Endothelial-Derived APT1-Mediated Macrophage-Endothelial Cell Interactions Participate in the Development of Atherosclerosis by Regulating the Ras/MAPK Signaling Pathway
Abstract
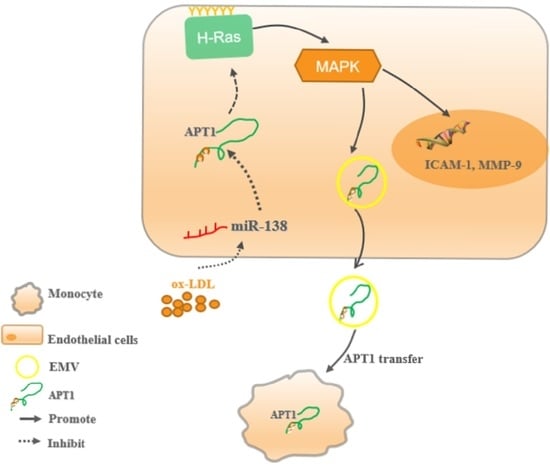
:1. Introduction
2. Results
2.1. APT1 Is a Crucial Factor in Atherosclerosis
2.2. miR-138 Is an Upstream Factor of APT1
2.3. H-Ras Is a Downstream Factor of APT1
2.4. APT1 Can Affect the MAPK Signaling Pathway and Related Factor Expression
2.5. Effect of Endothelium Derived APT1 on Macrophages
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Plasmids, and Adenoviral Vectors
4.2. Animal Experiment
4.3. Histology, Oil Red O, and Picro Sirius Red Staining
4.4. RNA Extraction and Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
4.5. Plasmids and Adenoviral Vectors
4.6. Western Blot Analysis
4.7. Luciferase Activity Assay
4.8. Fluorescence Microscopy Analysis of Monocyte Adhesion and Dil-Ox-LDL in HUVECs
4.9. BP Isolation and BP-Depleted FBS Preparations
4.10. Transmission Electron Microscopy (TEM)
4.11. BP Labeling and Cell Uptake Assay
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sugimoto, H.; Hayashi, H.; Yamashita, S. Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J. Biol. Chem. 1996, 271, 7705–7711. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Deems, R.A.; Dennis, E.A. Cloning, expression, and catalytic mechanism of murine lysophospholipase I. J. Biol. Chem. 1997, 272, 12723–12729. [Google Scholar] [CrossRef] [Green Version]
- Dekker, F.J.; Rocks, O.; Vartak, N.; Menninger, S.; Hedberg, C.; Balamurugan, R.; Wetzel, S.; Renner, S.; Gerauer, M.; Scholermann, B.; et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 2010, 6, 449–456. [Google Scholar] [CrossRef]
- Tian, L.; McClafferty, H.; Knaus, H.G.; Ruth, P.; Shipston, M.J. Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium channels. J. Biol. Chem. 2012, 287, 14718–14725. [Google Scholar] [CrossRef] [Green Version]
- Yeh, D.C.; Duncan, J.A.; Yamashita, S.; Michel, T. Depalmitoylation of endothelial nitric-oxide synthase by acyl-protein thioesterase 1 is potentiated by Ca(2+)-calmodulin. J. Biol. Chem. 1999, 274, 33148–33154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, J.A.; Gilman, A.G. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 1998, 273, 15830–15837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, T.; Kishi, M.; Sugimoto, H.; Taguchi, R.; Obinata, H.; Ohshima, N.; Tatei, K.; Izumi, T. Thioesterase activity and subcellular localization of acylprotein thioesterase 1/lysophospholipase 1. Biochim. Biophys. Acta 2009, 1791, 797–805. [Google Scholar] [CrossRef]
- Hallak, H.; Muszbek, L.; Laposata, M.; Belmonte, E.; Brass, L.F.; Manning, D.R. Covalent binding of arachidonate to G protein alpha subunits of human platelets. J. Biol. Chem. 1994, 269, 4713–4716. [Google Scholar] [CrossRef]
- Berg, V.; Rusch, M.; Vartak, N.; Jungst, C.; Schauss, A.; Waldmann, H.; Hedberg, C.; Pallasch, C.P.; Bastiaens, P.I.; Hallek, M.; et al. miRs-138 and -424 control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterases 1 and 2 in CLL. Blood 2015, 125, 2948–2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Runkle, K.B.; Terkowski, S.M.; Ekaireb, R.I.; Witze, E.S. Protein Depalmitoylation Is Induced by Wnt5a and Promotes Polarized Cell Behavior. J. Biol. Chem. 2015, 290, 15707–15716. [Google Scholar] [CrossRef] [Green Version]
