Potential Use of CTCs as Biomarkers in Renal Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients Enrolled in the Study
2.2. RNA-Microarray
2.3. Immunohistochemistry
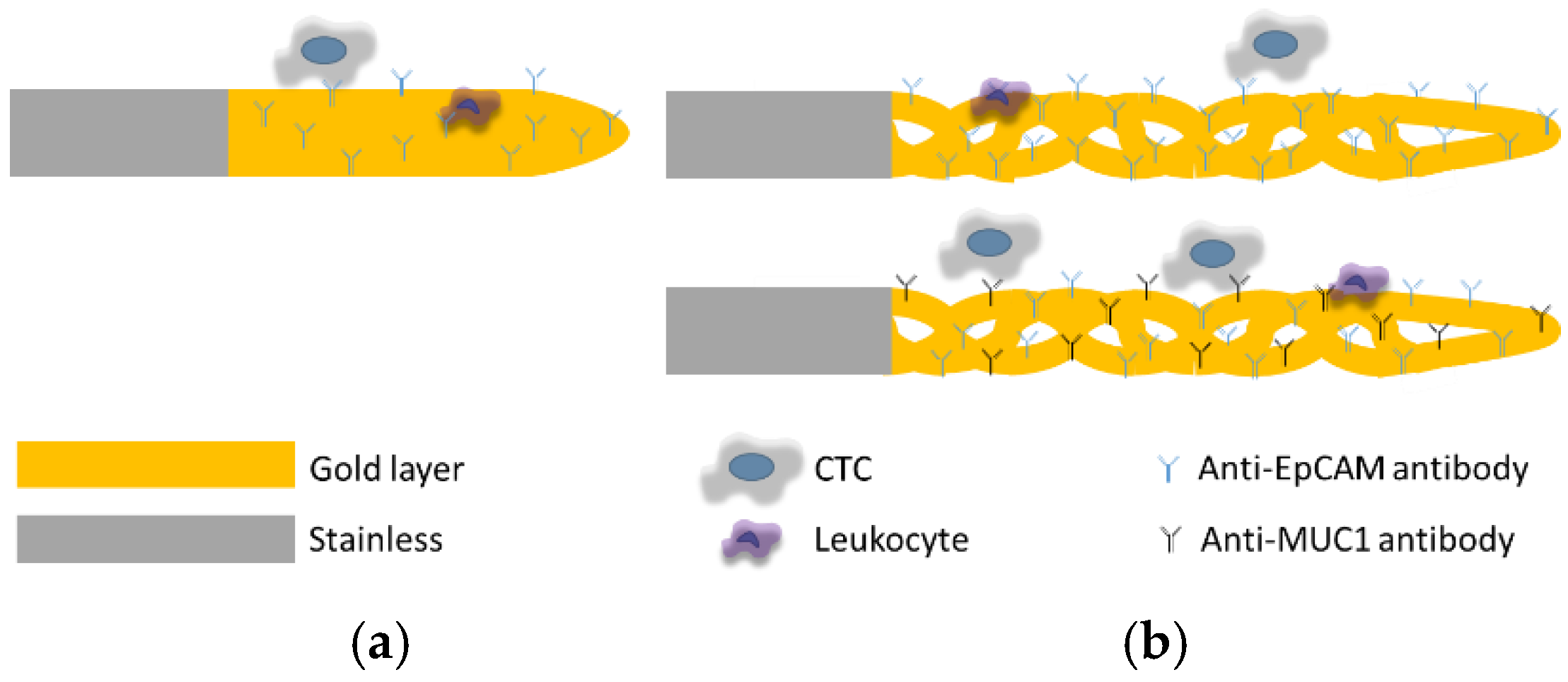
2.4. Antibody Functionalization of the CellCollector System
2.5. Cell Lines
2.6. Immunocytochemistry
2.7. Capture of CTC
2.7.1. Ex Vivo Experiments
2.7.2. In Vivo Experiments
2.8. Statistical Analysis
3. Results
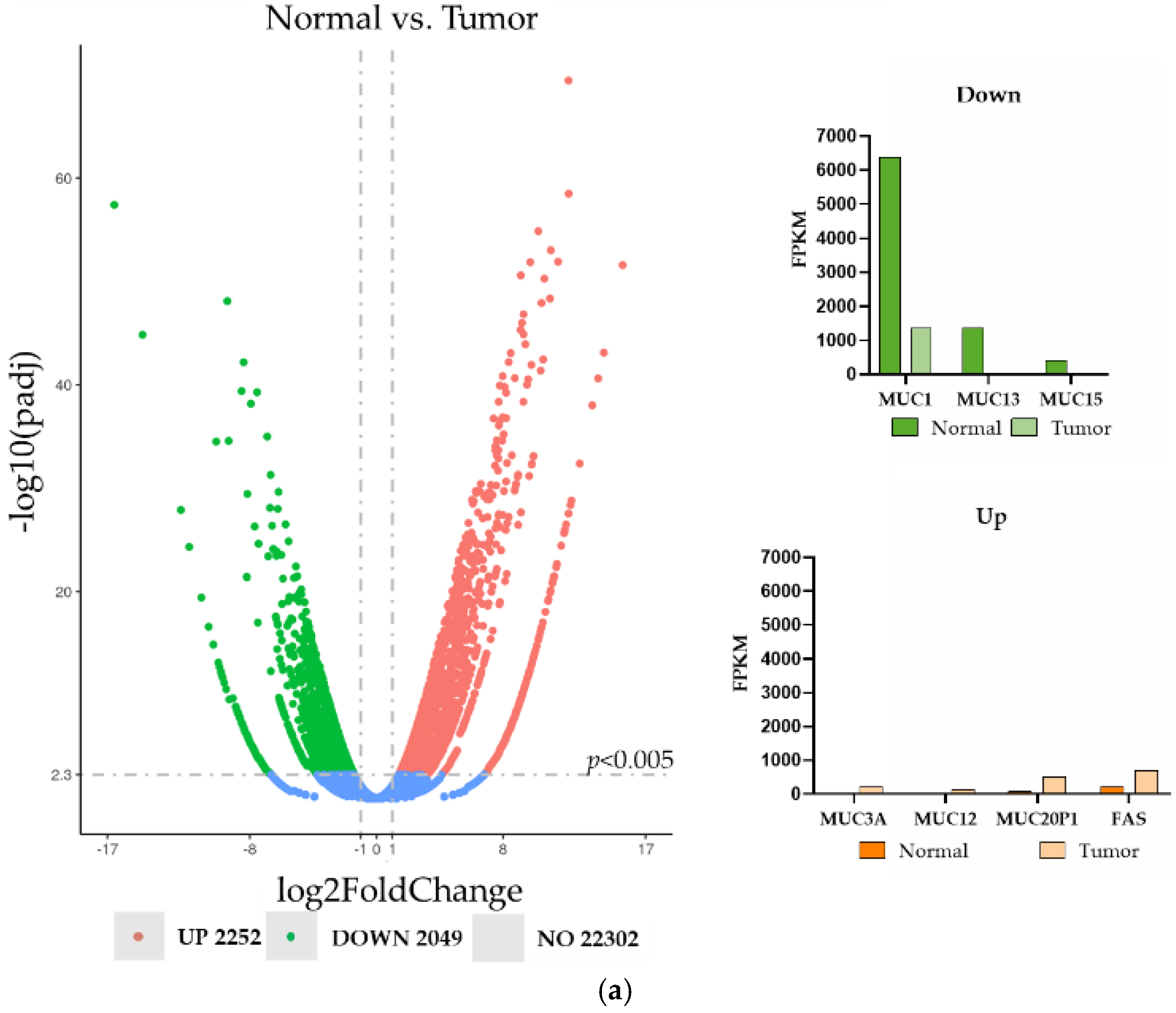
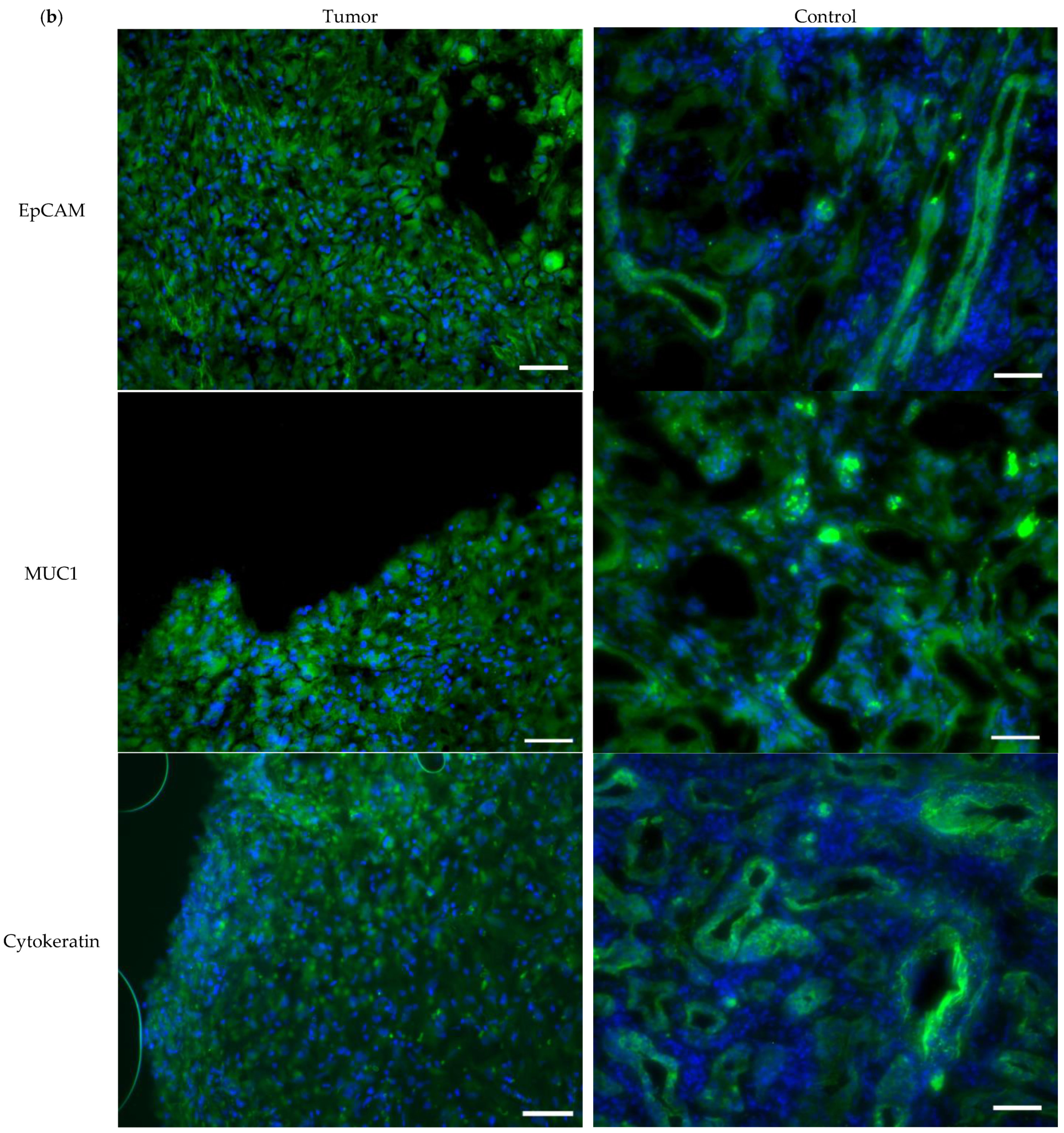
3.1. Tissue Expression of Cell Surface Proteins
3.2. Expression of EpCAM and MUC1 in Primary RCC Cell Lines
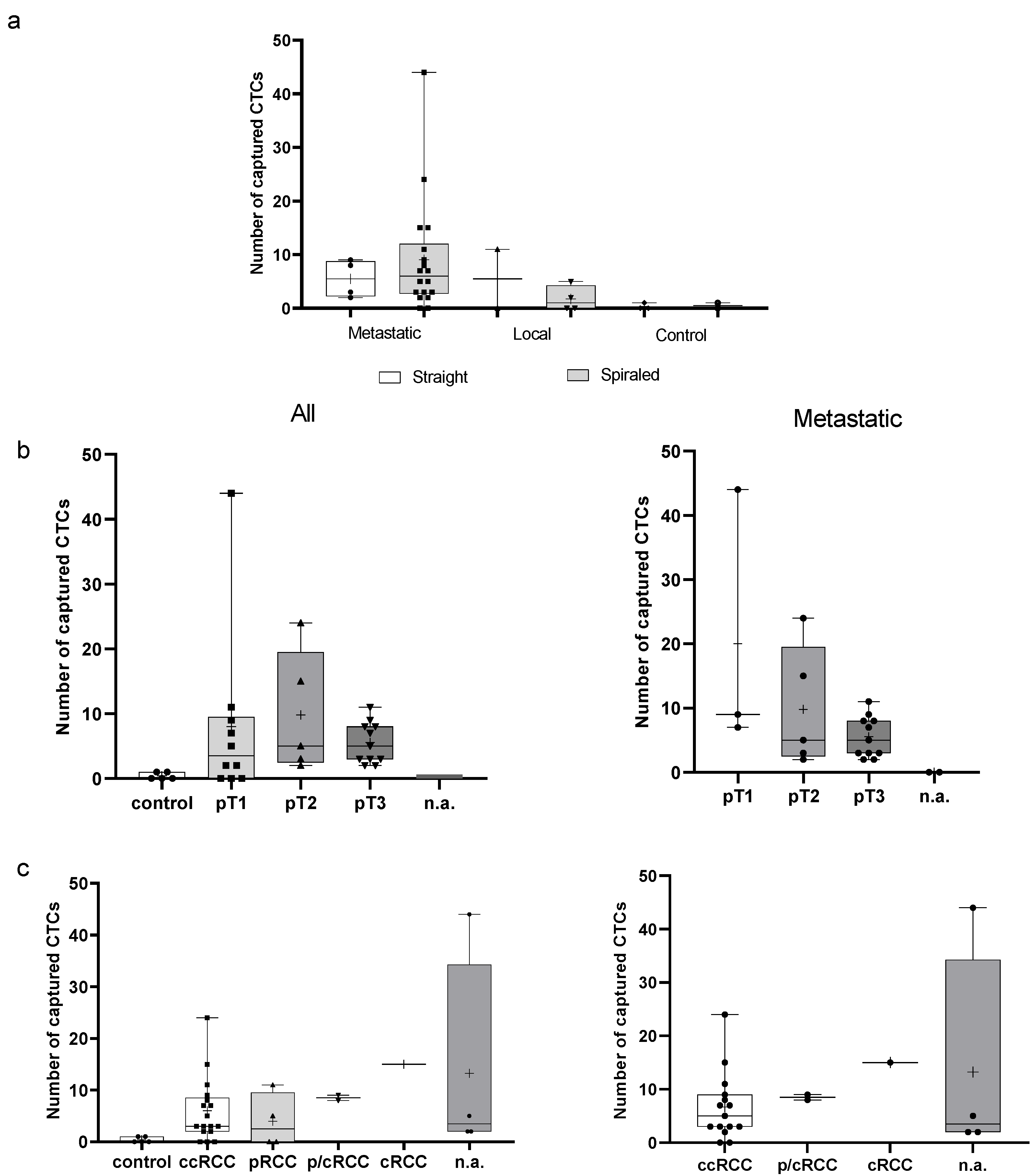
3.3. Ex Vivo EpCAM-Based Experiments Using a Hemodynamic System
3.4. In Vivo EpCAM-Based Experiments
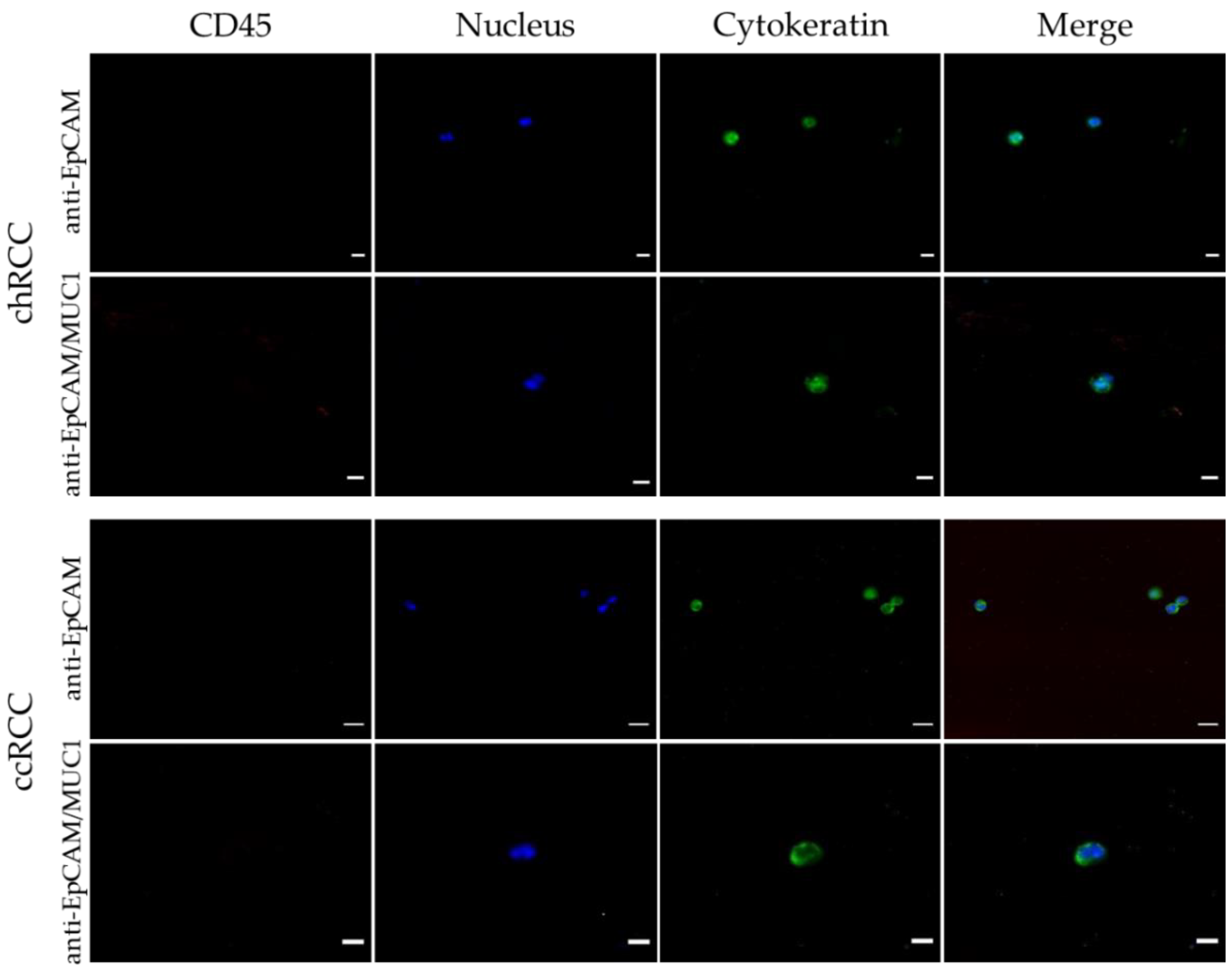
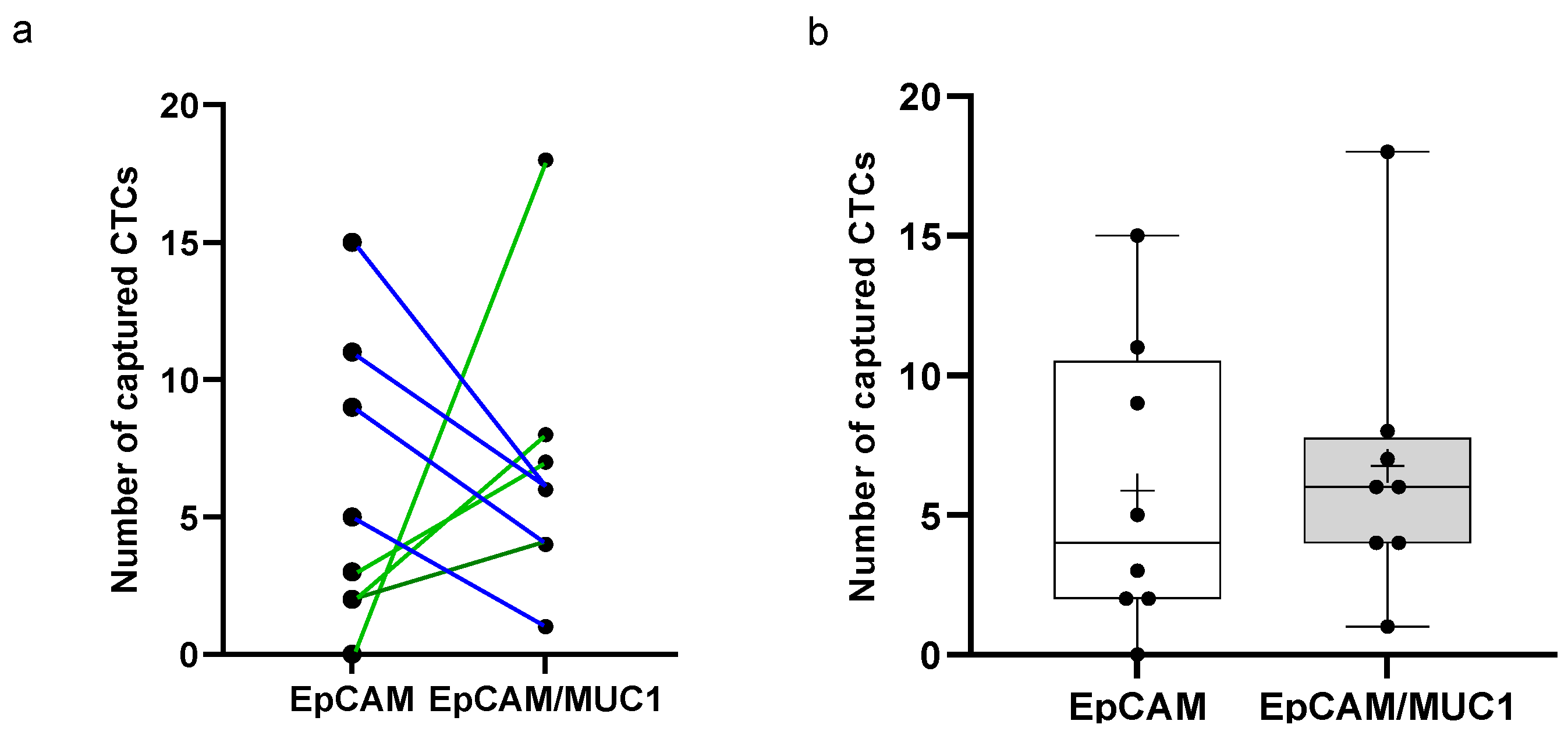
3.5. Anti-EpCAM/MUC1-Covered CellCollector
3.6. Limitations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol 2020, 11, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Ciarimboli, G.; Theil, G.; Bialek, J.; Edemir, B. Contribution and Expression of Organic Cation Transporters and Aquaporin Water Channels in Renal Cancer. In Reviews of Physiology Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Hanin, L.; Jandrig, B.; Pavlova, L.; Seidel, K. The natural history of renal cell carcinoma with pulmonary metastases illuminated through mathematical modeling. Math Biosci. 2019, 309, 118–130. [Google Scholar] [CrossRef]
- Theil, G.; Fornara, P.; Bialek, J. Position of Circulating Tumor Cells in the Clinical Routine in Prostate Cancer and Breast Cancer Patients. Cancers 2020, 12, 3782. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Morris, M.J.; Stadler, W.M.; Higano, C.; Basch, E.; Fizazi, K.; Antonarakis, E.S.; Beer, T.M.; Carducci, M.A.; Chi, K.N.; et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J. Clin. Oncol. 2016, 34, 1402–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giubellino, A.; Ricketts, C.J.; Moreno, V.; Linehan, W.M.; Merino, M.J. MicroRNA Profiling of Morphologically Heterogeneous Clear Cell Renal Cell Carcinoma. J. Cancer 2021, 12, 5375–5384. [Google Scholar] [CrossRef]
