Association between Usual Dietary Intake of Food Groups and DNA Methylation and Effect Modification by Metabotype in the KORA FF4 Cohort
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Habitual Dietary Intake
2.3. Metabotype
2.4. DNA Methylation Data
2.5. Statistical Analysis
2.6. Availability of Data and Materials
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHEI | Alternate Healthy Eating Index |
| CGI | CpG Island |
| DNMT | DNA methyltransferase |
| DR | Diabetic retinopathy |
| EWAS | Epigenome-wide association study |
| FDR | False Discovery Rate |
| FFQ | Food frequency questionnaire |
| MDS | Mediterranean Diet Score |
| QN | Quantile normalization |
| TSS | Transcription start site |
| UA | Uric acid |
References
- Kalea, A.Z.; Drosatos, K.; Buxton, J.L. Nutriepigenetics and cardiovascular disease. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmoud, A.M.; Ali, M.M. Methyl Donor Micronutrients that Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dugué, P.-A.; Chamberlain, J.A.; Bassett, J.K.; Hodge, A.M.; Brinkman, M.T.; Joo, J.E.; Jung, C.-H.; Wong, E.M.; Makalic, E.; Schmidt, D.F.; et al. Overall lack of replication of associations between dietary intake of folate and vitamin B-12 and DNA methylation in peripheral blood. Am. J. Clin. Nutr. 2020, 111, 228–230. [Google Scholar] [CrossRef]
- Mandaviya, P.R.; Joehanes, R.; Brody, J.; Castillo-Fernandez, J.E.; Dekkers, K.F.; Do, A.N.; Graff, M.; Hänninen, I.K.; Tanaka, T.; de Jonge, E.A.L.; et al. Association of dietary folate and vitamin B-12 intake with genome-wide DNA methylation in blood: A large-scale epigenome-wide association analysis in 5841 individuals. Am. J. Clin. Nutr. 2019, 110, 437–450. [Google Scholar] [CrossRef]
- Do, W.L.; Whitsel, E.A.; Costeira, R.; Masachs, O.M.; Le Roy, C.I.; Bell, J.T.; Staimez, L.R.; Stein, A.D.; Smith, A.K.; Horvath, S.; et al. Epigenome-wide association study of diet quality in the Women’s Health Initiative and TwinsUK cohort. Int. J. Epidemiol. 2020, 50, 675–684. [Google Scholar] [CrossRef]
- Karabegović, I.; Portilla-Fernandez, E.; Li, Y.; Ma, J.; Maas, S.C.E.; Sun, D.; Hu, E.A.; Kühnel, B.; Zhang, Y.; Ambatipudi, S.; et al. Epigenome-wide association meta-analysis of DNA methylation with coffee and tea consumption. Nat. Commun. 2021, 12, 2830. [Google Scholar] [CrossRef]
- Campanella, G.; Gunter, M.J.; Polidoro, S.; Krogh, V.; Palli, D.; Panico, S.; Sacerdote, C.; Tumino, R.; Fiorito, G.; Guarrera, S.; et al. Epigenome-wide association study of adiposity and future risk of obesity-related diseases. Int. J. Obes. 2018, 42, 2022–2035. [Google Scholar] [CrossRef] [Green Version]
- O’Donovan, C.B.; Walsh, M.C.; Gibney, M.J.; Gibney, E.R.; Brennan, L. Can metabotyping help deliver the promise of personalised nutrition? Proc. Nutr. Soc. 2015, 75, 106–114. [Google Scholar] [CrossRef] [Green Version]
- Dahal, C.; Wawro, N.; Meisinger, C.; Brandl, B.; Skurk, T.; Volkert, D.; Hauner, H.; Linseisen, J. Evaluation of the metabotype concept after intervention with oral glucose tolerance test and dietary fiber-enriched food: An enable study. Nutr. Metab. Cardiovasc. Dis. 2022, in press. [Google Scholar] [CrossRef]
- Lachat, C.; Hawwash, D.; Ocké, M.C.; Berg, C.; Forsum, E.; Hörnell, A.; Larsson, C.; Sonestedt, E.; Wirfält, E.; Åkesson, A.; et al. Strengthening the Reporting of Observational Studies in Epidemiology—Nutritional Epidemiology (STROBE-nut): An Extension of the STROBE Statement. PLoS Med. 2016, 13, e1002036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathmann, W.; Haastert, B.; Icks, A.; Löwel, H.; Meisinger, C.; Holle, R.; Giani, G. High prevalence of undiagnosed diabetes mellitus in Southern Germany: Target populations for efficient screening. The KORA survey 2000. Diabetologia 2003, 46, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Freese, J.; Feller, S.; Harttig, U.; Kleiser, C.; Linseisen, J.; Fischer, B.C.; Leitzmann, M.F.; Six-Merker, J.; Michels, K.B.; Nimptsch, K.; et al. Development and evaluation of a short 24-h food list as part of a blended dietary assessment strategy in large-scale cohort studies. Eur. J. Clin. Nutr. 2014, 68, 324–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illner, A.-K.; Harttig, U.; Tognon, G.; Palli, D.; Salvini, S.; Bower, E.; Amiano, P.; Kassik, T.; Metspalu, A.; Engeset, D.; et al. Feasibility of innovative dietary assessment in epidemiological studies using the approach of combining different assessment instruments. Public Health Nutr. 2011, 14, 1055–1063. [Google Scholar] [CrossRef]
- Mitry, P.; Wawro, N.; Six-Merker, J.; Zoller, D.; Jourdan, C.; Meisinger, C.; Thierry, S.; Nöthlings, U.; Knüppel, S.; Boeing, H.; et al. Usual Dietary Intake Estimation Based on a Combination of Repeated 24-H Food Lists and a Food Frequency Questionnaire in the KORA FF4 Cross-Sectional Study. Front. Nutr. 2019, 6, 145. [Google Scholar] [CrossRef]
- Slimani, N.; Deharveng, G.; Charrondière, R.U.; van Kappel, A.L.; Ocké, M.C.; Welch, A.; Lagiou, A.; van Liere, M.; Agudo, A.; Pala, V.; et al. Structure of the standardized computerized 24-h diet recall interview used as reference method in the 22 centers participating in the EPIC project. Comput. Methods Programs Biomed. 1999, 58, 251–266. [Google Scholar] [CrossRef]
- Max Rubner-Institut (MRI). Bundeslebensmittelschlüssel. Available online: https://www.blsdb.de/ (accessed on 2 August 2021).
