Incoherent Neutron Scattering and Terahertz Time-Domain Spectroscopy on Protein and Hydration Water
Abstract
:1. Introduction
2. Incoherent Neutron Scattering
2.1. Theory of Incoherent Neutron Scattering
2.2. Analysis of Incoherent Neutron Scattering
2.3. Dynamic Information in Incoherent Neutron Scattering
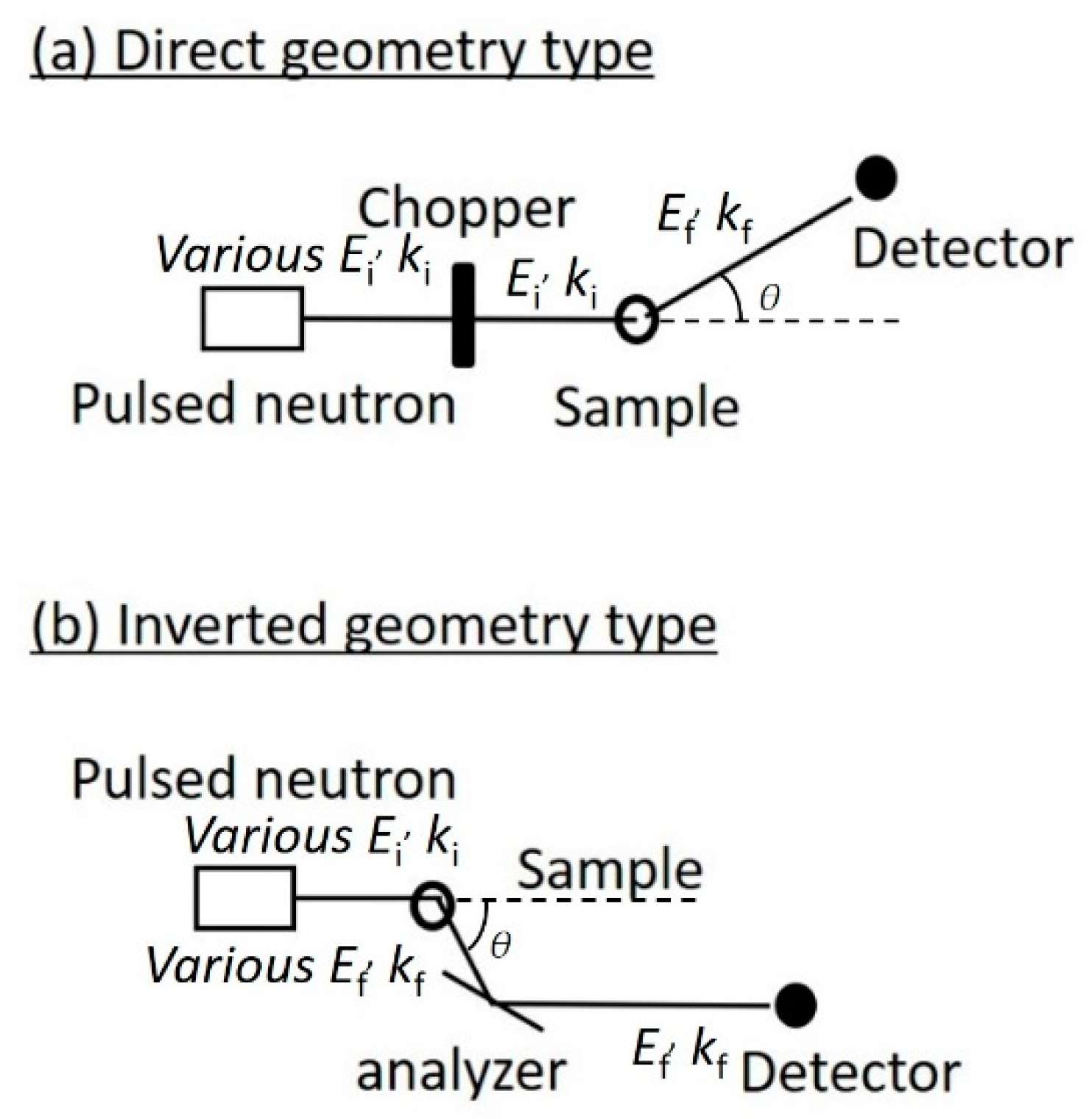
2.4. Experimental Details
3. Terahertz Time-Domain Spectroscopy
3.1. Theory of THz Spectroscopy
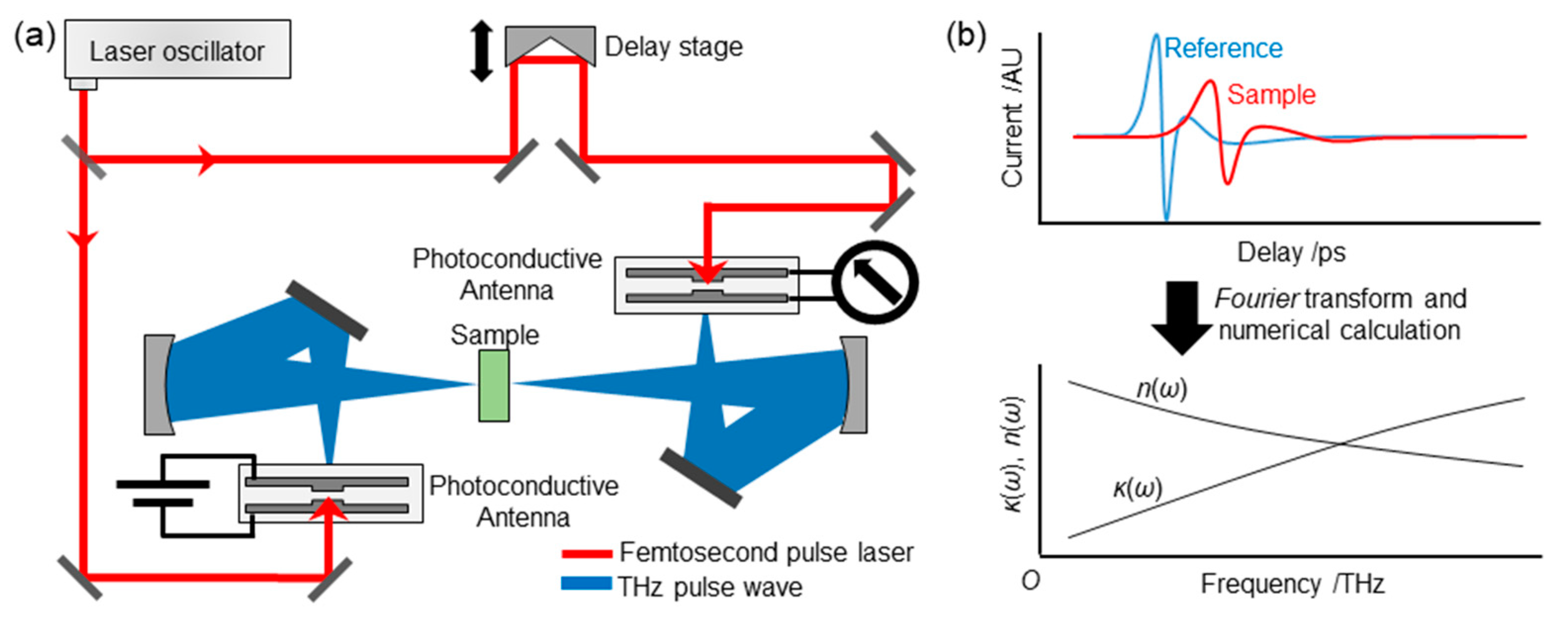
3.2. THz-TDS Equipment
3.3. Obtaining Spectra
3.4. Experimental Details
4. Computational Analysis
5. Applications by Combined Analysis
5.1. Effect of Freezable and Unfreezable Hydration Water on Protein Dynamics
5.2. Hydration Water of Biocompatible Polymer
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Careri, G. Cooperative charge fluctuations by migrating protons in globular proteins. Prog. Biophys. Mol. Biol. 1998, 70, 223–249. [Google Scholar] [CrossRef] [PubMed]
- Diehl, M.; Doster, W.; Petry, W.; Schober, H. Water-coupled low-frequency modes of myoglobin and lysozyme observed by inelastic neutron scattering. Biophys. J. 1997, 73, 2726–2732. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Joti, Y.; Kitao, A.; Kataoka, M. Hydration affects both harmonic and anharmonic nature of protein dynamics. Biophys. J. 2008, 95, 2916–2923. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Joti, Y.; Kitao, A.; Yamamuro, O.; Kataoka, M. Universality and Structural Implications of the Boson Peak in Proteins. Biophys. J. 2019, 117, 229–238. [Google Scholar] [CrossRef]
- Doster, W.; Cusack, S.; Petry, W. Dynamical Transition of Myoglobin Revealed by Inelastic Neutron-Scattering. Nature 1989, 337, 754–756. [Google Scholar] [CrossRef]
- Nakagawa, H.; Kataoka, M. Percolation of Hydration Water as a Control of Protein Dynamics. J. Phys. Soc. Jpn. 2010, 79, 083801. [Google Scholar] [CrossRef]