- Cappelletti, V.; Verzoni, E.; Ratta, R.; Vismara, M.; Silvestri, M.; Montone, R.; Miodini, P.; Reduzzi, C.; Claps, M.; Sepe, P.; et al. Analysis of Single Circulating Tumor Cells in Renal Cell Carcinoma Reveals Phenotypic Heterogeneity and Genomic Alterations Related to Progression. Int. J. Mol. Sci. 2020, 21, 1475. [Google Scholar] [CrossRef] [Green Version]
- Kufe, D.W. MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene 2013, 32, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- Rahn, J.J.; Dabbagh, L.; Pasdar, M.; Hugh, J.C. The importance of MUC1 cellular localization in patients with breast carcinoma: An immunohistologic study of 71 patients and review of the literature. Cancer 2001, 91, 1973–1982. [Google Scholar] [CrossRef]
- Lapointe, J.; Li, C.; Higgins, J.P.; van de Rijn, M.; Bair, E.; Montgomery, K.; Ferrari, M.; Egevad, L.; Rayford, W.; Bergerheim, U.; et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. USA 2004, 101, 811–816. [Google Scholar] [CrossRef] [Green Version]
- Nagata, K.; Horinouchi, M.; Saitou, M.; Higashi, M.; Nomoto, M.; Goto, M.; Yonezawa, S. Mucin expression profile in pancreatic cancer and the precursor lesions. J. Hepatobiliary Pancreat. Surg. 2007, 14, 243–254. [Google Scholar] [CrossRef]
- Wreesmann, V.B.; Sieczka, E.M.; Socci, N.D.; Hezel, M.; Belbin, T.J.; Childs, G.; Patel, S.G.; Patel, K.N.; Tallini, G.; Prystowsky, M.; et al. Genome-wide profiling of papillary thyroid cancer identifies MUC1 as an independent prognostic marker. Cancer Res. 2004, 64, 3780–3789. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Wang, L.; Chen, H.; Li, L.; Ma, Y.; Ni, J.; Li, Y. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer Metastasis Rev. 2013, 32, 535–551. [Google Scholar] [CrossRef]
