp. 51–101. [Google Scholar]
- Ahmed, W.; Zhang, Q.; Lobos, A.; Senkbeil, J.; Sadowsky, M.J.; Harwood, V.J.; Saeidi, N.; Marinoni, O.; Ishii, S. Precipitation influences pathogenic bacteria and antibiotic resistance gene abundance in storm drain outfalls in coastal sub-tropical waters. Environ. Int. 2018, 116, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Teranishi, K.; Matsuwaki, T.; Nukazawa, K.; Ogura, Y. Effects of bacterial pollution caused by a strong typhoon event and the restoration of a recreational beach: Transitions of fecal bacterial counts and bacterial flora in beach sand. Sci. Total Environ. 2018, 640, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Abreu, R.; Figueira, C.; Romão, D.; Brandão, J.; Freitas, M.C.; Andrade, C.; Calado, G.; Ferreira, C.; Campos, A.; Prada, S. Sediment characteristics and microbiological contamination of beach sand–A case–study in the archipelago of Madeira. Sci. Total Environ. 2016, 573, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Efstratiou, M.A. Microorganisms in beach sand: Health implications. In Encyclopedia of Environmental Health, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 385–389. [Google Scholar]
- Fang, H.; Chen, Y.; Huang, L.; He, G. Analysis of biofilm bacterial communities under different shear stresses using size-fractionated sediment. Sci. Rep. 2017, 7, 1299. [Google Scholar] [CrossRef] [Green Version]
- Weiskerger, C.J.; Brandão, J.; Ahmed, W.; Aslan, A.; Avolio, L.; Badgley, B.D.; Boehm, A.B.; Edge, T.A.; Fleisher, J.M.; Heaney, C.D. Impacts of a changing earth on microbial dynamics and human health risks in the continuum between beach water and sand. Water Res. 2019, 162, 456–470. [Google Scholar] [CrossRef]
- Korajkic, A.; McMinn, B.R.; Harwood, V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health 2018, 15, 2842. [Google Scholar] [CrossRef] [Green Version]
- Solo-Gabriele, H.M.; Harwood, V.J.; Kay, D.; Fujioka, R.S.; Sadowsky, M.J.; Whitman, R.L.; Wither, A.; Caniça, M.; Da Fonseca, R.C.; Duarte, A. Beach sand and the potential for infectious disease transmission: Observations and recommendations. J. Mar. Biol. Assoc. UK 2016, 96, 101–120. [Google Scholar] [CrossRef] [Green Version]
- Williamson, C.E.; Madronich, S.; Lal, A.; Zepp, R.G.; Lucas, R.M.; Overholt, E.P.; Rose, K.C.; Schladow, S.G.; Lee-Taylor, J. Climate change-induced increases in precipitation are reducing the potential for solar ultraviolet radiation to inactivate pathogens in surface waters. Sci. Rep. 2017, 7, 13033. [Google Scholar] [CrossRef]
- Ulrich, N.; Rosenberger, A.; Brislawn, C.; Wright, J.; Kessler, C.; Toole, D.; Solomon, C.; Strutt, S.; McClure, E.; Lamendella, R. Restructuring of the aquatic bacterial community by hydric dynamics associated with superstorm sandy. Appl. Environ. Microbiol. 2016, 82, 3525–3536. [Google Scholar] [CrossRef] [Green Version]
- McLellan, S.L.; Eren, A.M. Discovering new indicators of fecal pollution. Trends Microbiol. 2014, 22, 697–706. [Google Scholar] [CrossRef] [Green Version]
- National Standard of the People’s Republic of China. GB/T 12763.8; Specifications for Oceangraphic Survey—Part 8: Marine Gclogy and Geophysics Survey. China Standards Press of China National Standardization Administration: Bejing, China, 2007.
- National Standard of the People’s Republic of China. HJ 634-2012; Soil Determination of Ammonium, Nitrite and Nitrate by Extraction with Potassium Chloride Solution-Spectrophotometric Methods. China Environmental Science Press: Bejing, China, 2012.
