Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants
Abstract
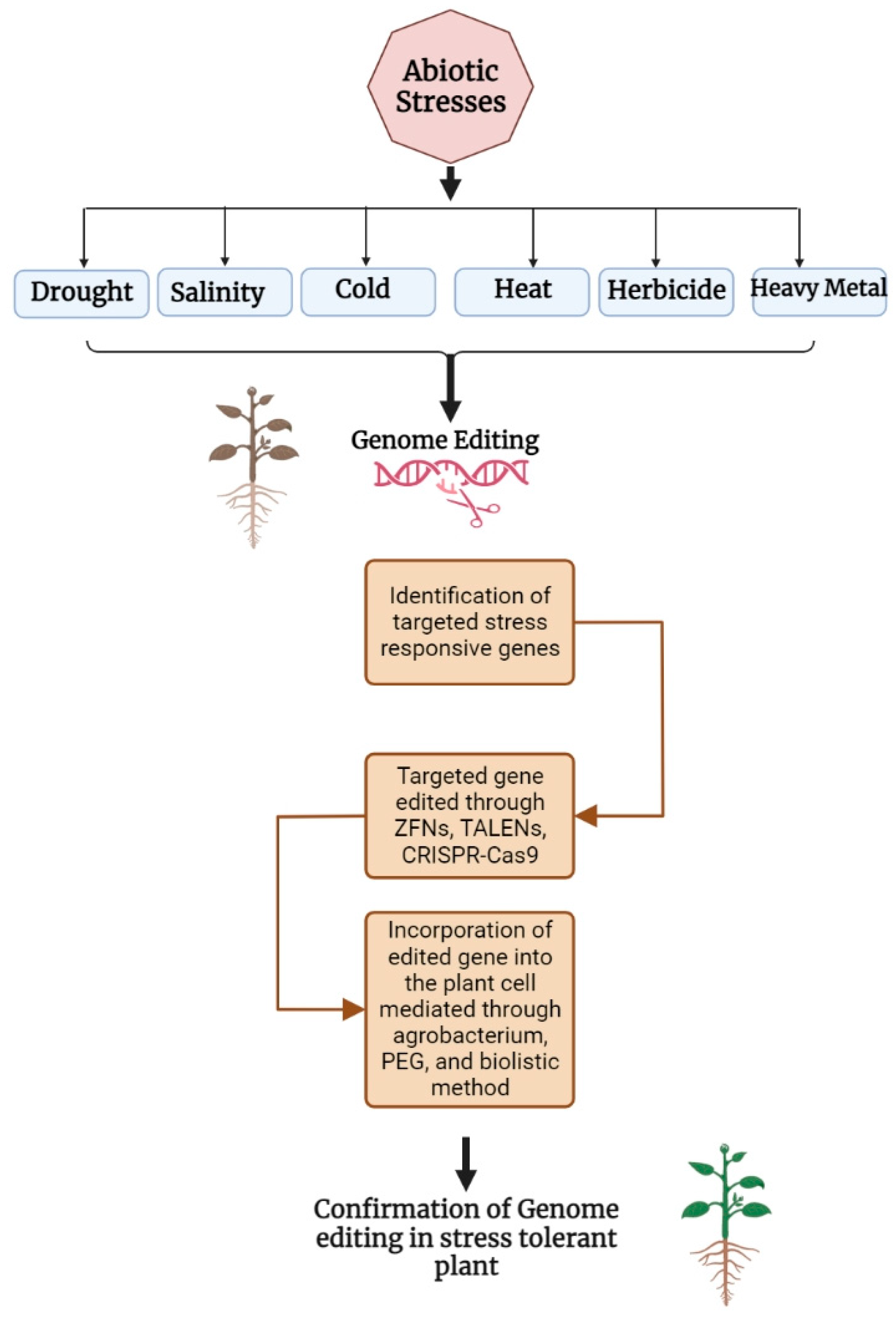
1. Introduction
2. Genome-Editing Strategy
2.1. Zinc-Finger Nucleases
2.2. Meganucleases
2.3. Transcription Activator-like Effector Nucleases (TALENs)
2.4. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-Associated Protein 9 (Cas9)
| Feature | ZFNs | Meganucleases | TALENs | CRISPR/Cas | References |
|---|---|---|---|---|---|
| Length of target sequence (bp) | 18–36 bp | 12–40 bp | 28–40 bp | 20–22 bp | [90,91] |
| Nuclease protein | FokI | I-SceI | FokI | Cas9 proteins | [91,92,93] |
| Dimerization | Required | Not required | Not required | Not required | [90,91,92] |
| Mode of action | Double-stranded break in target DNA | Direct conversions in targeted regions | Double-stranded break in target DNA | Double-stranded breaks or single-stranded nicks in target DNA | [94,95,96] |
| Repair events | NHEJ | HDR | HDR | NHEJ | [92,93,97] |
| Mutagenesis | High | Middle | Middle | Lower | [94] |
| Cloning | Necessary | Not necessary | Necessary | Not necessary | [91,98,99] |
| Creation of libraries and multiplexing | Challenging | Challenging | Challenging | Possible | [91,96,99] |
| Cost | Higher | Higher | Higher | Low | [100] |
| Types | One | One | One | Many | [101] |
| Specificity | Moderate | High | High | Low | [90,91] |
| Crop improvement | Low | Low | Low | High | [100] |
| Future use | Medium | Medium | Medium | High | [100] |
| Crop | Gene | Trait | Technique | References |
|---|---|---|---|---|
| Rice | OsQQR | Detection of safe harbor loci herbicide | ZFNs | [102] |
| OsBADH2, OsDEP1, OsSD1, OsCKX2 | Fragrance | TALEN | [103] | |
| Os11N3 | Bacterial blight resistance | TALEN | [67] | |
| OsCSA | Photoperiod sensitive male sterility | TALEN | [104] | |
| OsDERF1 | Drought tolerance | TALEN | [104] | |
| Wheat | TaMLO-A1, TaMLO-B1, TaMLO-D1 | Resistance to powdery mildew | TALEN | [105] |
| Maize | PAT | Herbicide resistance | ZFNs | [106] |
| ZmIPK1 | Herbicide tolerant and phytate reduced maize | ZFNs | [53] | |
| ZmTLP | Trait stacking | ZFNs | [107] | |
| ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4 | Biosynthesis of phytic acid | TALEN | [108] | |
| MS26 | Independent lines of male sterile plants | MNs | [109] | |
| Barley | HvPAPhy | Phytase reduction and seed development | TALEN | [110] |
| Soybean | DCL | Herbicide transmission | ZFNs | [111] |
| FAD2-1A, FAD2-1B | Low polyunsaturated fats | TALEN | [68,69] | |
| Tobacco | GUS: NPTII | Chromosome breaks | ZFNs | [112] |
| Endochitinase-50 gene (CHN50) | Emergence of resistance to herbicides | ZFNs | [113] | |
| Tomato | L1L4/NF-YB6 | Reduced contents of the anti-nutrient’s oxalic acid | ZFNs | [114] |
| Cotton | EPSPS | Herbicide tolerance | MNs | [115] |
| Hppd | Herbicide tolerance | MNs | [115] | |
| Potato | VInv | Sugar metabolism | TALEN | [116] |
2.5. DSB-Free Genome Editing
2.6. Base Editing
2.7. Prime Editing
2.8. Mobile CRISPR
3. Genome Editing Related to Abiotic Stresses
3.1. Drought Stress
3.2. Heat/Temperature Stress
3.3. Cold Stress
| Crops | Gene | Trait | Technique | References |
|---|---|---|---|---|
| Rice | OsDERF1 | Drought | CRISPR/Cas9 | [147] |
| Rice | SRL1, SRL2 | Drought | CRISPR/Cas9 | [149] |
| Rice | OsAAA-1, OsAAA-2 | Drought | CRISPR/Cas9 | [201] |
| Rice | OsNAC006 (transcription factor) | Drought and heat sensitivity | CRISPR/Cas9 | [202] |
| Rice | OsAOX1a | Drought resistance | CRISPR/Cas9 | [147] |
| Rice | OsDST | Drought and salinity | CRISPR/Cas9 | [151] |
| Rice | OsERA1, OsPYL9 | Drought | CRISPR/Cas9 | [148,150] |
| Rice | SAPK2 | Tolerance to salinity and drought | CRISPR/Cas9 | [131] |
| Rice | OsPMS3 | Photoperiod-sensitive male-sterile | CRISPR/Cas9 | [147] |
| Rice | Csa | Photosensitive-genic male-sterile | CRISPR/Cas9 | [203,204] |
| Rice | TMS5 | Thermo-sensitive genic male-sterile | CRISPR/Cas9 | [205] |
| Rice | OsNAC14 | Drought tolerance | CRISPR/Cas9 | [206] |
| Rice | OsPUB67 | Drought tolerance | CRISPR/Cas9 | [207] |
| Wheat | TaDREB2, TaERF3 | Tolerance to drought | CRISPR/Cas9 | [89] |
| Maize | ZmARGOS8 | Drought | CRISPR/Cas9 | [156] |
| Maize | ZmTMS5 | Creation of thermosensitive maize lines | CRISPR/Cas9 | [181] |
| Mustard | BnaA6.RGA | Drought tolerance | CRISPR/Cas9 | [163] |
| Soybean | Drb2a, Drb2b | Tolerance to drought and salinity stress | CRISPR/Cas9 | [208] |
| Soybean | GmMYB118 | Drought tolerance | CRISPR/Cas9 | [209] |
| Chickpea | 4CL, RVE7 | Drought tolerance | CRISPR/Cas9 | [152] |