- Saltos, A.; Khalil, F.; Smith, M.; Li, J.; Schell, M.; Antonia, S.J.; Gray, J.E. Clinical associations of mucin 1 in human lung cancer and precancerous lesions. Oncotarget 2018, 9, 35666–35675. [Google Scholar] [CrossRef] [PubMed]
- Kraus, S.; Abel, P.D.; Nachtmann, C.; Linsenmann, H.J.; Weidner, W.; Stamp, G.W.; Chaudhary, K.S.; Mitchell, S.E.; Franke, F.E.; Lalani, E.N. MUC1 mucin and trefoil factor 1 protein expression in renal cell carcinoma: Correlation with prognosis. Hum. Pathol. 2002, 33, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Leroy, X.; Zerimech, F.; Zini, L.; Copin, M.C.; Buisine, M.P.; Gosselin, B.; Aubert, J.P.; Porchet, N. MUC1 expression is correlated with nuclear grade and tumor progression in pT1 renal clear cell carcinoma. Am. J. Clin. Pathol. 2002, 118, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Denda, K.; Yamamoto, M.; Matsumoto, T.; Fujime, M.; Irimura, T. Expression of MUC1 mucins inversely correlated with post-surgical survival of renal cell carcinoma patients. Br. J. Cancer 1999, 80, 301–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theil, G.; Fischer, K.; Weber, E.; Medek, R.; Hoda, R.; Lücke, K.; Fornara, P. The Use of a New CellCollector to Isolate Circulating Tumor Cells from the Blood of Patients with Different Stages of Prostate Cancer and Clinical Outcomes—A Proof-of-Concept Study. PLoS ONE 2016, 11, e0158354. [Google Scholar] [CrossRef]
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G.; et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Gorges, T.M.; Penkalla, N.; Schalk, T.; Joosse, S.A.; Riethdorf, S.; Tucholski, J.; Lucke, K.; Wikman, H.; Jackson, S.; Brychta, N.; et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin. Cancer Res. 2016, 22, 2197–2206. [Google Scholar] [CrossRef] [Green Version]