- Fitter, J. A measure of conformational entropy change during thermal protein unfolding using neutron spectroscopy. Biophys. J. 2003, 84, 3924–3930. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Kamikubo, H.; Kataoka, M. Effect of conformational states on protein dynamical transition. Bba-Proteins Proteom. 2010, 1804, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Fitter, J.; Heberle, J. Structural equilibrium fluctuations in mesophilic and thermophilic alpha-amylase. Biophys. J. 2000, 79, 1629–1636. [Google Scholar] [CrossRef] [Green Version]
- Grimaldo, M.; Roosen-Runge, F.; Zhang, F.; Schreiber, F.; Seydel, T. Dynamics of proteins in solution. Q. Rev. Biophys. 2019, 52, 1–63. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Kataoka, M. How can we derive hydration water dynamics with incoherent neutron scattering and molecular dynamics simulation? Biophys. Physicobiol. 2019, 16, 213–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, K.; Grudinin, S.; Kessler, B.; Weik, M.; Johnson, M.; Kneller, G.R.; Oesterheit, D.; Zaccai, G. Dynamical heterogeneity of specific amino acids in bacteriorhodopsin. J. Mol. Biol. 2008, 380, 581–591. [Google Scholar] [CrossRef]
- Markelz, A.; Whitmire, S.; Hillebrecht, J.; Birge, R. THz time domain spectroscopy of biomolecular conformational modes. Phys. Med. Biol. 2002, 47, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Markelz, A.G.; Knab, J.R.; Chen, J.Y.; He, Y.F. Protein dynamical transition in terahertz dielectric response. Chem. Phys. Lett. 2007, 442, 413–417. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.; Tominaga, K.; Sasakawa, H.; Tamura, A.; Murakami, H.; Ohtake, H.; Sarukura, N. Terahertz time-domain spectroscopy of amino acids and polypeptides. Biophys. J. 2005, 89, L22–L24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, S.; Takeda, M.W.; Nishizawa, S. Terahertz time domain spectroscopy of complex dielectric constants of boson peaks. J. Mol. Struct. 2003, 651, 285–288. [Google Scholar] [CrossRef]
- Yamamoto, N.; Kofu, M.; Nakajima, K.; Nakagawa, H.; Shibayama, N. Freezable and Unfreezable Hydration Water: Distinct Contributions to Protein Dynamics Revealed by Neutron Scattering. J. Phys. Chem. Lett. 2021, 12, 2172–2176. [Google Scholar] [CrossRef]