- Willett, W.C.; Howe, G.R.; Kushi, L.H. Adjustment for total energy intake in epidemiologic studies. Am. J. Clin. Nutr. 1997, 65 (Suppl. S4), 1220S–1228S. [Google Scholar] [CrossRef]
- Chiuve, S.E.; Fung, T.T.; Rimm, E.B.; Hu, F.B.; McCullough, M.L.; Wang, M.; Stampfer, M.J.; Willett, W.C. Alternative Dietary Indices Both Strongly Predict Risk of Chronic Disease. J. Nutr. 2012, 142, 1009–1018. [Google Scholar] [CrossRef] [Green Version]
- Couto, E.; Boffetta, P.; Lagiou, P.; Ferrari, P.; Buckland, G.; Overvad, K.; Dahm, C.; Tjonneland, A.; Olsen, A.; Clavelchapelon, F.; et al. Mediterranean dietary pattern and cancer risk in the EPIC cohort. Br. J. Cancer 2011, 104, 1493–1499. [Google Scholar] [CrossRef] [Green Version]
- Lehne, B.; Drong, A.W.; Loh, M.; Zhang, W.; Scott, W.R.; Tan, S.-T.; Afzal, U.; Scott, J.; Jarvelin, M.-R.; Elliott, P.; et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015, 16, 37. [Google Scholar] [CrossRef] [Green Version]
- McCartney, D.L.; Walker, R.M.; Morris, S.W.; McIntosh, A.M.; Porteous, D.J.; Evans, K.L. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom. Data 2016, 9, 22–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.-Y.; Feng, Z.; Yi, X. A general introduction to adjustment for multiple comparisons. J. Thorac. Dis. 2017, 9, 1725–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Marioni, R.E.; Hedman, Å.K.; Pfeiffer, L.; Tsai, P.-C.; Reynolds, L.M.; Just, A.C.; Duan, Q.; Boer, C.G.; Tanaka, T.; et al. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry 2018, 23, 422–433. [Google Scholar] [CrossRef]
- Marzi, C.; Holdt, L.M.; Fiorito, G.; Tsai, P.-C.; Kretschmer, A.; Wahl, S.; Guarrera, S.; Teupser, D.; Spector, T.D.; Iacoviello, L.; et al. Epigenetic Signatures at AQP3 and SOCS3 Engage in Low-Grade Inflammation across Different Tissues. PLoS ONE 2016, 11, e0166015. [Google Scholar] [CrossRef] [Green Version]
- VanderWeele, T.J.; Shpitser, I. A New Criterion for Confounder Selection. Biometrics 2011, 67, 1406–1413. [Google Scholar] [CrossRef]
- van Iterson, M.; van Zwet, E.W.; Heijmans, B.T. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017, 18, 19. [Google Scholar] [CrossRef] [Green Version]
- Meisinger, C.; Thorand, B. Leisure time physical activity and the risk of type 2 diabetes in men and women from the general population. Diabetologia 2004, 48, 27–34. [Google Scholar] [CrossRef]
- Stöckl, D.; Peters, A.; Thorand, B.; Heier, M.; Koenig, W.; Seissler, J.; Thiery, J.; Rathmann, W.; Meisinger, C. Reproductive factors, intima media thickness and carotid plaques in a cross-sectional study of postmenopausal women enrolled in the population-based KORA F4 study. BMC Women Health 2014, 14, 17. [Google Scholar] [CrossRef] [Green Version]
- R: A Language and Environment for Statistical Computing. Vienna, Austria. 2021. Available online: https://www.R-project.org/ (accessed on 18 May 2021).
- Illumina Inc. Infinium MethylationEPIC Manifest Column Headings. 2020. Available online: https://emea.support.illumina.com/bulletins/2016/08/infinium-methylationepic-manifest-column-headings.html?langsel=/fo/ (accessed on 25 November 2021).
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Sturm, C.; Wagner, A.E. Brassica-Derived Plant Bioactives as Modulators of Chemopreventive and Inflammatory Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 1890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illingworth, R.S.; Bird, A.P. CpG islands—‘A rough guide’. FEBS Lett. 2009, 583, 1713–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes silencing. Oncotarget 2016, 8, 5629–5637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sueishi, T.; Akasaki, Y.; Goto, N.; Kurakazu, I.; Toya, M.; Kuwahara, M.; Uchida, T.; Hayashida, M.; Tsushima, H.; Bekki, H.; et al. GRK 5 Inhibition Attenuates Cartilage Degradation via Decreased NF -κB Signaling. Arthritis Rheumatol. 2019, 72, 620–631. [Google Scholar] [CrossRef]
- Lieu, M.; Koch, W.J. GRK2 and GRK5 as therapeutic targets and their role in maladaptive and pathological cardiac hypertrophy. Expert Opin. Ther. Targets 2019, 23, 201–214. [Google Scholar] [CrossRef]
- Zhao, T.L.; Gan, X.X.; Bao, Y.; Wang, W.P.; Liu, B.; Wang, L.H. GRK5 promotes tumor progression in renal cell carcinoma. Neoplasma 2019, 66, 261–270. [Google Scholar] [CrossRef]
- Yang, G.; Sun, S.; Wang, J.; Li, W.; Wang, X.; Yuan, L.; Li, S. S-Allylmercaptocysteine Targets Nrf2 in Osteoarthritis Treatment Through NOX4/NF-κB Pathway. Drug Des. Dev. Ther. 2020, 14, 4533–4546. [Google Scholar] [CrossRef]