- Mandair, D.; Vesely, C.; Ensell, L.; Lowe, H.; Spanswick, V.; Hartley, J.A.; Caplin, M.E.; Meyer, T. A comparison of CellCollector with CellSearch in patients with neuroendocrine tumours. Endocr. Relat. Cancer 2016, 23, L29–L32. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gong, S.; Liu, Y.; Liang, L.; He, S.; Zhang, Q.; Si, M.; Yu, Z. Enumeration and molecular characterization of circulating tumor cell using an in vivo capture system in squamous cell carcinoma of head and neck. Chin. J. Cancer Res. 2017, 29, 196–203. [Google Scholar] [CrossRef]
- Dizdar, L.; Fluegen, G.; van Dalum, G.; Honisch, E.; Neves, R.P.; Niederacher, D.; Neubauer, H.; Fehm, T.; Rehders, A.; Krieg, A.; et al. Detection of circulating tumor cells in colorectal cancer patients using the GILUPI CellCollector: Results from a prospective, single-center study. Mol. Oncol. 2019, 13, 1548–1558. [Google Scholar] [CrossRef] [Green Version]
- Kuske, A.; Gorges, T.M.; Tennstedt, P.; Tiebel, A.K.; Pompe, R.; Preißer, F.; Prues, S.; Mazel, M.; Markou, A.; Lianidou, E.; et al. Improved detection of circulating tumor cells in non-metastatic high-risk prostate cancer patients. Sci. Rep. 2016, 6, 39736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.B.; Geng, C.Z.; Yan, M.; Wang, Y.S.; Ouyang, Q.C.; Yin, Y.M.; Wu, L.N.; He, J.; Jiang, Z.F. Circulating tumor cells in patients with breast tumors were detected by a novel device: A multicenter, clinical trial in China. Zhonghua Yi Xue Za Zhi 2017, 97, 1857–1861. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Nayak, B.; Panaiyadiyan, S.; Singh, P.; Karmakar, S.; Kaushal, S.; Seth, A. Role of circulating tumor cells in patients with metastatic clear-cell renal cell carcinoma. Urol. Oncol. 2021, 39, 135.e9–135.e15. [Google Scholar] [CrossRef]
- Klezl, P.; Pospisilova, E.; Kolostova, K.; Sonsky, J.; Maly, O.; Grill, R.; Pawlak, I.; Bobek, V. Detection of Circulating Tumor Cells in Renal Cell Carcinoma: Disease Stage Correlation and Molecular Characterization. J. Clin. Med. 2020, 9, 1372. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Zhang, M.; Zheng, G.; Zhang, Y.; Li, Y.; Lin, P.; Li, H.; Jiao, W.; Niu, H. Clinical significance of phenotyping and karyotyping of detecting circulating tumor cells in renal cell carcinoma using subtraction enrichment and immunostaining-fluorescence in situ hybridization (SE-iFISH). Int. Urol. Nephrol. 2020, 52, 2281–2287. [Google Scholar] [CrossRef]