| Tomato | SIMAPK3 and SlNPR1 | Drought | CRISPR/Cas9 | [159,160] |
| Tomato | SlARF4 | Drought | CRISPR/Cas9 | [140] |
| Tomato | SIAGL6 | Heat stress | CRISPR/Cas9 | [210] |
| Crops | Gene | Trait | Technique | References |
|---|---|---|---|---|
| Rice | OsMYB30 | Cold tolerance | CRISPR/Cas9 | [198] |
| Rice | OsAnn3 | Cold tolerance | CRISPR/Cas9 | [211] |
| Rice | OsAnn5 | Cold tolerance | CRISPR/Cas9 | [211] |
| Rice | OsPRP1 | Cold tolerance | CRISPR/Cas9 | [212] |
| Tomato | SlCBF1 | Cold tolerance | CRISPR/Cas9 | [200] |
| Arabidopsis thaliana | AtCBF1, AtCBF2 | Cold tolerance | CRISPR/Cas9 | [213] |
3.4. Salinity Stress
3.5. Heavy Metals Stress
3.6. Herbicide Stress
| Crops | Gene | Trait | Technique | References |
|---|---|---|---|---|
| Rice | C287T | Herbicide resistance | CRISPR/Cas9 | [274] |
| Rice | BEL | Herbicide resistance | CRISPR/Cas9 | [71] |
| Rice | OsALS1 | Herbicide tolerance | CRISPR/Cas9 | [271] |
| Rice | EPSPS | Herbicide resistance | CRISPR/Cas9 | [203] |
| Rice | SF3B1 | Herbicide resistance | CRISPR/Cas9 | [72] |
| Wheat | ALS | Herbicide resistance | CRISPR/Cas9 | [275,276] |
| Maize | ALS1 and ALS2 | Herbicide resistance | CRISPR/Cas9 | [270] |
| Maize | MS26 | Herbicide resistance | CRISPR/Cas9 | [270] |
| Soybean | ALS1 | Resistant to Chlorsulfuron | CRISPR/Cas9 | [277] |
| Tomato | ALS | Resistant to Chlorsulfuron | CRISPR/Cas9 | [278] |
| Tomato | SlEPSPS | Herbicide resistance | CRISPR/Cas9 | [92] |
| Tomato | SlALS1, SlALS2 | Herbicide resistance | CRISPR/Cas9 | [92] |
| Tomato | Slpds1 | Herbicide resistance | CRISPR/Cas9 | [92] |
| Rice | OsTubA2 | Base editing | CRISPR/Cas9 | [279] |
| Rice | OsHAK1 | Low cesium accumulation | CRISPR/Cas9 | [263] |
| Rice | OsPRX2 | Potassium deficiency tolerance | CRISPR/Cas9 | [280] |
| Rice | OsARM1 | Increase tolerance to higharsenic | CRISPR/Cas9 | [260] |
| Rice | OsLCT1 | Less cadmium accumulation | CRISPR/Cas9 | [259] |
4. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Mohidem, N.A.J.; Hashim, N.; Shamsudin, R.; Che Man, H. Rice for Food Security: Revisiting Its Production, Diversity, Rice Milling Process and Nutrient Content. Agriculture 2022, 12, 741. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- Mahood, E.H.; Kruse, L.H.; Moghe, G.D. Machine Learning: A Powerful Tool for Gene Function Prediction in Plants. Appl. Plant Sci. 2020, 8, e11376. [Google Scholar] [CrossRef]
- Parkhi, V.; Bhattacharya, A.; Choudhary, S.; Pathak, R.; Gawade, V.; Palan, B.; Alamalakala, L.; Mikkilineni, V.; Char, B. Demonstration of CRISPR-cas9-mediated Pds Gene Editing in a Tomato Hybrid Parental Line. Ind. J. Gen. Plnt. Breed. 2018, 78, 132–137. [Google Scholar] [CrossRef]
- Ni, Z.; Han, Q.; He, Y.-Q.; Huang, S. Application of Genome-Editing Technology in Crop Improvement. Cereal Chem. 2018, 95, 35–48. [Google Scholar] [CrossRef]
- Rothstein, R.J. One-Step Gene Disruption in Yeast. Methods Enzymol. 1983, 101, 202–211. [Google Scholar] [CrossRef]
- Sharma, P.; Tiwari, S.; Tripathi, N.; Mehta, A.K. Polymorphism Analysis in Advanced Mutant Population of Oat (Avena sativa L.) Using ISSR Markers. Physiol. Mol. Biol. Plants 2016, 22, 115–120. [Google Scholar] [CrossRef]
- Thomas, K.R.; Folger, K.R.; Capecchi, M.R. High Frequency Targeting of Genes to Specific Sites in the Mammalian Genome. Cell 1986, 44, 419–428. [Google Scholar] [CrossRef]
- Brookes, G. Genetically Modified (GM) Crop Use 1996–2020: Environmental Impacts Associated with Pesticide Use Change. GM Crops Food 2022, 13, 262–289. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Tripathi, N.; Tiwari, S.; Mishra, N.; Sharma, A.; Tiwari, S.; Singh, S. Identification of Indian Soybean (Glycine max [L.] Merr.) Genotypes for Drought Tolerance and Genetic Diversity Analysis Using SSR Markers. Scientist 2023, 3, 31–46. [Google Scholar]
- Tripathi, N.; Tripathi, M.K.; Tiwari, S.; Payasi, D.K. Molecular Breeding to Overcome Biotic Stresses in Soybean: Update. Plants 2022, 11, 1967. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Asati, R.; Patel, V.; Sikarwar, R.S.; Payasi, D.K. Breeding and Genomic Approaches Towards Development of Fusarium Wilt Resistance in Chickpea. Life 2023, 13, 988. [Google Scholar] [CrossRef] [PubMed]
- Asati, R.; Tripathi, M.K.; Tiwari, S.; Yadav, R.K.; Tripathi, N. Molecular Breeding and Drought Tolerance in Chickpea. Life 2022, 12, 1846. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Lang, Z.; Botella, J.R.; Zhu, J.-K. Genome Editing—Principles and Applications for Functional Genomics Research and Crop Improvement. Crit. Rev. Plant Sci. 2017, 36, 291–309. [Google Scholar] [CrossRef]