- Mohammed, S.A.; Paramesha, B.; Kumar, Y.; Tariq, U.; Arava, S.K.; Banerjee, S.K. Allylmethylsulfide, a Sulfur Compound Derived from Garlic, Attenuates Isoproterenol-Induced Cardiac Hypertrophy in Rats. Oxidative Med. Cell. Longev. 2020, 2020, 7856318. [Google Scholar] [CrossRef]
- Min, K.-J.; Nam, J.-O.; Kwon, T.K. Fisetin Induces Apoptosis Through p53-Mediated Up-Regulation of DR5 Expression in Human Renal Carcinoma Caki Cells. Molecules 2017, 22, 1285. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, F.; Paredes-Gonzalez, X.; Kong, A.-N.T. Dietary Glucosinolates Sulforaphane, Phenethyl Isothiocyanate, Indole-3-Carbinol/3,3′-Diindolylmethane: Antioxidative Stress/Inflammation, Nrf2, Epigenetics/Epigenomics and In Vivo Cancer Chemopreventive Efficacy. Curr. Pharmacol. Rep. 2015, 1, 179–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.; Han, X.; Wu, C.; Keel, S.; Shang, X.; Zhang, L.; He, M. Does daily dietary intake affect diabetic retinopathy progression? 10-year results from the 45 and Up Study. Br. J. Ophthalmol. 2019, 104, 1774–1780. [Google Scholar] [CrossRef] [PubMed]
- Major, T.; Topless, R.K.; Dalbeth, N.; Merriman, T.R. Evaluation of the diet wide contribution to serum urate levels: Meta-analysis of population based cohorts. BMJ 2018, 363, k3951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Sun, X.; Zhao, X.; Liu, Y. Associations of serum uric acid and urinary albumin with the severity of diabetic retinopathy in individuals with type 2 diabetes. BMC Ophthalmol. 2020, 20, 467. [Google Scholar] [CrossRef]
- Thounaojam, M.C.; Montemari, A.; Powell, F.L.; Malla, P.; Gutsaeva, D.R.; Bachettoni, A.; Ripandelli, G.; Repossi, A.; Tawfik, A.; Martin, P.M.; et al. Monosodium Urate Contributes to Retinal Inflammation and Progression of Diabetic Retinopathy. Diabetes 2019, 68, 1014–1025. [Google Scholar] [CrossRef] [Green Version]
- Miao, A.; Lu, J.; Wang, Y.; Mao, S.; Cui, Y.; Pan, J.; Li, L.; Luo, Y. Identification of the aberrantly methylated differentially expressed genes in proliferative diabetic retinopathy. Exp. Eye Res. 2020, 199, 108141. [Google Scholar] [CrossRef]
- Timon, C.M.; O’Connor, A.; Bhargava, N.; Gibney, E.R.; Feeney, E.L. Dairy Consumption and Metabolic Health. Nutrients 2020, 12, 3040. [Google Scholar] [CrossRef]
- Fardet, A.; Rock, E. In vitro and in vivo antioxidant potential of milks, yoghurts, fermented milks and cheeses: A narrative review of evidence. Nutr. Res. Rev. 2017, 31, 52–70. [Google Scholar] [CrossRef] [Green Version]
- Guasch-Ferré, M.; Satija, A.; Blondin, S.; Janiszewski, M.; Emlen, E.; O’Connor, L.E.; Campbell, W.W.; Hu, F.B.; Willett, W.C.; Stampfer, M.J. Meta-Analysis of Randomized Controlled Trials of Red Meat Consumption in Comparison With Various Comparison Diets on Cardiovascular Risk Factors. Circulation 2019, 139, 1828–1845. [Google Scholar] [CrossRef]
- Braun, K.V.E.; Dhana, K.; de Vries, P.; Voortman, T.; Van Meurs, J.B.J.; Uitterlinden, A.G.; Hofman, A.; Hu, F.B.; Franco, O.H.; Dehghan, A. Epigenome-wide association study (EWAS) on lipids: The Rotterdam Study. Clin. Epigenetics 2017, 9, 15. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Huang, X.R.; You, Y.; Xue, L.; Wang, X.-J.; Meng, X.; Lin, X.; Shen, J.; Yu, X.; Lan, H.-Y.; et al. Latent TGF-β1 protects against diabetic kidney disease via Arkadia/Smad7 signaling. Int. J. Biol. Sci. 2021, 17, 3583–3594. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.A.E.; Averill, M.M.; Delaney, J.A.; Lemaitre, R.N.; Howard, B.V.; Fretts, A.M. Associations of diet quality and blood serum lipoprotein levels in a population at high risk for diabetes: The Strong Heart Family Study. Eur. J. Clin. Nutr. 2019, 74, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.J.; Midthune, D.; Subar, A.F.; Shumakovich, M.; Freedman, L.S.; Thompson, F.E.; Kipnis, V. Taking Advantage of the Strengths of 2 Different Dietary Assessment Instruments to Improve Intake Estimates for Nutritional Epidemiology. Am. J. Epidemiol. 2012, 175, 340–347. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.H.; Cho, E.R.; Lim, J.-E.; Jee, S.H. Association between serum persistent organic pollutants and DNA methylation in Korean adults. Environ. Res. 2017, 158, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Rafi, Z.; Greenland, S. Semantic and cognitive tools to aid statistical science: Replace confidence and significance by compatibility and surprise. BMC Med. Res. Methodol. 2020, 20, 244. [Google Scholar] [CrossRef]