- Martin, B.R.; Wang, C.; Adibekian, A.; Tully, S.E.; Cravatt, B.F. Global profiling of dynamic protein palmitoylation. Nat. Methods 2011, 9, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zheng, Y.; Coyaud, E.; Zhang, C.; Selvabaskaran, A.; Yu, Y.; Xu, Z.; Weng, X.; Chen, J.S.; Meng, Y.; et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science 2019, 366, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, A.; Green, W.N. Editorial: Role of Protein Palmitoylation in Synaptic Plasticity and Neuronal Differentiation. Front. Synaptic Neurosci. 2020, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qin, Z.; Sun, W.; Chu, F.; Zhou, F. Function of Protein S-Palmitoylation in Immunity and Immune-Related Diseases. Front. Immunol. 2021, 12, 661202. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.; Peng, S.; Chandra, G.; Sarkar, C.; Zhang, Z.; Bagh, M.B.; Mukherjee, A.B. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43. J. Biol. Chem. 2013, 288, 9112–9125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuiffo, B.; Ren, R. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood 2010, 115, 3598–3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.T.S.; Davis, N.G.; Conibear, E. Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem. Soc. Trans. 2017, 45, 913–921. [Google Scholar] [CrossRef]
- Tian, R.Q.; Wang, X.H.; Hou, L.J.; Jia, W.H.; Yang, Q.; Li, Y.X.; Liu, M.; Li, X.; Tang, H. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. J. Biol. Chem. 2011, 286, 25556–25563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, T.; Zhao, Z.; Li, G. Noncoding RNA in cardiac fibrosis. Int. J. Cardiol. 2015, 187, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.H.; Wang, D.D.; Chen, D.; Liu, S.W.; Wang, Z.; Yan, D.L.; Dong, S.C.; Feng, J.F. MiR-138: A promising therapeutic target for cancer. Tumor Biol. 2017, 39, 1010428317697575. [Google Scholar] [CrossRef] [Green Version]
- Morton, S.U.; Scherz, P.J.; Cordes, K.R.; Ivey, K.N.; Stainier, D.Y.; Srivastava, D. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. USA 2008, 105, 17830–17835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.S.; Yang, J.J.; Yu, F.Y.; Liu, B.H. Cardiotoxicity of mycotoxin citrinin and involvement of microRNA-138 in zebrafish embryos. Toxicol. Sci. 2013, 136, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Ran, Y.; Zhang, D.; Chen, J.; Li, S.; Zhu, D. MicroRNA-138 plays a role in hypoxic pulmonary vascular remodelling by targeting Mst1. Biochem. J. 2013, 452, 281–291. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Liu, P.; Jian, Z.; Li, J.; Zhu, Y.; Feng, Z.; Xiao, Y. miR-138 protects cardiomyocytes from hypoxia-induced apoptosis via MLK3/JNK/c-jun pathway. Biochem. Biophys. Res. Commun. 2013, 441, 763–769. [Google Scholar] [CrossRef]
- Lee, Y.T.; Laxton, V.; Lin, H.Y.; Chan, Y.W.F.; Fitzgerald-Smith, S.; To, T.L.O.; Yan, B.P.; Liu, T.; Tse, G. Animal models of atherosclerosis. Biomed. Rep. 2017, 6, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.T.; Lin, H.Y.; Chan, Y.W.; Li, K.H.; To, O.T.; Yan, B.P.; Liu, T.; Li, G.; Wong, W.T.; Keung, W.; et al. Mouse models of atherosclerosis: A historical perspective and recent advances. Lipids Health Dis. 2017, 16, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, S.; Adamiak, M.; Mathiyalagan, P.; Kenneweg, F.; Kafert-Kasting, S.; Thum, T. Therapeutic and Diagnostic Translation of Extracellular Vesicles in Cardiovascular Diseases: Roadmap to the Clinic. Circulation 2021, 143, 1426–1449. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pham, A.; Kang, L.; Walker, S.A.; Davidovich, I.; Iannotta, D.; TerKonda, S.P.; Shapiro, S.; Talmon, Y.; Pham, S.; et al. Effects of Adipose-Derived Biogenic Nanoparticle-Associated microRNA-451a on Toll-like Receptor 4-Induced Cytokines. Pharmaceutics 2021, 14, 16. [Google Scholar] [CrossRef]
- DIANA-microT. Available online: http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=MicroT_CDS/index (accessed on 10 March 2022).
- TargetScanHuman. Available online: https://www.targetscan.org/vert_80/ (accessed on 10 March 2022).
- miRDB Databases. Available online: http://mirdb.org/cgi-bin/search.cgi (accessed on 10 March 2022).