- Gradilone, A.; Iacovelli, R.; Cortesi, E.; Raimondi, C.; Gianni, W.; Nicolazzo, C.; Petracca, A.; Palazzo, A.; Longo, F.; Frati, L.; et al. Circulating tumor cells and “suspicious objects” evaluated through CellSearch® in metastatic renal cell carcinoma. Anticancer Res. 2011, 31, 4219–4221. [Google Scholar]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debien, V.; Thouvenin, J.; Lindner, V.; Barthélémy, P.; Lang, H.; Flippot, R.; Malouf, G.G. Sarcomatoid Dedifferentiation in Renal Cell Carcinoma: From Novel Molecular Insights to New Clinical Opportunities. Cancers 2019, 12, 99. [Google Scholar] [CrossRef] [Green Version]
- Jolly, M.K.; Ware, K.E.; Gilja, S.; Somarelli, J.A.; Levine, H. EMT and MET: Necessary or permissive for metastasis? Mol. Oncol. 2017, 11, 755–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.L.; Zhang, P.; Li, H.C.; Yang, X.J.; Zhang, Y.P.; Li, Z.L.; Xue, L.; Xue, Y.Q.; Li, H.L.; Chen, Q.; et al. Dynamic changes of different phenotypic and genetic circulating tumor cells as a biomarker for evaluating the prognosis of RCC. Cancer Biol. Ther. 2019, 20, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Basso, U.; Facchinetti, A.; Rossi, E.; Maruzzo, M.; Conteduca, V.; Aieta, M.; Massari, F.; Fraccon, A.P.; Mucciarini, C.; Sava, T.; et al. Prognostic Role of Circulating Tumor Cells in Metastatic Renal Cell Carcinoma: A Large, Multicenter, Prospective Trial. Oncologist 2021, 26, 740–750. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, J.M.; Buttyan, R.; Bander, N.H.; de la Taille, A.; Stifelman, M.D.; Emanuel, E.R.; Bagiella, E.; Rubin, M.A.; Katz, A.E.; Olsson, C.A.; et al. The detection of renal carcinoma cells in the peripheral blood with an enhanced reverse transcriptase-polymerase chain reaction assay for MN/CA9. Cancer 1999, 86, 492–497. [Google Scholar] [CrossRef]