- Wang, Z.F.; Hu, C.Y.; Lv, H.Y. A Method for Determining Total Nitrogen in Sea Water and Marine Sediments. J. Ocean Univ. Qingdao China 1999, 161–166. [Google Scholar]
- Hu, C.Y.; Wang, Z.F.; Lv, H.Y. A Method for Determining Total Phosphorus in Sea Water and Marine Sediments. Mar. Environ. Sci. China 1999, 8, 48–52. [Google Scholar]
- National Standard of the People’s Republic of China. GB 17378.5; The Specification for Marine Monitoring—Part 5: Sediment analysis. China Standards Press of China National Standardization Administration: Bejing, China, 2007.
- Ming, H.; Fan, J.; Li, D.; Su, J.; Wang, Y.; Shi, T.; Jin, Y.; Song, J.; Huang, H. A Method for the Identification of Enterococcus in Seawater and Marine Sediments. N202010177272.4, 13 March 2020. [Google Scholar]
- Newton, R.J.; McLellan, S.L.; Dila, D.K.; Vineis, J.H.; Morrison, H.G.; Eren, A.M.; Sogin, M.L. Sewage reflects the microbiomes of human populations. MBio 2015, 6, e02574-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanks, O.C.; Green, H.; Korajkic, A.; Field, K.G. Overview of microbial source tracking methods targeting human fecal pollution sources. In Manual of Environmental Microbiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 3–4. [Google Scholar]
- Brown, K.I.; Graham, K.E.; Boehm, A.B. Risk-based threshold of gull-associated fecal marker concentrations for recreational water. Environ. Sci. Technol. Lett. 2017, 4, 44–48. [Google Scholar] [CrossRef]
- Arnaud, E.; Best, A.; Parker, B.L.; Aravena, R.; Dunfield, K. Transport of Escherichia coli through a thick vadose zone. J. Environ. Qual. 2015, 44, 1424–1434. [Google Scholar] [CrossRef] [PubMed]
- Bolds, S.A.; Lockaby, B.G.; Ditchkoff, S.S.; Smith, M.D.; Vercauteren, K.C. Impacts of a large invasive mammal on water quality in riparian ecosystems. J. Environ. Qual. 2021, 50, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Genthe, B.; Momba, M.N.B. Quantitative microbial risk assessment (QMRA) shows increased public health risk associated with exposure to river water under conditions of riverbed sediment resuspension. Sci. Total Environ. 2016, 566, 1143–1151. [Google Scholar] [CrossRef]
- Butman, D.E.; Wilson, H.F.; Barnes, R.T.; Xenopoulos, M.A.; Raymond, P.A. Increased mobilization of ages carbon to rivers by human disturbance. Nat. Geosci. 2015, 8, 112–116. [Google Scholar] [CrossRef]
- Rudic, E.; Vujovi, B.; Boi, M.; Arizanović, T. Relationships between water and sediment quality parameters and faecal bacteria content in the Palic-Ludas canal, Serbia. Int. J. Environ. Res. Public Health 2018, 64, 292. [Google Scholar]
- Wu, M.Z.; O’Carroll, D.M.; Vogel, L.J.; Robinson, C.E. Effect of low energy waves on the accumulation and transport of fecal indicator bacteria in sand and pore water at freshwater beaches. Environ. Sci. Technol. 2017, 51, 2786–2794. [Google Scholar] [CrossRef]