| Overall | Male | ||||
|---|---|---|---|---|---|
| Overall | Overall | Metabotype 1 | Metabotype 2 | Metabotype 3 | |
| n | 1261 | 595 | 122 | 393 | 80 |
| Age in years (median [IQR]) | 58.0 [49.0, 66.0] | 59.0 [49.0, 68.0] | 55.0 [49.0, 65.0] | 58.0 [48.0, 67.0] | 66.0 [61.0, 73.0] |
| BMI (WHO-Class.) (%) | |||||
| Underweight (x < 18.5) | 5 (0.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Normal weight (18.5 ≥ x < 25) | 407 (32.3) | 142 (23.9) | 75 (61.5) | 59 (15.0) | 8 (10.0) |
| Pre-obesity (25 ≥ x < 30) | 520 (41.2) | 288 (48.4) | 43 (35.2) | 224 (57.0) | 21 (26.2) |
| Obesity class I (30 ≥ x < 35) | 230 (18.2) | 124 (20.8) | 3 (2.5) | 96 (24.4) | 25 (31.2) |
| Obesity class II (35 ≥ x < 40) | 67 (5.3) | 29 (4.9) | 0 (0.0) | 12 (3.1) | 17 (21.2) |
| Obesity class III (x > 40) | 32 (2.5) | 12 (2.0) | 1 (0.8) | 2 (0.5) | 9 (11.2) |
| Total energy intake in Kcal/d (median [IQR]) | 1825.5 [1551.1, 2117.3] | 2094.2 [1889.1, 2337.1] | 2159.7 [1931.1, 2407.7] | 2080.8 [1859.8, 2308.1] | 2100.5 [1954.8, 2391.6] |
| Alcohol g/day (median [IQR]) | 5.0 [2.4, 13.9] | 13.1 [5.1, 24.6] | 15.6 [7.4, 26.7] | 12.4 [4.6, 23.5] | 13.3 [3.9, 23.4] |
| Smoking behavior (%) | |||||
| Regular smoker | 178 (14.1) | 89 (15.0) | 20 (16.4) | 62 (15.8) | 7 (8.8) |
| Former smoker | 486 (38.5) | 273 (45.9) | 42 (34.4) | 172 (43.8) | 59 (73.8) |
| Never smoker | 597 (47.3) | 233 (39.2) | 60 (49.2) | 159 (40.5) | 14 (17.5) |
| Physical activity = Active (%) | 777 (61.6) | 350 (58.8) | 87 (71.3) | 231 (58.8) | 32 (40.0) |
| Education in years = < 13 years (%) | 790 (62.6) | 347 (58.3) | 61 (50.0) | 235 (59.8) | 51 (63.7) |
| Overall | Female | ||||
|---|---|---|---|---|---|
| Overall | Overall | Metabotype 1 | Metabotype 2 | Metabotype 3 | |
| n | 666 | 459 | 167 | 40 | |
| Age in years (median [IQR]) | 58.0 [48.2, 66.0] | 56.0 [47.0, 63.0] | 63.0 [55.0, 72.0] | 64.0 [60.0, 71.2] | |
| BMI (WHO-Class.) (%) | |||||
| Underweight (x < 18.5) | 5 (0.8) | 5 (1.1) | 0 (0.0) | 0 (0.0) | |
| Normal weight (18.5 ≥ x < 25) | 265 (39.8) | 244 (53.2) | 20 (12.0) | 1 (2.5) | |
| Pre-obesity (25 ≥ x < 30) | 232 (34.8) | 161 (35.1) | 68 (40.7) | 3 (7.5) | |
| Obesity class I (30 ≥ x < 35) | 106 (15.9) | 38 (8.3) | 53 (31.7) | 15 (37.5) | |
| Obesity class II (35 ≥ x < 40) | 38 (5.7) | 10 (2.2) | 21 (12.6) | 7 (17.5) | |
| Obesity class III (x > 40) | 20 (3.0) | 1 (0.2) | 5 (3.0) | 14 (35.0) | |
| Total energy intake in Kcal/d (median [IQR]) | 1578.2 [1428.6, 1793.6] | 1607.7 [1441.0, 1816.8] | 1534.2 [1419.8, 1716.8] | 1526.9 [1397.4, 1752.2] | |
| Alcohol g/day (median [IQR]) | 2.7 [1.7, 5.3] | 3.4 [2.0, 6.1] | 1.8 [1.3, 3.5] | 1.3 [1.0, 2.3] | |
| Smoking behavior (%) | |||||
| Regular smoker | 89 (13.4) | 64 (13.9) | 24 (14.4) | 1 (2.5) | |
| Former smoker | 213 (32.0) | 146 (31.8) | 50 (29.9) | 17 (42.5) | |
| Never smoker | 364 (54.7) | 249 (54.2) | 93 (55.7) | 22 (55.0) | |
| Physical activity = Active (%) | 427 (64.1) | 320 (69.7) | 92 (55.1) | 15 (37.5) | |
| Education in years =< 13 years (%) | 443 (66.5) | 283 (61.7) | 127 (76.0) | 33 (82.5) | |
| Overall | Male | ||||
|---|---|---|---|---|---|
| Overall | Overall | Metabotype 1 | Metabotype 2 | Metabotype 3 | |
| n | 1261 | 595 | 122 | 393 | 80 |
| Median [Interquartilerange] | |||||
| Protein | 67.8 [58.8, 78.6] | 76.9 [69.2, 86.5] | 77.7 [70.2, 86.8] | 75.9 [68.2, 86.0] | 80.2 [72.4, 90.2] |
| Carbohydrates | 193.0 [162.0, 228.7] | 218.5 [188.6, 250.8] | 227.5 [193.9, 263.3] | 216.5 [187.2, 251.0] | 206.6 [179.6, 238.7] |
| Fats | 75.9 [65.4, 88.6] | 87.2 [77.9, 97.7] | 87.5 [77.9, 97.6] | 86.2 [77.6, 96.3] | 92.7 [82.3, 101.5] |
| Potatoes (g/day) | 54.7 [44.4, 68.5] | 59.5 [49.1, 73.8] | 58.0 [48.3, 68.7] | 59.5 [49.2, 74.1] | 63.2 [52.6, 82.4] |
| Total Vegetables (g/day) | 163.3 [132.7, 204.0] | 147.4 [121.2, 184.3] | 159.4 [127.3, 195.1] | 145.6 [121.6, 181.6] | 142.5 [112.9, 168.4] |
| Leafy Vegetables (g/day) | 23.1 [15.3, 31.7] | 24.0 [15.7, 31.7] | 25.0 [15.6, 31.7] | 23.9 [16.0, 32.6] | 20.6 [14.7, 30.1] |
| Fruit vegetables (g/day) | 71.6 [54.5, 96.4] | 62.6 [49.0, 84.2] | 69.7 [56.0, 96.2] | 60.8 [48.6, 81.1] | 54.9 [41.5, 78.9] |
| Root vegetables (g/day) | 15.4 [10.7, 25.2] | 12.4 [9.5, 19.4] | 14.7 [10.0, 19.7] | 12.7 [9.7, 19.5] | 9.6 [8.1, 14.1] |
| Cruciferous vegetables (g/day) | 14.5 [11.3, 19.2] | 14.1 [11.3, 18.7] | 13.4 [11.5, 17.3] | 13.9 [10.7, 18.2] | 16.6 [13.5, 21.9] |