- Ashida, S.; Okuda, H.; Chikazawa, M.; Tanimura, M.; Sugita, O.; Yamamoto, Y.; Nakamura, S.; Moriyama, M.; Shuin, T. Detection of circulating cancer cells with von hippel-lindau gene mutation in peripheral blood of patients with renal cell carcinoma. Clin. Cancer Res. 2000, 6, 3817–3822. [Google Scholar]
- Huang, Q.; Sun, Y.; Ma, X.; Gao, Y.; Li, X.; Niu, Y.; Zhang, X.; Chang, C. Androgen receptor increases hematogenous metastasis yet decreases lymphatic metastasis of renal cell carcinoma. Nat. Commun. 2017, 8, 918. [Google Scholar] [CrossRef]
- Chen, Y.L.; Ge, G.J.; Qi, C.; Wang, H.; Wang, H.L.; Li, L.Y.; Li, G.H.; Xia, L.Q. A five-gene signature may predict sunitinib sensitivity and serve as prognostic biomarkers for renal cell carcinoma. J. Cell Physiol. 2018, 233, 6649–6660. [Google Scholar] [CrossRef]
- Agrawal, B.; Krantz, M.J.; Reddish, M.A.; Longenecker, B.M. Cancer-associated MUC1 mucin inhibits human T-cell proliferation, which is reversible by IL-2. Nat. Med. 1998, 4, 43–49. [Google Scholar] [CrossRef]
- Irimura, T.; McIsaac, A.M.; Carlson, D.A.; Yagita, M.; Grimm, E.A.; Menter, D.G.; Ota, D.M.; Clary, K.R. Soluble factor in normal tissues that stimulates high-molecular-weight sialoglycoprotein production by human colon carcinoma cells. Cancer Res. 1990, 50, 3331–3338. [Google Scholar]
- Ligtenberg, M.J.; Buijs, F.; Vos, H.L.; Hilkens, J. Suppression of cellular aggregation by high levels of episialin. Cancer Res. 1992, 52, 2318–2324. [Google Scholar] [PubMed]
- Jamil, A.; Kasi, A. Lung Metastasis. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