- Elmir, S.M.; Wright, M.E.; Abdelzaher, A.; Solo-Gabriele, H.M.; Fleming, L.E.; Miller, G.; Rybolowik, M.; Shih, M.T.P.; Pillai, S.P.; Cooper, J.A. Quantitative evaluation of bacteria released by bathers in a marine water. Water Res. 2007, 41, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Bakar, A.; Ahmadian, R.; Falconer, R.A. Modelling the transport and decay processes of microbial tracers in a macro-tidal estuary. Water Res. 2017, 123, 802–824. [Google Scholar] [CrossRef]
- Cloutier, D.D.; Alm, E.W.; McLellan, S.L. Influence of land use, nutrients, and geography on microbial communities and fecal indicator abundance at Lake Michigan beaches. Appl. Environ. Microbiol. 2015, 81, 4904–4913. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.I.; Boehm, A.B. Comparative decay of Catellicoccus marimmalium and enterococci in beach sand and seawater. Water Res. 2015, 83, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Reniers, A.; Haus, B.K.; Solo-Gabriele, H.M.; Kelly, E.A. Wave energy level and geographic setting correlate with Florida beach water quality. Mar. Pollut. Bull. 2016, 104, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassard, F.; Andrews, A.; Jones, D.L.; Parsons, L.; Jones, V.; Cox, B.A.; Daldorph, P.; Brett, H.; McDonald, J.E.; Malham, S.K. Physicochemical factors influence the abundance and culturability of human enteric pathogens and fecal indicator organisms in estuarine water and sediment. Front. Microbiol. 2017, 8, 1996. [Google Scholar] [CrossRef]
- Hu, W.; Murata, K.; Horikawa, Y.; Naganuma, A.; Zhang, D.Z. Bacterial community composition in rainwater associated with synoptic weather in an area downwind of the Asian continent. Sci. Total Environ. 2017, 601, 1775–1784. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Losey, N.A.; Lawson, P. Thermoanaerobaculaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 1–3. [Google Scholar]
- Li, D.; Werfhorst, L.C.V.D.; Rugh, M.B.; Feraud, M.; Hung, W.C.; Jay, J.; Cao, Y.P.; Parker, E.A.; Grant, S.B.; Holden, P.A. Limited Bacterial Removal in Full-Scale Stormwater Biofilters as Evidenced by Community Sequencing Analysis. Environ. Sci. Technol. 2021, 55, 9199–9208. [Google Scholar] [CrossRef]
- Kan, J. Storm events restructured bacterial community and their biogeochemical potentials. J. Geophys. Res. Biogeosci. 2018, 123, 1–34. [Google Scholar] [CrossRef]
- Yannarell, A.C.; Steppe, T.F.; Paerl, H.W. Disturbance and recovery of microbial community structure and function following Hurricane Frances. Environ. Microbiol. 2007, 9, 576–583. [Google Scholar] [CrossRef]
- Haller, L.; Amedegnato, E.; Poté, J.; Wildi, W. Influence of Freshwater Sediment Characteristics on Persistence of Fecal Indicator Bacteria. Water Air Soil Poll. 2009, 203, 217–227. [Google Scholar] [CrossRef] [Green Version]



| Samples | Water Temperature (°C) | DO (mg/L) | Conductivity (µS/m) | Salinity | pH | ORP (mV) |