| Mushrooms (g/day) | 2.3 [1.6, 3.7] | 2.2 [1.4, 3.4] | 2.4 [2.0, 4.2] | 2.1 [1.5, 3.4] | 1.8 [1.1, 2.5] |
| Onions & garlic (g/day) | 6.4 [4.4, 9.0] | 6.1 [4.0, 8.7] | 4.9 [3.3, 7.3] | 6.1 [4.1, 8.9] | 7.3 [5.8, 9.9] |
| Legumes (g/day) | 4.8 [3.6, 6.8] | 4.1 [3.3, 6.1] | 4.2 [3.5, 6.0] | 4.0 [3.2, 6.1] | 4.4 [3.5, 6.2] |
| Total fruit (g/day) | 141.2 [87.9, 201.6] | 133.9 [75.2, 196.3] | 136.9 [76.6, 190.3] | 132.4 [71.9, 199.1] | 135.6 [87.8, 215.1] |
| Nuts & seeds (g/day) | 4.2 [3.0, 8.9] | 4.5 [3.3, 8.9] | 5.1 [3.4, 9.0] | 4.3 [3.2, 8.8] | 4.8 [3.3, 11.0] |
| Milk (g/day) | 73.8 [27.6, 140.5] | 59.0 [19.5, 122.2] | 61.0 [20.5, 125.4] | 62.2 [19.8, 130.8] | 45.5 [17.3, 83.7] |
| Yogurt (g/day) | 30.7 [14.0, 66.8] | 21.1 [11.9, 52.8] | 31.6 [12.5, 64.7] | 20.8 [11.9, 49.2] | 15.6 [11.8, 43.1] |
| Cheese (g/day) | 30.2 [21.4, 41.9] | 30.4 [21.4, 42.5] | 33.0 [22.1, 47.4] | 29.6 [21.0, 40.5] | 31.4 [22.1, 44.9] |
| Cream (g/day) | 1.4 [1.2, 2.2] | 1.4 [1.2, 1.8] | 1.4 [1.1, 1.6] | 1.4 [1.2, 1.9] | 1.4 [1.2, 1.7] |
| Grain products (g/day) | 161.6 [133.5, 195.4] | 187.8 [162.2, 218.6] | 203.5 [168.5, 239.7] | 184.7 [159.5, 215.0] | 178.3 [163.8, 213.6] |
| Whole grain products (g/day) | 16.5 [7.3, 34.5] | 14.2 [6.9, 36.5] | 17.6 [7.6, 38.7] | 14.2 [6.8, 36.5] | 10.6 [5.4, 27.6] |
| Total meat (g/day) | 107.2 [83.4, 142.6] | 142.0 [119.2, 166.3] | 131.2 [105.9, 152.1] | 143.1 [120.6, 165.1] | 158.7 [132.0, 198.4] |
| Fresh red meat (g/day) | 42.6 [33.2, 54.6] | 54.0 [46.5, 64.6] | 51.8 [45.0, 59.0] | 55.1 [47.6, 65.9] | 54.9 [44.3, 67.0] |
| Processed meat (g/day) | 42.5 [29.3, 62.7] | 61.0 [47.0, 79.2] | 53.9 [37.2, 71.0] | 60.3 [47.8, 77.7] | 74.5 [57.2, 102.1] |
| Total fish (g/day) | 16.3 [11.8, 24.6] | 18.2 [13.1, 28.0] | 19.8 [13.6, 29.3] | 18.0 [13.0, 27.0] | 19.0 [13.0, 28.3] |
| Eggs (g/day) | 13.4 [10.2, 19.2] | 14.1 [10.5, 20.9] | 13.0 [10.5, 18.7] | 13.8 [10.3, 20.9] | 16.9 [12.3, 24.3] |
| Plant oils (g/day) | 5.3 [3.6, 8.0] | 5.6 [3.6, 8.5] | 5.4 [3.6, 8.2] | 5.5 [3.5, 8.3] | 6.2 [4.1, 9.6] |
| Butter (g/day) | 13.7 [7.8, 17.4] | 16.5 [9.1, 21.8] | 18.7 [10.6, 23.1] | 16.0 [8.9, 21.4] | 16.2 [8.4, 21.3] |
| Margarine (g/day) | 0.6 [0.3, 1.8] | 0.8 [0.5, 2.8] | 0.7 [0.4, 2.0] | 0.8 [0.5, 2.7] | 0.9 [0.7, 3.9] |
| Total sweets (g/day) | 35.1 [25.9, 46.0] | 37.7 [27.0, 49.9] | 44.2 [31.8, 54.2] | 37.5 [27.2, 49.9] | 29.4 [22.2, 40.3] |
| Cakes (g/day) | 48.8 [38.6, 63.8] | 53.9 [40.2, 70.3] | 58.5 [41.0, 73.0] | 53.5 [39.9, 68.5] | 52.4 [40.5, 71.0] |
| Sugar sweetened beverages (g/day) | 6.7 [3.6, 24.6] | 10.8 [6.1, 67.2] | 8.0 [4.7, 19.0] | 11.4 [6.7, 64.4] | 14.7 [6.8, 104.2] |
| Coffee (g/day) | 435.0 [365.1, 478.3] | 445.1 [375.0, 497.7] | 443.4 [389.9, 503.7] | 445.1 [371.9, 494.0] | 450.6 [369.5, 501.2] |
| Tea (g/day) | 63.4 [27.6, 322.5] | 35.7 [22.0, 223.3] | 64.1 [25.2, 364.3] | 34.6 [22.0, 201.1] | 30.4 [18.9, 199.8] |
| Wine (g/day) | 17.6 [11.9, 39.4] | 18.4 [12.7, 44.6] | 24.4 [17.5, 63.9] | 17.5 [12.7, 38.3] | 11.8 [8.2, 37.3] |
| Beer (g/day) | 39.7 [6.5, 204.4] | 208.2 [50.8, 482.6] | 223.6 [55.4, 560.6] | 204.4 [51.5, 472.2] | 210.4 [43.3, 474.1] |
| Spirits (g/day) | 0.3 [0.2, 0.5] | 0.4 [0.3, 0.7] | 0.5 [0.3, 0.8] | 0.4 [0.3, 0.7] | 0.3 [0.2, 0.4] |
| Alcohol (g/day) | 5.0 [2.4, 13.9] | 13.1 [5.1, 24.6] | 15.6 [7.4, 26.7] | 12.4 [4.6, 23.5] | 13.3 [3.9, 23.4] |
| AHEI | 42.5 [36.2, 48.9] | 41.1 [34.7, 46.8] | 42.8 [37.1, 49.6] | 40.8 [34.8, 45.9] | 40.4 [33.6, 46.6] |
| MDS | 4.0 [3.0, 6.0] | 5.0 [3.0, 6.0] | 5.0 [4.0, 6.0] | 4.0 [3.0, 6.0] | 5.0 [3.8, 6.0] |
| Folic acid (µg/d) | 200.1 [169.7, 237.7] | 212.6 [179.2, 249.5] | 223.6 [185.6, 257.2] | 208.6 [176.7, 245.6] | 215.1 [180.7, 257.5] |
| Overall | Female | ||||
| Overall | Overall | Metabotype 1 | Metabotype 2 | Metabotype 3 | |
| n | 666 | 459 | 167 | 40 | |
| Median [Interquartilerange] | |||||
| Protein | 76.9 [69.2, 86.5] | 77.7 [70.2, 86.8] | 75.9 [68.2, 86.0] | 80.2 [72.4, 90.2] | |
| Carbohydrates | 218.5 [188.6, 250.8] | 227.5 [193.9, 263.3] | 216.5 [187.2, 251.0] | 206.6 [179.6, 238.7] | |
| Fats | 87.2 [77.9, 97.7] | 87.5 [77.9, 97.6] | 86.2 [77.6, 96.3] | 92.7 [82.3, 101.5] | |
| Potatoes (g/day) | 50.4 [40.6, 63.9] | 49.0 [39.5, 61.0] | 55.8 [42.1, 71.6] | 52.1 [43.0, 66.9] | |
| Total Vegetables (g/day) | 178.1 [146.2, 218.7] | 182.1 [150.4, 224.4] | 176.3 [139.1, 215.8] | 163.4 [146.1, 191.0] | |
| Leafy Vegetables (g/day) | 22.8 [14.9, 32.0] | 22.9 [14.8, 32.4] | 22.8 [15.3, 31.5] | 21.9 [14.8, 26.7] | |