- Rausch, S.; Beermann, J.; Scharpf, M.; Hennenlotter, J.; Fend, F.; Stenzl, A.; Schollenberger, D.; Bedke, J.; Kruck, S. Differential expression and clinical relevance of MUC1 in renal cell carcinoma metastasis. World J. Urol. 2016, 34, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, V.; Appierto, V.; Tiberio, P.; Fina, E.; Callari, M.; Daidone, M.G. Circulating Biomarkers for Prediction of Treatment Response. J. Natl. Cancer Inst. Monogr. 2015, 2015, 60–63. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.P.; Chakraborty, S.; Souchek, J.; Batra, S.K. Mucin-based targeted pancreatic cancer therapy. Curr. Pharm. Des. 2012, 18, 2472–2481. [Google Scholar] [CrossRef]
- Rittig, S.M.; Haentschel, M.; Weimer, K.J.; Heine, A.; Müller, M.R.; Brugger, W.; Horger, M.S.; Maksimovic, O.; Stenzl, A.; Hoerr, I.; et al. Long-term survival correlates with immunological responses in renal cell carcinoma patients treated with mRNA-based immunotherapy. Oncoimmunology 2016, 5, e1108511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, T.; Cen, Q.; Lei, H. A review on development of MUC1-based cancer vaccine. Biomed. Pharmacother. 2020, 132, 110888. [Google Scholar] [CrossRef] [PubMed]

| In Vitro | In Vivo | |||
|---|---|---|---|---|
| Sex | n | % | n | % |
| Male | 19 | 67.86 | 4 | 100 |
| Female | 9 | 32.14 | 0 | 0 |
| Histology | n | % | n | % |
| ccRCC | 14 | 50.00 | 1 | 25 |
| pRCC | 4 | 14.29 | 1 | 25 |
| c/pRCC | 1 | 3.57 | ||
| chRCC | 1 | 3.57 | ||
| p.d.RCC | 1 | 25 | ||
| control | 5 | 17.86 | ||
| n.a. | 3 | 17.86 | 1 | 25 |
| pT stage | n | % | n | % |
| pT1 | 10 | 35.71 | 1 | 25 |
| pT2 | 4 | 14.29 | 2 | 50 |
| pT3 | 8 | 28.57 | 1 | 25 |
| n.a. | 1 | 3.575 | ||
| control | 5 | 17.86 | ||
| Wire | n | % | n | % |
| Smooth | 8 | 28.57 | 4 | 100 |
| Spiral | 20 | 71.43 | ||
| Tumor | n | % | n | % |
| Local | 6 | 19.23 | 2 | 50 |
| Metastasis (all) | 17 | 73.08 | 2 | 50 |
| Bone | 12 | 46.15 a | 1 | 25 |
| Lung | 9 | 34.62 a | 1 | 25 |
| Liver | 3 | 11.54 a | 1 | 25 |
| Adrenal gland | 4 | 15.38 a | 2 | 50 |
| Lymph nodes | 5 | 19.23 a | ||
| Other | 2 | 7.69 a | ||
| Timepoint of wire application | n | % | n | % |
| >2 years before nephrectomy | 14 | 50 | ||
| Shortly before nephrectomy | 14 | 50 | 4 | 100 |