- Mishra, R.; Zhao, K. Genome Editing Technologies and Their Applications in Crop Improvement. Plant Biotechnol. Rep. 2018, 12, 57–68. [Google Scholar] [CrossRef]
- Imran, M.; Butt, M.; Hannan, A.; Manzoor, A.; Qaisar, U. Gene Editing: A Potential Tool to Enhance Field Crop Production. Int. J. Biotech Trends Technol. 2020, 10, 72–82. [Google Scholar] [CrossRef]
- Tan, Y.Y.; Du, H.; Wu, X.; Liu, Y.H.; Jiang, M.; Song, S.Y.; Wu, L.; Shu, Q.Y. Gene Editing: An Instrument for Practical Application of Gene Biology to Plant Breeding. J. Zhejiang Univ. Sci. B 2020, 21, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Abdurakhmonov, I.Y.; Ayubov, M.S.; Ubaydullaeva, K.A.; Buriev, Z.T.; Shermatov, S.E.; Ruziboev, H.S.; Shapulatov, U.M.; Saha, S.; Ulloa, M.; Yu, J.Z.; et al. RNA interference for Functional Genomics and Improvement of Cotton (Gossypium spp.). Front. Plant Sci. 2016, 7, 202. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Bae, H. Genome editing tools in plants. Genes 2017, 8, 399. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN and CRISPR/Cas-Based Methods for Genome Engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Chen, K.; Gao, C. Targeted Genome Modification Technologies and Their Applications in Crop Improvements. Plant Cell Rep. 2014, 33, 575–583. [Google Scholar] [CrossRef]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and Potential of Genome Editing in Crop Improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef]
- Rathore, M.S.; Tiwari, S.; Tripathi, N.; Tripathi, M.K.; Tiwari, S. Status and Scenario of Genome Editing Device CRISPR-Cas9 in Crop Advancement. Curr. Appl. Sci. Technol. 2021, 40, 8–20. [Google Scholar] [CrossRef]
- Tripathi, L.; Dhugga, K.S.; Ntui, V.O.; Runo, S.; Syombua, E.D.; Muiruri, S.; Wen, Z.; Tripathi, J.N. Genome Editing for Sustainable Agriculture in Africa. Front. Genome Ed. 2022, 4, 876697. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Buell, C.R.; Voytas, D.F.; Starker, C.G.; Douches, D.S. Genome Editing for Crop Improvement—Applications in Clonally Propagated Polyploids with a Focus on Potato (Solanum tuberosum L.). Front. Plant Sci. 2018, 9, 1607. [Google Scholar] [CrossRef] [PubMed]
- Pixley, K.V.; Falck-Zepeda, J.B.; Paarlberg, R.L.; Phillips, P.W.B.; Slamet-Loedin, I.H.; Dhugga, K.S.; Campos, H.; Gutterson, N. Genome-Edited Crops for Improved Food Security of Smallholder Farmers. Nat. Genet. 2022, 54, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, M.R. High Efficiency Transformation by Direct Microinjection of DNA into Cultured Mammalian Cells. Cell 1980, 22 Pt 2, 479–488. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Parkhi, V.; Char, B. Genome Editing for Crop Improvement: A Perspective from India. In Vitro Cell. Dev. Biol. Plant 2021, 57, 565–573. [Google Scholar] [CrossRef]
- Ferreira, S.S.; Reis, R.S. Using CRISPR/Cas to Enhance Gene Expression for Crop Trait Improvement by Editing miRNA Targets. J. Exp. Bot. 2023, 74, 2208–2212. [Google Scholar] [CrossRef]
- Jansing, J.; Schiermeyer, A.; Schillberg, S.; Fischer, R.; Bortesi, L. Genome Editing in Agriculture: Technical and Practical Considerations. Int. J. Mol. Sci. 2019, 20, 2888. [Google Scholar] [CrossRef]
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56. [Google Scholar] [CrossRef]
- Upadhyay, S.; Singh, A.K.; Tripathi, M.K.; Tiwari, S.; Tripathi, N. Biotechnological Interventions to Combat Against Charcoal Rot and Rhizoctonia Root Rot Diseases of Soybean [Glycine max (L.) Merrill]. Curr. Top. Agric. Sci. 2022, 6, 1–18. [Google Scholar] [CrossRef]
- Rajpoot, P.; Tripathi, M.K.; Tiwari, S.; Bimal, S.S.; Tripathi, N.; Parihar, P.; Pandya, R.K.; Satyavathi, C.T. Characterization of Pearl Millet [Pennisetum glaucum (L.) R Br.] Genotypes Against Blast Disease Employing Disease Scoring and Gene Specific SSR Markers. Scientist 2023, 3, 16–30. [Google Scholar]
- Mishra, N.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Gupta, N.; Sharma, A.; Solanki, R.S.; Tiwari, S. Characterization of Soybean Genotypes on the Basis of Yield Attributing Traits and SSR Molecular Markers. In Innovations in Science and Technology; B P International: Hong Kong, China, 2022; Volume 3, pp. 87–106. [Google Scholar] [CrossRef]
- Yadav, P.K.; Singh, A.K.; Tripathi, M.K.; Tiwari, S.; Yadav, S.K.; Tripathi, N. Morphophysiological and Molecular Characterization of Maize (Zea mays L.) Genotypes for Drought Tolerance. Eur. J. Appl. Sci. 2022, 10, 65–87. [Google Scholar]