| Fruit vegetables (g/day) | 81.2 [61.6, 106.7] | 82.8 [63.2, 110.9] | 77.1 [58.1, 104.0] | 75.9 [54.5, 90.5] | |
| Root vegetables (g/day) | 18.9 [13.1, 30.6] | 20.9 [14.1, 33.3] | 15.1 [11.4, 26.1] | 12.2 [10.4, 16.7] | |
| Cruciferous vegetables (g/day) | 14.8 [11.4, 20.1] | 14.1 [10.9, 18.8] | 16.4 [12.4, 21.8] | 15.9 [12.3, 22.6] | |
| Mushrooms (g/day) | 2.4 [1.7, 3.9] | 2.6 [2.0, 4.5] | 2.1 [1.2, 2.7] | 1.9 [1.4, 2.4] | |
| Onions & garlic (g/day) | 6.7 [4.7, 9.3] | 6.3 [4.5, 8.4] | 7.4 [5.2, 9.9] | 9.5 [7.2, 12.9] | |
| Legumes (g/day) | 5.2 [4.2, 7.5] | 5.3 [4.2, 8.0] | 5.3 [4.2, 7.1] | 5.0 [3.7, 6.4] | |
| Total fruit (g/day) | 145.4 [96.5, 203.3] | 143.0 [93.3, 201.6] | 154.4 [100.2, 212.3] | 144.3 [95.8, 192.5] | |
| Nuts & seeds (g/day) | 4.0 [2.6, 8.7] | 4.3 [2.8, 9.4] | 3.3 [2.4, 6.0] | 3.5 [2.3, 7.4] | |
| Milk (g/day) | 86.8 [42.8, 150.6] | 92.6 [45.0, 160.3] | 80.5 [37.3, 129.8] | 83.4 [26.8, 122.2] | |
| Yogurt (g/day) | 38.6 [17.9, 76.1] | 40.4 [18.7, 79.9] | 36.4 [15.9, 70.8] | 23.4 [17.0, 49.9] | |
| Cheese (g/day) | 29.8 [21.5, 41.8] | 30.5 [21.8, 42.0] | 27.5 [20.1, 39.8] | 26.1 [20.1, 38.8] | |
| Cream (g/day) | 1.5 [1.2, 2.5] | 1.5 [1.2, 2.6] | 1.5 [1.2, 2.5] | 1.4 [1.2, 2.1] | |
| Grain products (g/day) | 138.1 [121.1, 163.9] | 143.0 [123.6, 169.3] | 129.6 [117.0, 153.3] | 129.8 [109.8, 141.2] | |
| Whole grain products (g/day) | 18.0 [8.3, 34.1] | 19.2 [8.7, 35.2] | 15.7 [7.0, 29.3] | 16.2 [9.7, 29.5] | |
| Total meat (g/day) | 86.0 [72.9, 101.7] | 81.9 [69.9, 96.6] | 90.6 [78.5, 108.8] | 104.8 [93.2, 134.2] | |
| Fresh red meat (g/day) | 34.0 [29.5, 40.1] | 33.7 [29.5, 39.4] | 35.6 [29.0, 41.0] | 33.5 [30.4, 43.0] | |
| Processed meat (g/day) | 31.0 [24.0, 41.6] | 29.1 [22.8, 37.4] | 34.4 [27.4, 46.8] | 49.7 [40.2, 69.4] | |
| Total fish (g/day) | 14.2 [10.9, 21.8] | 13.7 [10.7, 21.7] | 15.0 [11.9, 22.1] | 12.8 [11.1, 19.0] | |
| Eggs (g/day) | 13.0 [9.9, 17.9] | 12.9 [9.9, 18.0] | 13.2 [10.1, 17.9] | 13.0 [9.5, 16.4] | |
| Plant oils (g/day) | 5.2 [3.6, 7.6] | 5.1 [3.5, 7.6] | 5.3 [3.7, 7.7] | 4.4 [3.6, 6.3] | |
| Butter (g/day) | 12.0 [7.0, 15.3] | 12.5 [7.4, 15.4] | 11.0 [6.2, 15.0] | 9.9 [6.1, 14.5] | |
| Margarine (g/day) | 0.4 [0.2, 1.0] | 0.3 [0.2, 0.8] | 0.5 [0.3, 1.6] | 0.8 [0.4, 2.3] | |
| Total sweets (g/day) | 33.4 [24.9, 43.1] | 34.3 [25.8, 44.8] | 31.7 [23.7, 39.8] | 30.2 [23.1, 40.4] | |
| Cakes (g/day) | 46.2 [37.5, 57.9] | 47.4 [38.5, 58.8] | 43.9 [36.1, 55.3] | 39.4 [34.5, 47.9] | |
| Sugar sweetened beverages (g/day) | 4.2 [2.8, 8.4] | 3.9 [2.6, 7.1] | 4.5 [3.0, 14.2] | 6.6 [4.4, 65.6] | |
| Coffee (g/day) | 419.5 [356.7, 465.6] | 412.0 [351.8, 467.0] | 430.0 [366.8, 464.2] | 430.4 [365.5, 455.1] | |
| Tea (g/day) | 135.7 [38.2, 372.5] | 151.4 [41.8, 377.5] | 124.5 [34.2, 343.8] | 53.5 [27.6, 278.4] | |
| Wine (g/day) | 17.0 [11.0, 36.1] | 19.4 [14.4, 43.4] | 11.6 [8.2, 19.8] | 7.4 [5.3, 9.9] | |
| Beer (g/day) | 6.7 [5.7, 8.2] | 7.2 [6.0, 8.8] | 6.0 [5.0, 7.0] | 5.2 [4.6, 6.1] | |
| Spirits (g/day) | 0.2 [0.1, 0.3] | 0.2 [0.2, 0.3] | 0.1 [0.1, 0.2] | 0.1 [0.1, 0.1] | |
| Alcohol (g/day) | 2.7 [1.7, 5.3] | 3.4 [2.0, 6.1] | 1.8 [1.3, 3.5] | 1.3 [1.0, 2.3] | |
| AHEI | 43.9 [37.7, 50.5] | 45.2 [39.4, 51.7] | 42.0 [35.6, 48.0] | 36.2 [31.3, 40.8] | |
| MDS | 4.0 [3.0, 6.0] | 4.0 [3.0, 6.0] | 4.0 [3.0, 5.0] | 3.0 [3.0, 4.0] | |
| Folic acid (µg/d) | 190.3 [162.7, 224.7] | 194.3 [166.8, 230.5] | 182.0 [155.2, 216.8] | 179.3 [155.0, 199.4] | |
| ProbeID | Sample Size | Effect Size ** | Standard Error | p-Value | Foodgroup | Chr | RefGene Name | RefGene Group | Relation to CpG Island |
|---|---|---|---|---|---|---|---|---|---|
| cg01838728 | 1319 | −8.91 × 10−4 | 1.60 × 10−4 | 0.0268 | Leafy vegetables | 15 | N/A | N/A | N/A |
| cg15351590 | 1321 | −1.82 × 10−4 | 3.16 × 10−5 | 0.00809 | Root vegetables | 6 | KIFC1 | TSS1500 | N_Shore |
| cg14698575 | 1319 | 8.51 × 10−4 | 1.37 × 10−4 | 6.27 × 10−4 | Cruciferous vegetables | 9 | N/A | N/A | S_Shore |
| cg23709902 | 1310 | 4.40 × 10−4 | 7.90 × 10−5 | 0.0243 | Cruciferous vegetables | 17 | SRCIN1 | Body | Island |
| cg06102690 | 1319 | 6.72 × 10−4 | 1.24 × 10−4 | 0.0494 | Cruciferous vegetables | 4 | CCDC149 | TSS200 | N/A |
| cg10399824 | 1322 | −6.43 × 10−4 | 1.11 × 10−4 | 0.00596 | Onions-garlic | 10 | GRK5 | Body | N/A |
| cg06690548 | 1277 | −1.04 × 10−4 | 1.88 × 10−5 | 0.0269 | Wine | 4 | SLC7A11 | Body | N/A |
| cg06690548 | 1277 | −5.10 × 10−5 | 5.21 × 10−6 | 6.01 × 10−16 | Beer | 4 | SLC7A11 | Body | N/A |