- Yamamoto, N.; Ohta, K.; Tamura, A.; Tominaga, K. Broadband Dielectric Spectroscopy on Lysozyme in the Sub-Gigahertz to Terahertz Frequency Regions: Effects of Hydration and Thermal Excitation. J. Phys. Chem. B 2016, 120, 4743–4755. [Google Scholar] [CrossRef]
- Yamamoto, N.; Ito, S.; Nakanishi, M.; Chatani, E.; Inoue, K.; Kandori, H.; Tominaga, K. Effect of Temperature and Hydration Level on Purple Membrane Dynamics Studied Using Broadband Dielectric Spectroscopy from Sub-GHz to THz Regions. J. Phys. Chem. B 2018, 122, 1367–1377. [Google Scholar] [CrossRef]
- Tarek, M.; Martyna, G.J.; Tobias, D.J. Amplitudes and frequencies of protein dynamics: Analysis of discrepancies between neutron scattering and molecular dynamics simulations. J. Am. Chem. Soc. 2000, 122, 10450–10451. [Google Scholar] [CrossRef]
- Tarek, M.; Tobias, D.J. The dynamics of protein hydration water: A quantitative comparison of molecular dynamics simulations and neutron-scattering experiments. Biophys. J. 2000, 79, 3244–3257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joti, Y.; Kitao, A. Cancellation between auto- and mutual correlation contributions of protein/water dynamics in terahertz time-domain spectra. Biophys. Physicobiol. 2019, 16, 240–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tominaga, T.; Hishida, M.; Murakami, D.; Fujii, Y.; Tanaka, M.; Seto, H. Experimental Evidence of Slow Mode Water in the Vicinity of Poly(ethylene oxide) at Physiological Temperature. J. Phys. Chem. B 2022, 126, 1758–1767. [Google Scholar] [CrossRef]
- Go, N. A Theorem on Amplitudes of Thermal Atomic Fluctuations in Large Molecules Assuming Specific Conformations Calculated by Normal Mode Analysis. Biophys. Chem. 1990, 35, 105–112. [Google Scholar] [CrossRef]
- Roh, J.H.; Novikov, V.N.; Gregory, R.B.; Curtis, J.E.; Chowdhuri, Z.; Sokolov, A.P. Onsets of anharmonicity in protein dynamics. Phys. Rev. Lett. 2005, 95, 038101. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C. Low-Frequency Vibrations of Helical Structures in Protein Molecules. Biochem. J. 1983, 209, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, M.; Kamikubo, H.; Yunoki, J.; Tokunaga, F.; Kanaya, T.; Izumi, Y.; Shibata, K. Low energy dynamics of globular proteins studied by inelastic neutron scattering. J. Phys. Chem. Solids 1999, 60, 1285–1289. [Google Scholar] [CrossRef]