- Hoang, T.M.L.; Tran, T.N.; Nguyen, T.K.T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of Salinity Stress Tolerance in Rice: Challenges and Opportunities. Agronomy 2016, 6, 54. [Google Scholar] [CrossRef]
- Zaidi, S.S.-e.-A.; Mahas, A.; Vanderschuren, H.; Mahfouz, M.M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020, 21, 289. [Google Scholar] [CrossRef] [PubMed]
- Schaart, J.G.; van de Wiel, C.C.M.; Lotz, L.A.P.; Smulders, M.J.M. Opportunities for Products of New Plant Breeding Techniques. Trends Plant Sci. 2016, 21, 438–449. [Google Scholar] [CrossRef]
- Hazman, M.Y. Are CRISPR/Cas genome editing techniques the future of plant breeding? Egypt. J. Agril. Res. 2022, 101, 1–13. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.; Kamoun, S. Targeted Mutagenesis in the Model Plant Nicotiana benthamiana Using Cas9 RNA-Guided Endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and Homologous Recombination-Mediated Genome Editing in Arabidopsis and Nicotiana benthamiana Using Guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Chilcoat, D.; Liu, Z.B.; Sander, J. Use of CRISPR/Cas9 for Crop Improvement in Maize and Soybean. Prog Mol Biol Transl Sci. 2017, 149, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Showalter, A.M. CRISPR/Cas9 genome editing technology: A valuable tool for understanding plant cell wall biosynthesis and function. Front. Plant Sci. 2020, 11, 589517. [Google Scholar] [CrossRef]
- Gao, C. Genome Engineering for Crop Improvement and Future Agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Carroll, D. Genome Engineering with Zinc-Finger Nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome Editing with Engineered Zinc Finger Nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Bibikova, M.; Carroll, D.; Segal, D.J.; Trautman, J.K.; Smith, J.; Kim, Y.G.; Chandrasegaran, S. Stimulation of Homologous Recombination Through Targeted Cleavage by Chimeric Nucleases. Mol. Cell. Biol. 2001, 21, 289–297. [Google Scholar] [CrossRef]
- Puchta, H. The Repair of Double-Strand Breaks in Plants: Mechanisms and Consequences for Genome Evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef]
- Lusser, M.; Parisi, C.; Plan, D.; Rodriguez-Cerezo, E. New Plant Breeding Techniques. In En State-of-the-Art and Prospects for Commercial Development 1-220; Joint Research Centre-Institute for Prospective Technological Studies, Publications Office of the European Union: Luxembourg, 2011; p. 24760. [Google Scholar]
- Araki, M.; Nojima, K.; Ishii, T. Caution Required for Handling Genome Editing Technology. Trends Biotechnol. 2014, 32, 234–237. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise Genome Modification in the Crop Species Zea mays Using Zinc-Finger Nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-Frequency Modification of Plant Genes Using Engineered Zinc-Finger Nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Voytas, D.F. Targeted Mutagenesis in Arabidopsis Using Zinc-Finger Nucleases. In Plant Chromosome Engineering; Humana Press: Totowa, NJ, USA, 2011; Volume 701, pp. 167–177. [Google Scholar] [CrossRef]
- Curtin, S.J.; Anderson, J.E.; Starker, C.G.; Baltes, N.J.; Mani, D.; Voytas, D.F.; Stupar, R.M. Targeted Mutagenesis for Function Analysis of Gene Duplication in Legumes. In Legume Genomics; Humana Press: Totowa, NJ, USA, 2013; pp. 25–42. [Google Scholar]
- Petolino, J.F. Genome Editing in Plants via Designed Zinc Finger Nucleases. In Vitro Cell. Dev. Biol. Plant 2015, 51, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M. The genome editing revolution: Review. J. Gernt. Engin. Biotechnol. 2020, 18, 68. [Google Scholar] [CrossRef]
- Arnould, S.; Delenda, C.; Grizot, S.; Desseaux, C.; Pâques, F.; Silva, G.H.; Smith, J. The I-CreI Meganuclease and Its Engineered Derivatives: Applications from Cell Modification to Gene Therapy. Protein Eng. Des. Sel. 2011, 24, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Poirot, L.; Galetto, R.; Smith, J.; Montoya, G.; Duchateau, P.; Pâques, F. Mega Nucleases and Other Tools for Targeted Genome Engineering: Perspectives and Challenges for Gene Therapy. Curr. Gene Ther. 2011, 11, 11–27. [Google Scholar] [CrossRef]
- Belfort, M.; Bonocora, R.P. Homing Endonucleases: From Genetic Anomalies to Programmable Genomic Clippers. Methods Mol. Biol. 2014, 1123, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Sultan, L.D.; Mileshina, D.; Grewe, F.; Rolle, K.; Abudraham, S.; Głodowicz, P.; Niazi, A.K.; Keren, I.; Shevtsov, S.; Klipcan, L.; et al. The Reverse Transcriptase/RNA Maturase Protein MatR Is Required for the Splicing of Various Group II Introns in Brassicaceae Mitochondria. Plant Cell. 2016, 28, 2805–2829. [Google Scholar] [CrossRef]