| During therapy * | 2 | 50 | ||
| Cell Line | Gene Name | Log2 Fold Change | p Value | p Adj |
|---|---|---|---|---|
| ccRCC31 | EpCAM | 5.36 | 5.40 × 10−13 | 2.06 × 10−11 |
| 5.30 | 1.26 × 10−12 | 4.90 × 10−11 | ||
| ccRCC122 | EpCAM | 5.68 | 5.69 × 10−15 | 2.98 × 10−13 |
| 5.61 | 1.59 × 10−14 | 7.83 × 10−13 | ||
| ccRCC162 | EpCAM | 2.46 | 1.44 × 106 | 1.34 × 107 |
| 2.51 | 7.69 × 105 | 7.45 × 106 | ||
| ccRCC31 | MUC1 | 1.39 | 0.00 | 0.01 |
| 1.81 | 0.00 | 0.00 |
| Straight Wire—Ex Vivo | Spiraled Wire—Ex Vivo | |||
|---|---|---|---|---|
| Captured CTCs as Median (Range) | CTC Positive (%) | Captured CTCs as Median (Range) | CTC Positive (%) | |
| Metastatic | ||||
| pT1 | 9 (7–44) | 100 | ||
| pT2 | 5 (2–24) | 100 | ||
| pT3 | 5.5 (2–9) | 100 | 5 (2–11) | 100 |
| n.a. | 0 | 0 | ||
| all | 5.5 (2–9) | 100 | 6 (0–44) | 98.82 |
| ccRCC | 2.5 (2–3) | 100 | 7 (0–24) | 84.62 |
| p/cRCC | 8.5 (8–9) | 100 | ||
| cRCC | 12 | 100 | ||
| n.a. | 3.5 (2–44) | 100 | ||
| Local | ||||
| pT1 | 5.5 (0–11) | 50 | 2 (0–5) | 50 |
| Control | ||||
| 0 (0–1) | 33.33 | 0.5 (0–1) | 50 | |
| Straight Wire—In Vivo | ||||
| Captured CTCs (range) | CTC positive (%) | |||
| Metastatic | ||||
| pT1 | 2 a | 100 | ||
| pT2 | 7 (10–4) b | 100 | ||
| Local | ||||
| pT2 | 2 a | 100 | ||
| pT3 | 5 (10–0) b | 50 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bialek, J.; Wencker, A.; Kawan, F.; Yankulov, S.; Fornara, P.; Theil, G. Potential Use of CTCs as Biomarkers in Renal Cancer Patients. Life 2022, 12, 89. https://doi.org/10.3390/life12010089
Bialek J, Wencker A, Kawan F, Yankulov S, Fornara P, Theil G. Potential Use of CTCs as Biomarkers in Renal Cancer Patients. Life. 2022; 12(1):89. https://doi.org/10.3390/life12010089
Chicago/Turabian StyleBialek, Joanna, Andreas Wencker, Felix Kawan, Stefan Yankulov, Paolo Fornara, and Gerit Theil. 2022. "Potential Use of CTCs as Biomarkers in Renal Cancer Patients" Life 12, no. 1: 89. https://doi.org/10.3390/life12010089