| cg26457483 | 1319 | −6.03 × 10−5 | 8.37 × 10−6 | 7.99 × 10−7 | Beer | 1 | PHGDH | Body | S_Shore |
| cg14476101 | 1320 | −6.32 × 10−5 | 9.30 × 10−6 | 1.31 × 10−5 | Beer | 1 | PHGDH | Body | S_Shore |
| cg06088069 | 1319 | −2.71 × 10−5 | 4.32 × 10−6 | 3.74 × 10−4 | Beer | 14 | JDP2 * | 5′UTR * | S_Shore |
| cg16246545 | 1320 | −4.68 × 10−5 | 7.85 × 10−6 | 0.00250 | Beer | 1 | PHGDH | Body | S_Shore |
| cg15837522 | 1322 | −6.45 × 10−5 | 1.09 × 10−5 | 0.00324 | Beer | 8 | N/A | N/A | N/A |
| cg18120259 | 1320 | −3.23 × 10−5 | 5.59 × 10−6 | 0.00755 | Beer | 6 | LOC100132354 | Body | N/A |
| cg08228578 | 1322 | −2.39 × 10−5 | 4.21 × 10−6 | 0.0125 | Beer | 12 | SHMT2 * | Body * | S_Shore |
| cg10223198 | 1322 | −2.88 × 10−5 | 5.27 × 10−6 | 0.0427 | Beer | 11 | N/A | N/A | N/A |
| ProbeID | Effect Size ** | Standard Error | p-Value | p-Value (Bacon) | Foodgroup | Cluster | Chr | RefGene Name | RefGene Group | Relation to CpG Island |
|---|---|---|---|---|---|---|---|---|---|---|
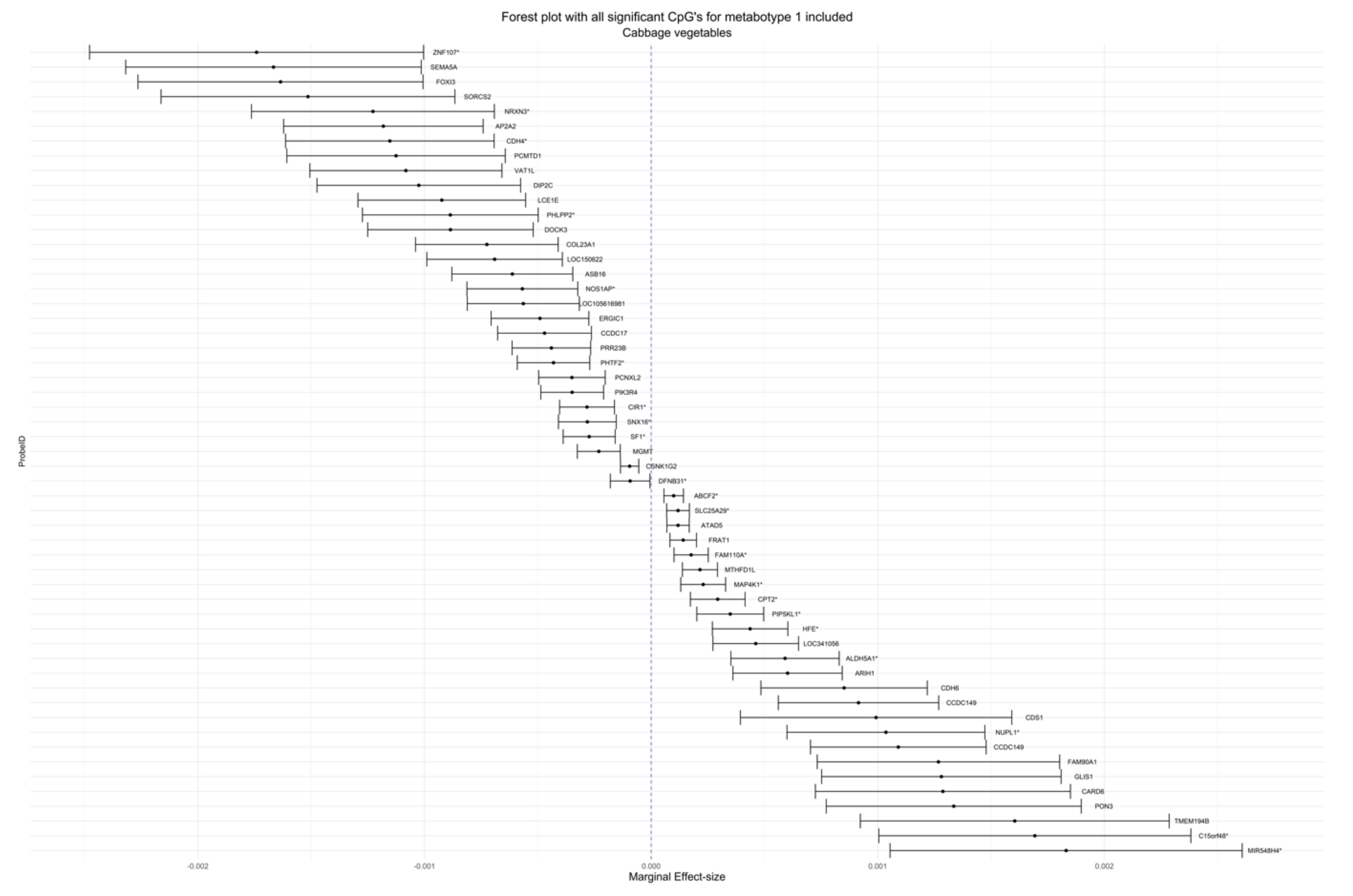
| cg00067414 | 2.15 × 10−4 | 3.94 × 10−5 | 0.01538 | 0.03974 | Cruciferous | Metabotype 1 | 6 | MTHFD1L | Body | Island |
| cg20561564 | −1.31 × 10−3 | 2.40 × 10−4 | 0.01538 | 0.03974 | Cruciferous | Metabotype 1 | 9 | N/A | N/A | N/A |
| cg11945292 | 1.09 × 10−3 | 1.98 × 10−4 | 0.01538 | 0.03974 | Cruciferous | Metabotype 1 | 4 | CCDC149 | TSS200 | N/A |
| cg22614518 | −4.31 × 10−4 | 8.16 × 10−5 | 0.02687 | 0.06638 | Cruciferous | Metabotype 1 | 7 | PHTF2 * | Body * | N/A |
| cg04183158 | −1.18 × 10−3 | 2.25 × 10−4 | 0.02687 | 0.06638 | Cruciferous | Metabotype 1 | 11 | AP2A2 | 3′UTR | S_Shore |
| cg06892726 | 4.37 × 10−4 | 8.50 × 10−5 | 0.04280 | 0.09608 | Cruciferous | Metabotype 1 | 6 | HFE * | 1stExon * | N/A |
| cg23160569 | −3.49 × 10−4 | 7.07 × 10−5 | 0.04454 | 0.09608 | Cruciferous | Metabotype 1 | 3 | PIK3R4 | Body | N/A |
| cg23923117 | −8.52 × 10−4 | 1.74 × 10−4 | 0.04454 | 0.09608 | Cruciferous | Metabotype 1 | 2 | N/A | N/A | N/A |
| cg01841471 | −1.21 × 10−3 | 2.43 × 10−4 | 0.04454 | 0.09608 | Cruciferous | Metabotype 1 | 13 | N/A | N/A | S_Shelf |
| cg08921926 | 6.02 × 10−4 | 1.23 × 10−4 | 0.04454 | 0.09608 | Cruciferous | Metabotype 1 | 15 | ARIH1 | TSS1500 | N_Shore |
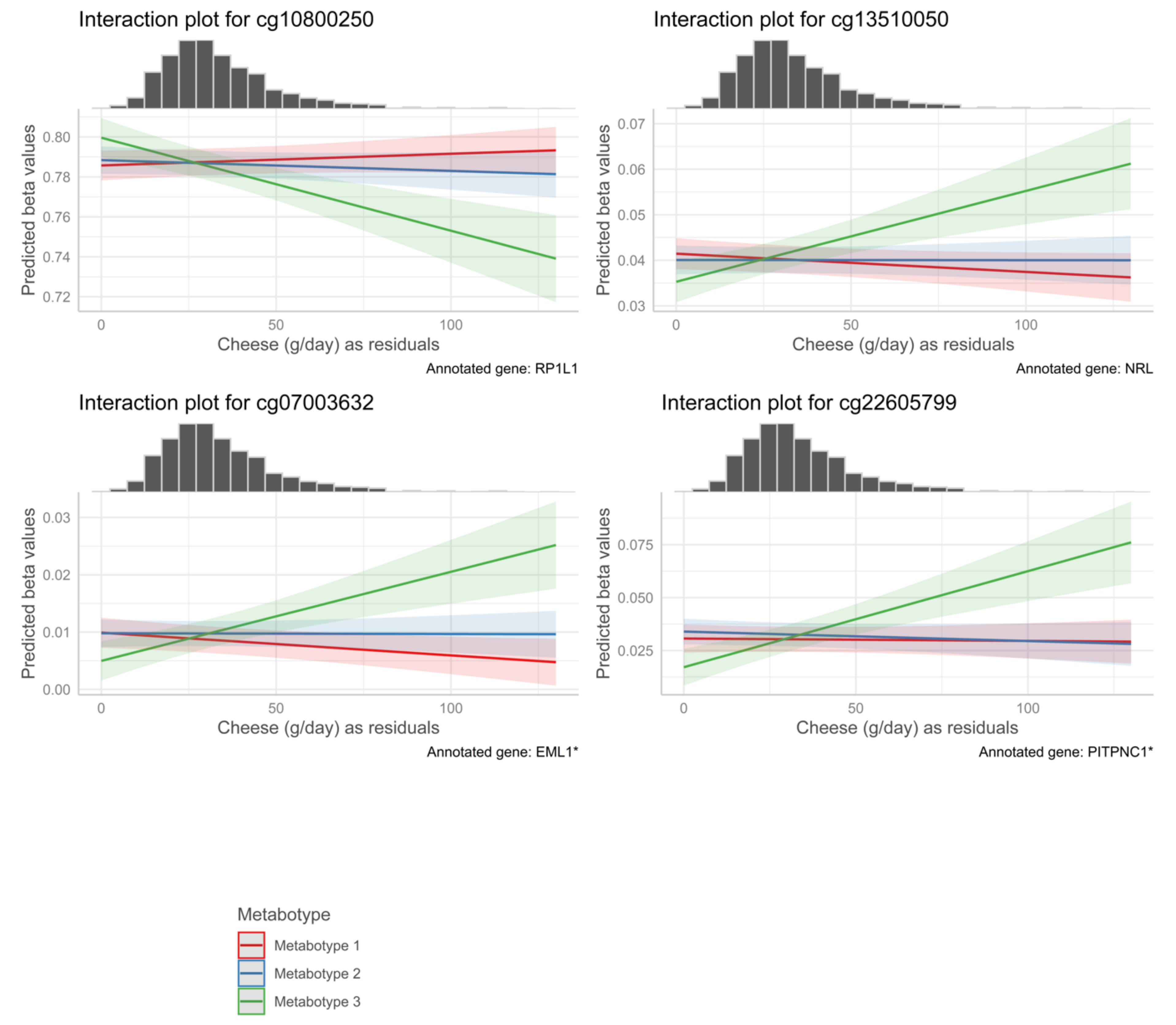
| cg00073181 | −1.21 × 10−3 | 2.14 × 10−4 | 0.00116 | 0.00167 | Cheese | Metabotype 3 | 1 | TLR5 | 5′UTR | N/A |
| cg23795938 | −1.24 × 10−3 | 2.00 × 10−4 | 0.00249 | 0.00350 | Cheese | Metabotype 3 | 1 | TMEM88B | TSS200 | N_Shore |
| cg04045906 | −6.27 × 10−4 | 1.08 × 10−4 | 0.00555 | 0.00766 | Cheese | Metabotype 3 | 4 | N/A | N/A | N/A |
| cg10888278 | −8.36 × 10−4 | 1.49 × 10−4 | 0.01856 | 0.02485 | Cheese | Metabotype 3 | 11 | NTM | Body | N/A |
| cg15379294 | −5.78 × 10−4 | 1.16 × 10−4 | 0.02049 | 0.03083 | Cheese | Metabotype 3 | 3 | POLR2H * | TSS1500 * | N_Shore |
| cg00741624 | 8.41 × 10−4 | 1.61 × 10−4 | 0.02049 | 0.04099 | Cheese | Metabotype 3 | 14 | KIAA1409 | 5′UTR | Island |
| cg18244100 | −4.92 × 10−4 | 1.00 × 10−4 | 0.02049 | 0.03083 | Cheese | Metabotype 3 | 6 | SKIV2L | Body | N_Shelf |
| cg21880900 | −7.15 × 10−4 | 1.60 × 10−4 | 0.02049 | 0.02728 | Cheese | Metabotype 3 | 3 | N/A | N/A | N/A |
| cg12274082 | −5.76 × 10−4 | 1.17 × 10−4 | 0.02423 | 0.03569 | Cheese | Metabotype 3 | 6 | CYP21A2 * | Body * | N/A |
| cg05531689 | −6.16 × 10−4 | 1.22 × 10−4 | 0.03207 | 0.04485 | Cheese | Metabotype 3 | 2 | OTOF | Body | S_Shelf |
| cg00039945 | −7.68 × 10−4 | 1.24 × 10−4 | 0.05176 | 0.03773 | Whole grain | Metabotype 3 | 1 | LGR6 * | Body * | N/A |
| cg12515635 | −7.85 × 10−4 | 1.79 × 10−4 | 0.05176 | 0.03773 | Whole grain | Metabotype 3 | 15 | KLF13 | Body | N_Shelf |
| cg16687213 | −1.78 × 10−3 | 3.74 × 10−4 | 0.05176 | 0.03773 | Whole grain | Metabotype 3 | 7 | TRIM4 * | TSS1500 * | S_Shore |
| cg07268926 | −6.94 × 10−4 | 1.50 × 10−4 | 0.05351 | 0.05357 | Whole grain | Metabotype 3 | 11 | IGSF9B | Body | N/A |
| cg04395306 | 2.21 × 10−4 | 5.12 × 10−5 | 0.05351 | 0.06912 | Whole grain | Metabotype 3 | 20 | PREX1 | Body | Island |
| cg10143811 | 4.40 × 10−4 | 1.06 × 10−4 | 0.05351 | 0.07192 | Whole grain | Metabotype 3 | 12 | LMO3 * | 5′UTR * | N/A |
| cg10762466 | 7.30 × 10−4 | 1.42 × 10−4 | 0.06360 | 0.07745 | Whole grain | Metabotype 3 | 19 | N/A | N/A | N_Shore |
| cg01755100 | −8.44 × 10−4 | 1.82 × 10−4 | 0.07429 | 0.06912 | Whole grain | Metabotype 3 | 17 | N/A | N/A | S_Shelf |
| cg15200604 | −7.81 × 10−4 | 1.84 × 10−4 | 0.07429 | 0.06912 | Whole grain | Metabotype 3 | 13 | N/A | N/A | N/A |