- Kaur, N.; Sharma, S.; Hasanuzzaman, M.; Pati, P.K. Genome Editing: A Promising Approach for Achieving Abiotic Stress Tolerance in Plants. Int. J. Genom. 2022, 2022, 5547231. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A Widely Applicable Technology for Targeted Genome Editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Martínez-Fortún, J.; Phillips, D.W.; Jones, H.D. Potential Impact of Genome Editing in World Agriculture. Emerg. Top. Life Sci. 2017, 1, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Malzahn, A.; Lowder, L.; Qi, Y. Plant Genome Editing with TALEN and CRISPR. Cell Biosci. 2017, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-Efficiency TALEN-Based Gene Editing Produces Disease-Resistant Rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Haun, W.; Coffman, A.; Clasen, B.M.; Demorest, Z.L.; Lowy, A.; Ray, E.; Retterath, A.; Stoddard, T.; Juillerat, A.; Cedrone, F.; et al. Improved Soybean Oil Quality by Targeted Mutagenesis of the Fatty Acid Desaturase 2 Gene Family. Plant Biotechnol. J. 2014, 12, 934–940. [Google Scholar] [CrossRef]
- Demorest, Z.L.; Coffman, A.; Baltes, N.J.; Stoddard, T.J.; Clasen, B.M.; Luo, S.; Retterath, A.; Yabandith, A.; Gamo, M.E.; Bissen, J.; et al. Direct Stacking of Sequence-Specific Nuclease-Induced Mutations to Produce High Oleic and Low Linolenic Soybean Oil. BMC Plant Biol. 2016, 16, 225. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and Function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Schornack, S.; Lahaye, T. TAL Effectors: Finding Plant Genes for Disease and Defense. Curr. Opin. Plant Biol. 2010, 13, 394–401. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zhou, J.; Li, D.; Wang, G.; Wang, F.; Kunjal, M.; Joldersma, D.; Liu, Z. Application and Future Perspective of CRISPR/Cas9 Genome Editing in Fruit Crops. J. Integr. Plant Biol. 2020, 62, 269–286. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.G. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.6.1–31.6.21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, Z.; Li, J.F. Targeted Gene Manipulation in Plants Using the CRISPR/Cas Technology. J. Genet. Genom. 2016, 43, 251–262. [Google Scholar] [CrossRef]
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR Systems in Plant Genome Editing: Toward New Opportunities in Agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR–Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Zhang, Y.; Yin, H. Genome Editing with mRNA Encoding ZFN, TALEN, and Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 System for Plant Genome Editing and Beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 Nuclease with Expanded Targeting Space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- Ming, M.; Ren, Q.; Pan, C.; He, Y.; Zhang, Y.; Liu, S.; Zhong, Z.; Wang, J.; Malzahn, A.A.; Wu, J.; et al. CRISPR-Cas12benables Efficient Plant Genome Engineering. Nat. Plants 2020, 6, 202–208. [Google Scholar] [CrossRef]
- Cong, L.; Zhang, F. Genome Engineering Using CRISPR-Cas9 System. Methods Mol. Biol. 2015, 1239, 197–217. [Google Scholar] [CrossRef]
- Tan, J.; Zhao, Y.; Wang, B.; Hao, Y.; Wang, Y.; Li, Y.; Luo, W.; Zong, W.; Li, G.; Chen, S.; et al. Efficient CRISPR/Cas9-Based Plant Genomic Fragment Deletions by Microhomology-Mediated End Joining. Plant Biotechnol. J. 2020, 18, 2161–2163. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR-Cpf1 System for Efficient Genome Editing and Transcriptional Repression in Plants. Nat. Plants 2017, 3, 17103. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, Z.; Gosavi, G.; Ren, B.; Cao, Y.; Kuang, Y.; Zhou, C.; Spetz, C.; Yan, F.; Zhou, X.; et al. Targeted Base Editing in Rice with CRISPR/ScCas9 System. Plant Biotechnol. J. 2020, 18, 1645–1647. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, M.F.; Karlson, C.K.S.; Teoh, E.Y.; Lau, S.E.; Tan, B.C. Genome Editing for Sustainable Crop Improvement and Mitigation of Biotic and Abiotic Stresses. Plants 2022, 11, 2625. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome Editing in Rice and Wheat Using the CRISPR/Cas System. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 Genome Editing in Wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Guha, T.K.; Edgell, D.R. Applications of Alternative Nucleases in the Age of CRISPR/Cas9. Int. J. Mol. Sci. 2017, 18, 2565. [Google Scholar] [CrossRef]