|---|---|---|---|---|---|---|
| 11 August 2018 W1AS | 27.7 | 7.97 | 50,798 | 31.43 | 8.09 | 206 |
| 21 August 2018 W1AS | 25.8 | 6.57 | 48,317 | 30.94 | 7.94 | 149 |
| 15 September 2018 W1AS | 23.7 | 6.46 | 46,530 | 31.07 | 8.08 | 217 |
| 11 August 2018 W1OS | 27.2 | 8.63 | 50,444 | 31.49 | 8.13 | 265 |
| 21 August 2018 W1OS | 25.7 | 6.30 | 48,265 | 30.96 | 7.91 | 185 |
| 15 September 2018 W1OS | 23.6 | 6.43 | 46,535 | 31.17 | 8.09 | 210 |
| 11 August 2018 W2AS | 27.0 | 7.40 | 50,122 | 31.40 | 8.03 | 144 |
| 21 August 2018 W2AS | 25.5 | 6.91 | 46,919 | 30.15 | 7.92 | 125 |
| 15 September 2018 W2AS | 23.6 | 6.31 | 46,650 | 31.20 | 8.07 | 204 |
| 11 August 2018 W2OS | 26.8 | 7.88 | 50,049 | 31.47 | 8.06 | 244 |
| 21 August 2018 W2OS | 25.5 | 6.95 | 47,163 | 30.29 | 7.93 | 151 |
| 15 September 2018 W2OS | 23.6 | 6.18 | 46,614 | 31.19 | 8.07 | 207 |
| 11 August 2018 W3AS | 29.4 | 7.57 | 52,546 | 31.46 | 8.02 | 182 |
| 21 August 2018 W3AS | 25.8 | 7.17 | 27,905 | 16.95 | 7.87 | 67 |
| 15 September 2018 W3AS | 23.7 | 6.60 | 46,528 | 31.05 | 7.99 | 190 |
| 11 August 2018 W3OS | 29.0 | 10.26 | 52,180 | 31.52 | 8.22 | 213 |
| 21 August 2018 W3OS | 26.2 | 6.67 | 39,792 | 24.72 | 7.89 | 101 |
| 15 September 2018 W3OS | 23.4 | 6.22 | 46,420 | 31.20 | 8.07 | 205 |
| Samples | Sediment Grain Size (%) | Median Size (Mdφ) | Average Particle Size (Mzφ) | Water Content (%) | NO3– (10–3 µg/g) | NH4+ (10–3 µg/g) | TP (10–3 µg/g) | TN (10–3 µg/g) | TOC (10–3 µg/g) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gravel | Sand | Silt | Clay | |||||||||
| 11 August 2018 W1DS | 0.00 | 99.90 | 0.10 | 0.00 | 0.67 | 0.68 | 0.96 | 1.744 | 15.4 | 140 | 16.7 | 0.059 |
| 21 August 2018 W1DS | 20.05 | 79.95 | 0.00 | 0.00 | 0.32 | 0.05 | 1.19 | 1.668 | 10.8 | 209 | 12.5 | 0.027 |
| 15 September 2018 W1DS | 0.00 | 99.55 | 0.28 | 0.17 | 0.99 | 1.01 | 0.18 | 2.458 | 9.1 | 185 | 10.7 | 0.057 |
| 11 August 2018 W2DS | 0.00 | 99.50 | 0.29 | 0.21 | 1.17 | 1.22 | 3.22 | 1.693 | 12.5 | 131 | 13.7 | 0.593 |
| 21 August 2018 W2DS | 0.00 | 99.91 | 0.09 | 0.00 | 0.81 | 0.82 | 1.26 | 1.617 | 13.1 | 103 | 15.8 | 0.210 |
| 15 September 2018 W2DS | 0.00 | 100.00 | 0.00 | 0.00 | 0.94 | 0.94 | 0.02 | 1.234 | 9.7 | 204 | 10.4 | 0.210 |
| 11 August 2018W3DS | 0.00 | 99.58 | 0.26 | 0.16 | 0.79 | 0.80 | 2.20 | 1.923 | 14.9 | 204 | 16.0 | 0.026 |
| 21 August 2018 W3DS | 0.00 | 100.00 | 0.00 | 0.00 | 0.51 | 0.51 | 1.55 | 1.617 | 12.5 | 149 | 13.8 | 0.262 |
| 15 September 2018 W3DS | 0.00 | 100.00 | 0.00 | 0.00 | 0.96 | 0.96 | 0.19 | 1.081 | 13.1 | 170 | 37.4 | 0.223 |
| 11 August 2018W1WS | 24.71 | 75.29 | 0.00 | 0.00 | −0.08 | −0.23 | 2.02 | 1.744 | 13.1 | 138 | 14.6 | 0.040 |
| 21 August 2018 W1WS | 0.00 | 100.00 | 0.00 | 0.00 | 0.31 | 0.30 | 15.74 | 1.489 | 10.8 | 189 | 11.1 | 0.044 |
| 15 September 2018 W1WS | 0.00 | 99.60 | 0.23 | 0.17 | 1.10 | 1.17 | 16.86 | 2.535 | 10.8 | 243 | 13.9 | 0.303 |