| cg00880872 | −5.80 × 10−4 | 1.34 × 10−4 | 0.07429 | 0.06912 | Whole grain | Metabotype 3 | 9 | N/A | N/A | N_Shore |
| cg18029285 | 2.67 × 10−4 | 6.16 × 10−5 | 0.00771 | 0.43405 | Total meat | Metabotype 2 | 17 | KRTAP9-6 | TSS1500 | N/A |
| cg06713760 | 1.37 × 10−4 | 4.30 × 10−5 | 0.02080 | 0.81005 | Total meat | Metabotype 2 | 10 | N/A | N/A | S_Shelf |
| cg05581388 | 2.19 × 10−4 | 5.30 × 10−5 | 0.03327 | 0.95588 | Total meat | Metabotype 2 | 17 | KRTAP9-6 | TSS1500 | N/A |
| cg08991742 | 6.48 × 10−5 | 1.72 × 10−5 | 0.04204 | 0.95588 | Total meat | Metabotype 2 | 2 | ARHGAP25 * | 5′UTR * | N/A |
| cg27582585 | 6.24 × 10−5 | 2.93 × 10−5 | 0.07862 | 0.95588 | Total meat | Metabotype 2 | 1 | KLHDC9 * | Body * | S_Shore |
| cg05831315 | 1.15 × 10−4 | 3.55 × 10−5 | 0.08613 | 0.95588 | Total meat | Metabotype 2 | 8 | N/A | N/A | N/A |
| cg10919344 | 1.35 × 10−4 | 4.72 × 10−5 | 0.08613 | 0.95588 | Total meat | Metabotype 2 | 11 | OR5A1 | TSS200 | N/A |
| cg07454320 | 3.95 × 10−4 | 7.43 × 10−5 | 0.08825 | 0.68647 | Eggs | Metabotype 3 | 1 | WNT2B * | TSS200 * | Island |
| cg17634390 | −1.74 × 10−3 | 3.24 × 10−4 | 0.08825 | 0.68647 | Eggs | Metabotype 3 | 4 | COX7B2 | 5′UTR | N/A |
| cg13202871 | −2.15 × 10−3 | 4.30 × 10−4 | 0.08825 | 0.68647 | Eggs | Metabotype 3 | 12 | SLCO1B7 * | ExonBnd * | N/A |
| cg23049758 | −7.36 × 10−4 | 1.62 × 10−4 | 0.08825 | 0.68647 | Eggs | Metabotype 3 | 17 | SPAG9 * | Body * | N/A |
| cg09034467 | −1.83 × 10−3 | 4.23 × 10−4 | 0.08825 | 0.68647 | Eggs | Metabotype 3 | 21 | N/A | N/A | N/A |
| cg00857137 | −1.06 × 10−3 | 2.46 × 10−4 | 0.09857 | 0.73530 | Eggs | Metabotype 3 | 19 | TLE2 * | Body * | Island |
| cg16181002 | 3.62 × 10−3 | 5.97 × 10−4 | 0.01779 | 0.01344 | Margarine | Metabotype 3 | 6 | PARK2 * | Body * | N/A |
| cg05534678 | 4.58 × 10−4 | 1.25 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 16 | ZNF688 * | 5′UTR * | Island |
| cg23229016 | 1.10 × 10−3 | 2.05 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 1 | RPS6KA1 * | 1stExon * | N/A |
| cg08027748 | −9.80 × 10−4 | 2.01 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 3 | UROC1 * | TSS1500 * | N/A |
| cg07199337 | 2.10 × 10−3 | 4.67 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 11 | PRMT3 * | TSS1500 * | N_Shore |
| cg25141008 | 1.67 × 10−3 | 4.51 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 20 | C20orf27 * | TSS1500 * | S_Shore |
| cg08644318 | 5.61 × 10−4 | 1.60 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 3 | YEATS2 | TSS1500 | N_Shore |
| cg02958895 | 1.92 × 10−3 | 4.41 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 1 | N/A | N/A | S_Shore |
| cg25356086 | 6.57 × 10−4 | 1.39 × 10−4 | 0.07021 | 0.06355 | Margarine | Metabotype 3 | 21 | C21orf119 * | TSS1500 * | N_Shore |
| cg26536849 | −6.74 × 10−4 | 2.06 × 10−4 | 0.07021 | 0.07213 | Margarine | Metabotype 3 | 20 | DDX27 | Body | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hellbach, F.; Baumeister, S.-E.; Wilson, R.; Wawro, N.; Dahal, C.; Freuer, D.; Hauner, H.; Peters, A.; Winkelmann, J.; Schwettmann, L.; et al. Association between Usual Dietary Intake of Food Groups and DNA Methylation and Effect Modification by Metabotype in the KORA FF4 Cohort. Life 2022, 12, 1064. https://doi.org/10.3390/life12071064
Hellbach F, Baumeister S-E, Wilson R, Wawro N, Dahal C, Freuer D, Hauner H, Peters A, Winkelmann J, Schwettmann L, et al. Association between Usual Dietary Intake of Food Groups and DNA Methylation and Effect Modification by Metabotype in the KORA FF4 Cohort. Life. 2022; 12(7):1064. https://doi.org/10.3390/life12071064
Chicago/Turabian StyleHellbach, Fabian, Sebastian-Edgar Baumeister, Rory Wilson, Nina Wawro, Chetana Dahal, Dennis Freuer, Hans Hauner, Annette Peters, Juliane Winkelmann, Lars Schwettmann, and et al. 2022. "Association between Usual Dietary Intake of Food Groups and DNA Methylation and Effect Modification by Metabotype in the KORA FF4 Cohort" Life 12, no. 7: 1064. https://doi.org/10.3390/life12071064
APA StyleHellbach, F., Baumeister, S.-E., Wilson, R., Wawro, N., Dahal, C., Freuer, D., Hauner, H., Peters, A., Winkelmann, J., Schwettmann, L., Rathmann, W., Kronenberg, F., Koenig, W., Meisinger, C., Waldenberger, M., & Linseisen, J. (2022). Association between Usual Dietary Intake of Food Groups and DNA Methylation and Effect Modification by Metabotype in the KORA FF4 Cohort. Life, 12(7), 1064. https://doi.org/10.3390/life12071064