- Sauer, N.J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Walker, K.A.; Beetham, P.R.; Schöpke, C.R.; Gocal, G.F. Oligonucleotide Directed Mutagenesis for Precision Gene Editing. Plant Biotechnol. J. 2016, 14, 496–502. [Google Scholar] [CrossRef]
- Yang, S.H.; Kim, E.; Park, H.; Koo, Y. Selection of the High Efficient sgRNA for CRISPR-Cas9 to Edit Herbicide Related Genes, PDS, ALS, and EPSPS in Tomato. Appl. Biol. Chem. 2022, 65, 13. [Google Scholar] [CrossRef]
- Janik, E.; Niemcewicz, M.; Ceremuga, M.; Krzowski, L.; Saluk-Bijak, J.; Bijak, M. Various Aspects of a Gene Editing System crispr–cas 9. Int. J. Mol. Sci. 2020, 21, 9604. [Google Scholar] [CrossRef]
- Eş, I.; Gavahian, M.; Marti-Quijal, F.J.; Lorenzo, J.M.; Mousavi Khaneghah, A.M.; Tsatsanis, C.; Kampranis, S.C.; Barba, F.J. The Application of the CRISPR-Cas9 Genome Editing Machinery in Food and Agricultural Science: Current Status, Future Perspectives, and Associated Challenges. Biotechnol. Adv. 2019, 37, 410–421. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; He, S. CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing. Front. Plant Sci. 2016, 7, 1740. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.K. Application of the CRISPR-Cas System for Efficient Genome Engineering in Plants. Mol. Plant 2013, 6, 2008–2011. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Mahfouz, M.M. Genome Editing: The Road of CRISPR/Cas9 from Bench to Clinic. Exp. Mol. Med. 2016, 48, e265. [Google Scholar] [CrossRef]
- Weeks, D.P.; Spalding, M.H.; Yang, B. Use of Designer Nucleases for Targeted Gene and Genome Editing in Plants. Plant Biotechnol. J. 2016, 14, 483–495. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted Genome Engineering in Human Cells with the Cas9 RNA-Guided Endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Prajapat, R.K.; Mathur, M.; Upadhyay, T.K.; Lal, D.; Maloo, S.; Sharma, D. Genome Editing for Crop Improvement. In Crop Improvement; CRC Press: Boca Raton, FL, USA, 2021; pp. 111–123. [Google Scholar]
- Khandagale, K.; Nadaf, A. Genome Editing for Targeted Improvement of Plants. Plant Biotechnol. Rep. 2016, 10, 327–343. [Google Scholar] [CrossRef]
- Cantos, C.; Francisco, P.; Trijatmiko, K.R.; Slamet-Loedin, I.; Chadha-Mohanty, P.K. Identification of “Safe Harbor” Loci in Indica Rice Genome by Harnessing the Property of Zinc-Finger Nucleases to Induce DNA Damage and Repair. Front. Plant Sci. 2014, 5, 302. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Chen, K.; Liang, Z.; Li, J.; Zhang, Y.; Zhang, K.; Liu, J.; Voytas, D.F.; Zheng, X.; et al. Rapid and Efficient Gene Modification in Rice and Brachypodium Using TALENs. Mol. Plant. 2013, 6, 1365–1368. [Google Scholar] [CrossRef]
- Zhang, H.; Gou, F.; Zhang, J.; Liu, W.; Li, Q.; Mao, Y.; Botella, J.R.; Zhu, J.K. TALEN-Mediated Targeted Mutagenesis Produces a Large Variety of Heritable Mutations in Rice. Plant Biotechnol. J. 2016, 14, 186–194. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Schornack, S.; Meyer, A.; Römer, P.; Jordan, T.; Lahaye, T. Gene-for-Gene-Mediated Recognition of Nuclear-Targeted AvrBs3-Like Bacterial Effector Proteins. J. Plant Physiol. 2006, 163, 256–272. [Google Scholar] [CrossRef]
- Ainley, W.M.; Sastry-Dent, L.; Welter, M.E.; Murray, M.G.; Zeitler, B.; Amora, R.; Corbin, D.R.; Miles, R.R.; Arnold, N.L.; Strange, T.L.; et al. Trait Stacking via Targeted Genome Editing. Plant Biotechnol. J. 2013, 11, 1126–1134. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D.; et al. Male-Sterile Maize Plants Produced by Targeted Mutagenesis of the Cytochrome P450-Like Gene (MS26) Using a Re-designed I-CreI Homing Endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Wendt, T.; Holm, P.B.; Starker, C.G.; Christian, M.; Voytas, D.F.; Brinch-Pedersen, H.; Holme, I.B. TAL Effector Nucleases Induce Mutations at a Pre-selected Location in the Genome of Primary Barley Transformants. Plant Mol. Biol. 2013, 83, 279–285. [Google Scholar] [CrossRef]
- Curtin, S.J.; Zhang, F.; Sander, J.D.; Haun, W.J.; Starker, C.; Baltes, N.J.; Reyon, D.; Dahlborg, E.J.; Goodwin, M.J.; Coffman, A.P.; et al. Targeted Mutagenesis of Duplicated Genes in Soybean with Zinc-Finger Nucleases. Plant Physiol. 2011, 156, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.A.; Townsend, J.A.; Winfrey, R.J., Jr.; Irwin, P.A.; Rajagopal, J.; Lonosky, P.M.; Hall, B.D.; Jondle, M.D.; Voytas, D.F. High-Frequency Homologous Recombination in Plants Mediated by Zinc-Finger Nucleases. Plant J. 2005, 44, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Novak, S. Plant Biotechnology Applications of Zinc Finger Technology. In Transgenic Plants; Humana Press: Totowa, NJ, USA, 2019; pp. 295–310. [Google Scholar] [CrossRef]