- Ellis, R.J. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr. Opin. Struc. Biol. 2001, 11, 114–119. [Google Scholar] [CrossRef]
- Roosen-Runge, F.; Hennig, M.; Zhang, F.J.; Jacobs, R.M.J.; Sztucki, M.; Schober, H.; Seydel, T.; Schreiber, F. Protein self-diffusion in crowded solutions. Proc. Natl. Acad. Sci. USA 2011, 108, 11815–11820. [Google Scholar] [CrossRef] [Green Version]
- Wood, K.; Frolich, A.; Paciaroni, A.; Moulin, M.; Hartlein, M.; Zaccai, G.; Tobias, D.J.; Weik, M. Coincidence of dynamical transitions in a soluble protein and its hydration water: Direct measurements by neutron scattering and MD simulations. J. Am. Chem. Soc. 2008, 130, 4586–4587. [Google Scholar] [CrossRef]
- Svergun, D.I.; Richard, S.; Koch, M.H.J.; Sayers, Z.; Kuprin, S.; Zaccai, G. Protein hydration in solution: Experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. USA 1998, 95, 2267–2272. [Google Scholar] [CrossRef] [Green Version]
- Oroguchi, T.; Hashimoto, H.; Shimizu, T.; Sato, M.; Ikeguchi, M. Intrinsic Dynamics of Restriction Endonuclease EcoO109I Studied by Molecular Dynamics Simulations and X-Ray Scattering Data Analysis. Biophys. J. 2009, 96, 2808–2822. [Google Scholar] [CrossRef] [Green Version]
- Shiraga, K.; Ogawa, Y.; Kondo, N. Hydrogen Bond Network of Water around Protein Investigated with Terahertz and Infrared Spectroscopy. Biophys. J. 2016, 111, 2629–2641. [Google Scholar] [CrossRef] [Green Version]
- Yada, H.; Nagai, M.; Tanaka, K. Origin of the fast relaxation component of water and heavy water revealed by terahertz time-domain attenuated total reflection spectroscopy. Chem. Phys. Lett. 2008, 464, 166–170. [Google Scholar] [CrossRef]
- Yada, H.; Nagai, M.; Tanaka, K. The intermolecular stretching vibration mode in water isotopes investigated with broadband terahertz time-domain spectroscopy. Chem. Phys. Lett. 2009, 473, 279–283. [Google Scholar] [CrossRef]