- Yang, Z.; Bian, C.; Zhou, H.; Huang, S.; Wang, S.; Liao, L.; Zhao, R.C. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011, 20, 259–267. [Google Scholar] [CrossRef]
- Zeidman, R.; Jackson, C.S.; Magee, A.I. Protein acyl thioesterases (Review). Mol. Membr. Biol. 2009, 26, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Ao, L.; Shi, Y.; Johnson, T.R.; Fullerton, D.A.; Meng, X. Oxidized low density lipoprotein induces bone morphogenetic protein-2 in coronary artery endothelial cells via Toll-like receptors 2 and 4. J. Biol. Chem. 2011, 286, 12213–12220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satou, M.; Nishi, Y.; Yoh, J.; Hattori, Y.; Sugimoto, H. Identification and characterization of acyl-protein thioesterase 1/lysophospholipase I as a ghrelin deacylation/lysophospholipid hydrolyzing enzyme in fetal bovine serum and conditioned medium. Endocrinology 2010, 151, 4765–4775. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, X.; Chen, C.; Li, S.; Shen, J.; Tse, G.; Chan, M.T.V.; Wu, W.K.K. Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2018, 51, e12483. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, X.; Jiang, C.; Qian, W.; Tse, G.; Chan, M.T.V.; Wu, W.K.K. Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. 2018, 51, e12404. [Google Scholar] [CrossRef]
- Yu, X.; Zheng, H.; Tse, G.; Chan, M.T.; Wu, W.K. Long non-coding RNAs in melanoma. Cell Prolif. 2018, 51, e12457. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.J.; Bie, Z.D.; Sun, C.F. Long noncoding RNA AK088388 regulates autophagy through miR-30a to affect cardiomyocyte injury. J. Cell. Biochem. 2019, 120, 10155–10163. [Google Scholar] [CrossRef] [PubMed]
- Fasolo, F.; Jin, H.; Winski, G.; Chernogubova, E.; Pauli, J.; Winter, H.; Li, D.Y.; Glukha, N.; Bauer, S.; Metschl, S.; et al. Long Noncoding RNA MIAT Controls Advanced Atherosclerotic Lesion Formation and Plaque Destabilization. Circulation 2021, 144, 1567–1583. [Google Scholar] [CrossRef] [PubMed]
- Vacante, F.; Rodor, J.; Lalwani, M.K.; Mahmoud, A.D.; Bennett, M.; De Pace, A.L.; Miller, E.; Van Kuijk, K.; de Bruijn, J.; Gijbels, M.; et al. CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice. Circ. Res. 2021, 128, 1258–1275. [Google Scholar] [CrossRef]
- Hopkins, P.N. Molecular biology of atherosclerosis. Physiol. Rev. 2013, 93, 1317–1542. [Google Scholar] [CrossRef]
- Lee, W.R.; Kim, A.; Kim, K.S.; Park, Y.Y.; Park, J.H.; Kim, K.H.; Kim, S.J.; Park, K.K. Alpha-lipoic acid attenuates atherosclerotic lesions and inhibits proliferation of vascular smooth muscle cells through targeting of the Ras/MEK/ERK signaling pathway. Mol. Biol. Rep. 2012, 39, 6857–6866. [Google Scholar] [CrossRef] [PubMed]
- Celi, A.; Lorenzet, R.; Furie, B.C.; Furie, B. Microparticles and a P-selectin-mediated pathway of blood coagulation. Dis. Markers 2004, 20, 347–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.C.; Tseng, C.C.; Hsiao, C.C.; Chang, H.C.; Chang, L.T.; Fang, W.F.; Leu, S.; Wang, Y.H.; Tsai, T.H.; Yang, C.T.; et al. Circulating endothelial-derived activated microparticle: A useful biomarker for predicting one-year mortality in patients with advanced non-small cell lung cancer. Biomed. Res. Int. 2014, 2014, 173401. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, D.; Williams, K.J.; Liu, M.L. Tobacco smoke induces the generation of procoagulant microvesicles from human monocytes/macrophages. Arter. Thromb. Vasc. Biol. 2010, 30, 1818–1824. [Google Scholar] [CrossRef]
- Lo, S.K.; Bovis, L.; Matura, R.; Zhu, B.; He, S.; Lum, H.; Turco, S.J.; Ho, J.L. Leishmania lipophosphoglycan reduces monocyte transendothelial migration: Modulation of cell adhesion molecules, intercellular junctional proteins, and chemoattractants. J. Immunol. 1998, 160, 1857–1865. [Google Scholar] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Cheng, L.; Fu, H.; Chan, C.Z.Y.; Tse, G.; Liu, T.; Li, G. Endothelial-Derived APT1-Mediated Macrophage-Endothelial Cell Interactions Participate in the Development of Atherosclerosis by Regulating the Ras/MAPK Signaling Pathway. Life 2022, 12, 551. https://doi.org/10.3390/life12040551
Wang X, Cheng L, Fu H, Chan CZY, Tse G, Liu T, Li G. Endothelial-Derived APT1-Mediated Macrophage-Endothelial Cell Interactions Participate in the Development of Atherosclerosis by Regulating the Ras/MAPK Signaling Pathway. Life. 2022; 12(4):551. https://doi.org/10.3390/life12040551
Chicago/Turabian StyleWang, Xinghua, Lijun Cheng, Huaying Fu, Calista Zhuo Yi Chan, Gary Tse, Tong Liu, and Guangping Li. 2022. "Endothelial-Derived APT1-Mediated Macrophage-Endothelial Cell Interactions Participate in the Development of Atherosclerosis by Regulating the Ras/MAPK Signaling Pathway" Life 12, no. 4: 551. https://doi.org/10.3390/life12040551
APA StyleWang, X., Cheng, L., Fu, H., Chan, C. Z. Y., Tse, G., Liu, T., & Li, G. (2022). Endothelial-Derived APT1-Mediated Macrophage-Endothelial Cell Interactions Participate in the Development of Atherosclerosis by Regulating the Ras/MAPK Signaling Pathway. Life, 12(4), 551. https://doi.org/10.3390/life12040551