- Gago, C.; Drosou, V.; Paschalidis, K.; Guerreiro, A.; Miguel, G.; Antunes, D.; Hilioti, Z. Targeted Gene Disruption Coupled with Metabolic Screen Approach to Uncover the LEAFY COTYLEDON1-LIKE4 (L1L4) Function in Tomato Fruit Metabolism. Plant Cell Rep. 2017, 36, 1065–1082. [Google Scholar] [CrossRef]
- D’Halluin, K.; Vanderstraeten, C.; Van Hulle, J.; Rosolowska, J.; Van Den Brande, I.; Pennewaert, A.; D’Hont, K.; Bossut, M.; Jantz, D.; Ruiter, R.; et al. Targeted Molecular Trait Stacking in Cotton through Targeted Double-Strand Break Induction. Plant Biotechnol. J. 2013, 11, 933–941. [Google Scholar] [CrossRef]
- Clasen, B.M.; Stoddard, T.J.; Luo, S.; Demorest, Z.L.; Li, J.; Cedrone, F.; Tibebu, R.; Davison, S.; Ray, E.E.; Daulhac, A.; et al. Improving Cold Storage and Processing Traits in Potato Through Targeted Gene Knockout. Plant Biotechnol. J. 2016, 14, 169–176. [Google Scholar] [CrossRef]
- Geurts, M.H.; Clevers, H. CRISPR Engineering in Organoids for Gene Repair and Disease Modelling. Nat. Rev. Bioeng. 2023, 1, 32–45. [Google Scholar] [CrossRef]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved Base Excision Repair Inhibition and Bacteriophage Mu Gam Protein Yields C:G-to-T:A Base Editors with Higher Eficiency and Product Purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef] [PubMed]
- Cascalho, M. Advantages and disadvantages of cytidine deamination. J. Immunol. 2004, 172, 6513–6518. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.; Yeh, W.H.; Pendse, N.; Davis, J.R.; Hennessey, E.; Butcher, R.; Koblan, L.W.; Comander, J.; Liu, Q.; Liu, D.R. Cytosine and Adenine Base Editing of the Brain, Liver, Retina, Heart and Skeletal Muscle of Mice via Adeno-Associated Viruses. Nat. Biomed. Eng. 2020, 4, 97–110. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2022, 40, 731–740. [Google Scholar] [CrossRef]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs Improve Prime Editing Eficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M.; et al. Enhanced Prime Editing Systems by Manipulating Cellular Determinants of Editing Outcomes. Cell 2021, 184, 5635–5652. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; Hou, L.; Wu, X.; Wang, D.; Zhang, L.; Liu, P. Exogenous SA Affects Rice Seed Germination Under Salt Stress by Regulating Na(+)/K(+) Balance and Endogenous GAs and ABA Homeostasis. Int. J. Mol. Sci. 2022, 23, 3293. [Google Scholar] [CrossRef]
- Ningwal, R.; Tripathi, M.K.; Tiwari, S.; Yadav, R.K.; Tripathi, N.; Solanki, R.S.; Asati, R.; Yasin, M. Assessment of Genetic Variability, Correlation and Path Coefficient Analysis for Yield and Its Attributing Traits in Chickpea (Cicer arietinum L.) The Pharma. Innov. J. 2023, 12, 4851–4859. [Google Scholar]
- Sharma, S.; Tripathi, M.K.; Tiwari, S.; Solanki, R.S.; Chauhan, S.; Tripathi, N.; Dwivedi, N.; Kandalkar, V.S. The Exploitation of Genetic Variability and Trait Association Analysis for Diverse Quantitative Traits in Bread Wheat (Triticum aestivum L.). Curr. J. Appl. Sci. Technol. 2023, 42, 19–33. [Google Scholar] [CrossRef]
- Shyam, C.; Tripathi, M.K.; Tripathi, N.; Tiwari, S.; Sikarwar, R.S. Identification of Low and High Erucic Acid Containing Genotype(S) in Indian Mustard Employing Molecular Markers. In Recent Progress in Plant and Soil Research; B P International: Hong Kong, China, 2022; Volume 5, pp. 18–36. [Google Scholar] [CrossRef]
- Yadav, S.; Tiwari, S.; Tripathi, M.K.; Tripathi, N.; Gupta, N.; Tiwari, S. Evaluation of High Oleic Acid Content in a Set of 96 Genotypes of Arachis hypogaea L. Scientist 2023, 2, 132–143. [Google Scholar]
- Sandhu, K.S.; Mihalyov, P.D.; Lewien, M.J.; Pumphrey, M.O.; Carter, A.H. Genomic Selection and Genome-Wide Association Studies for Grain Protein Content Stability in a Nested Association Mapping Population of Wheat. Agronomy 2021, 11, 2528. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Patil, S.S.; Aoun, M.; Carter, A.H. Multi-trait Multi-environment Genomic Prediction for End-Use Quality Traits in Winter Wheat. Front. Genet. 2022, 13, 831020. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. Ossapk2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic Stresses: General Defenses of Land Plants and Chances for Engineering Multistress Tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef]