- Shiraga, K.; Tanaka, K.; Arikawa, T.; Saito, S.; Ogawa, Y. Reconsideration of the relaxational and vibrational line shapes of liquid water based on ultrabroadband dielectric spectroscopy. Phys. Chem. Chem. Phys. 2018, 20, 26200–26209. [Google Scholar] [CrossRef] [PubMed]
- Ebbinghaus, S.; Kim, S.J.; Heyden, M.; Yu, X.; Heugen, U.; Gruebele, M.; Leitner, D.M.; Havenith, M. An extended dynamical hydration shell around proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 20749–20752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.F.; Ku, P.I.; Knab, J.R.; Chen, J.Y.; Markelz, A.G. Protein Dynamical Transition Does Not Require Protein Structure. Phys. Rev. Lett. 2008, 101, 178103. [Google Scholar] [CrossRef] [PubMed]
- Lipps, F.; Levy, S.; Markelz, A.G. Hydration and temperature interdependence of protein picosecond dynamics. Phys. Chem. Chem. Phys. 2012, 14, 6375–6381. [Google Scholar] [CrossRef]
- Roh, J.H.; Curtis, J.E.; Azzam, S.; Novikov, V.N.; Peral, I.; Chowdhuri, Z.; Gregory, R.B.; Sokolov, A.P. Influence of hydration on the dynamics of lysozyme. Biophys. J. 2006, 91, 2573–2588. [Google Scholar] [CrossRef] [Green Version]
- Panagopoulou, A.; Kyritsis, A.; Aravantinou, A.M.; Nanopoulos, D.; Serra, R.S.I.; Ribelles, J.L.G.; Shinyashiki, N.; Pissis, P. Glass Transition and Dynamics in Lysozyme-Water Mixtures Over Wide Ranges of Composition. Food Biophys. 2011, 6, 199–209. [Google Scholar] [CrossRef]
- Kitao, A.; Hayward, S.; Go, N. Energy landscape of a native protein: Jumping-among-minima model. Proteins 1998, 33, 496–517. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakagawa, H.; Yamamoto, N. Incoherent Neutron Scattering and Terahertz Time-Domain Spectroscopy on Protein and Hydration Water. Life 2023, 13, 318. https://doi.org/10.3390/life13020318
Nakagawa H, Yamamoto N. Incoherent Neutron Scattering and Terahertz Time-Domain Spectroscopy on Protein and Hydration Water. Life. 2023; 13(2):318. https://doi.org/10.3390/life13020318
Chicago/Turabian StyleNakagawa, Hiroshi, and Naoki Yamamoto. 2023. "Incoherent Neutron Scattering and Terahertz Time-Domain Spectroscopy on Protein and Hydration Water" Life 13, no. 2: 318. https://doi.org/10.3390/life13020318