| 11 August 2018W2WS | 42.66 | 57.16 | 0.11 | 0.07 | −0.09 | −0.60 | 3.70 | 1.642 | 9.1 | 268 | 13.5 | 0.436 |
| 21 August 2018 W2WS | 0.00 | 100.00 | 0.00 | 0.00 | 0.95 | 0.95 | 7.57 | 1.489 | 7.34 | 129 | 8.13 | 0.222 |
| 15 September 2018 W2WS | 0.00 | 99.63 | 0.21 | 0.17 | 1.07 | 1.09 | 9.09 | 1.132 | 10.2 | 197 | 11.3 | 0.225 |
| 11 August 2018W3WS | 64.75 | 35.25 | 0.00 | 0.00 | −1.22 | −1.24 | 4.75 | 1.719 | 12.5 | 242 | 13.2 | 0 |
| 21 August 2018 W3WS | 0.00 | 100.00 | 0.00 | 0.00 | 0.94 | 0.83 | 9.55 | 1.591 | 10.8 | 221 | 11.2 | 0.241 |
| 15 September 2018 W3WS | 0.00 | 99.67 | 0.18 | 0.16 | 1.06 | 1.06 | 10.89 | 1.336 | 9.1 | 160 | 19.4 | 0.228 |
| 11 August 2018W1SS | 0.00 | 98.87 | 0.71 | 0.42 | 2.20 | 2.07 | 7.63 | 1.770 | 39.2 | 177 | 45.9 | 0.070 |
| 21 August 2018 W1SS | 63.72 | 36.26 | 0.02 | 0.00 | −0.97 | −0.86 | 9.66 | 1.668 | 9.7 | 213 | 13.3 | 0 |
| 15 September 2018 W1SS | 52.96 | 46.84 | 0.14 | 0.06 | 1.86 | 1.72 | 19.77 | 1.540 | 14.3 | 262 | 15.9 | 0.263 |
| 11 August 2018 W2SS | 0.00 | 98.58 | 0.99 | 0.43 | 2.10 | 1.94 | 20.28 | 1.668 | 7.3 | 169 | 13.7 | 0.324 |
| 21 August 2018 W2SS | 0.00 | 100.00 | 0.00 | 0.00 | 0.12 | 0.12 | 19.11 | 1.489 | 21.8 | 200 | 11.6 | 0.268 |
| 15 September 2018 W2SS | 0.00 | 99.23 | 0.45 | 0.31 | −1.04 | −0.85 | 13.91 | 1.795 | 11.4 | 256 | 11.9 | 0.180 |
| 11 August 2018 W3SS | 0.00 | 100.00 | 0.00 | 0.00 | 1.57 | 1.57 | 13.59 | 1.566 | 14.3 | 158 | 44.0 | 0.045 |
| 21 August 2018 W3SS | 0.00 | 100.00 | 0.00 | 0.00 | 1.19 | 1.19 | 21.34 | 1.821 | 11.4 | 252 | 13.4 | 0.196 |
| 15 September 2018 W3SS | 0.00 | 100.00 | 0.00 | 0.00 | 1.15 | 1.14 | 22.75 | 0.903 | 16.6 | 152 | 22.7 | 0.227 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ming, H.; Wang, Y.; Su, J.; Fu, Y.; Xu, J.; Shi, T.; Ren, K.; Jin, Y.; Fan, J. The Effects of a Typhoon on the Dynamic of Microbial Community Structure and Water Quality of the Marine Bathing Beach. Water 2022, 14, 1631. https://doi.org/10.3390/w14101631
Ming H, Wang Y, Su J, Fu Y, Xu J, Shi T, Ren K, Jin Y, Fan J. The Effects of a Typhoon on the Dynamic of Microbial Community Structure and Water Quality of the Marine Bathing Beach. Water. 2022; 14(10):1631. https://doi.org/10.3390/w14101631
Chicago/Turabian StyleMing, Hongxia, Yantao Wang, Jie Su, Yunhan Fu, Jianrong Xu, Tingting Shi, Kaijia Ren, Yuan Jin, and Jingfeng Fan. 2022. "The Effects of a Typhoon on the Dynamic of Microbial Community Structure and Water Quality of the Marine Bathing Beach" Water 14, no. 10: 1631. https://doi.org/10.3390/w14101631
APA StyleMing, H., Wang, Y., Su, J., Fu, Y., Xu, J., Shi, T., Ren, K., Jin, Y., & Fan, J. (2022). The Effects of a Typhoon on the Dynamic of Microbial Community Structure and Water Quality of the Marine Bathing Beach. Water, 14(10), 1631. https://doi.org/10.3390/w14101631