- Joshi, R.K.; Bharat, S.S.; Mishra, R. Engineering Drought Tolerance in Plants Through CRISPR/Cas Genome Editing. 3 Biotech 2020, 10, 400. [Google Scholar] [CrossRef]
- Kumar, S. Abiotic Stresses and Their Effects on Plant Growth, Yield and Nutritional Quality of Agricultural Produce. Int. J. Food Sci. Agric. 2020, 4, 367–378. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global Synthesis of Drought Effects on Maize and Wheat Production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.-A. Global Synthesis of Drought Effects on Cereal, Legume, Tuber and Root Crops Production: A Review. Agric. Water Manag. 2017, 179, 18–33. [Google Scholar] [CrossRef]
- Mafakheri, A.; Siosemardeh, A.; Bahramnejad, B.; Struik, P.C.; Sohrabi, Y. Effect of Drought Stress on Yield, Proline and Chlorophyll Contents in Three Chickpea Cultivars. Aust. J. Crop Sci. 2010, 4, 580–585. [Google Scholar]
- Farooq, M.; Gogoi, N.; Barthakur, S.; Baroowa, B.; Bharadwaj, N.; Alghamdi, S.S.; Siddique, K.H.M. Drought Stress in Grain Legumes During Reproduction and Grain Filling. J. Agron. Crop Sci. 2017, 203, 81–102. [Google Scholar] [CrossRef]
- Maleki, A.; Naderi, A.; Naseri, R.; Fathi, A.; Bahamin, S.; Maleki, R. Physiological Performance of Soybean Cultivars Under Drought Stress. Bull. Environ. Pharmacol. Life Sci. 2013, 2, 38–44. [Google Scholar]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the Entire Family of AITR Genes in Arabidopsis Leads to Enhanced Drought and Salinity Tolerance Without Fitness Costs. BMC Plant Biol. 2021, 21, 137. [Google Scholar] [CrossRef]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A Novel Family of Transcription Factors Conserved in Angiosperms Is Required for ABA Signalling. Plant Cell Environ. 2017, 40, 2958–2971. [Google Scholar] [CrossRef]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 Genome Editing to Modify Abiotic Stress Responses in Plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Liu, W.; Gao, W.; Liu, C.; Song, G.; Li, W.X.; Mao, L.; Chen, B.; Xu, Y.; et al. An Alternative Strategy for Targeted Gene Replacement in Plants Using a Dual-sgRNA/Cas9 Design. Sci. Rep. 2016, 6, 23890. [Google Scholar] [CrossRef]
- Park, J.J.; Dempewolf, E.; Zhang, W.; Wang, Z.Y. RNA-Guided Transcriptional Activation via CRISPR/dCas9 Mimics Overexpression Phenotypes in Arabidopsis. PLoS ONE 2017, 12, e0179410. [Google Scholar] [CrossRef]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.d.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved Drought Stress Tolerance in Arabidopsis by CRISPR/dCas9 Fusion with a Histone Acetyltransferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef]
- Nuñez-Muñoz, L.; Vargas-Hernández, B.; Hinojosa-Moya, J.; Ruiz-Medrano, R.; Xoconostle-Cázares, B. Plant Drought Tolerance Provided Through Genome Editing of the Trehalose Gene. Plant Signal. Behav. 2021, 16, 1877005. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 System Produces Specific and Homozygous Targeted Gene Editing in Rice in One Generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-Targeted Mutagenesis of OsERA1 Confers Enhanced Responses to Abscisic Acid and Drought Stress and Increased Primary Root Growth Under Nonstressed Conditions in Rice. PLoS ONE 2020, 15, e0243376. [Google Scholar] [CrossRef]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-rolled leaf1, 2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Liu, Y.; Li, R. Precise Editing of the OsPYL9 Gene by RNA-Guided Cas9 Nuclease Confers Enhanced Drought Tolerance and Grain Yield in Rice (Oryza sativa L.) by Regulating Circadian Rhythm and Abiotic Stress Responsive Proteins. Int. J. Mol. Sci. 2020, 21, 7854. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 Mediated Genome Editing of Drought and Salt Tolerance (OsDST) Gene in Indica Mega Rice Cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Badhan, S.; Ball, A.S.; Mantri, N. First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Akhter, M.; Ahmad, R.M.; Cheema, K.L.; Hina, A.; Karikari, B.; Raza, G.; Xing, G.; Gai, J.; Khurshid, M. CRISPR-Cas9 Based Stress Tolerance: New Hope for Abiotic Stress Tolerance in Chickpea (Cicer arietinum). Mol. Biol. Rep. 2022, 49, 8977–8985. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Wei, J.; Winkler, C.; Goncalves-Butruille, M.; Weers, B.P.; Cerwick, S.F.; Dieter, J.A.; Duncan, K.E.; Howard, R.J.; et al. Maize ARGOS1 (ZAR1) Transgenic Alleles Increase Hybrid Maize Yield. J. Exp. Bot. 2014, 65, 249–260. [Google Scholar] [CrossRef]
- Shi, J.; Habben, J.E.; Archibald, R.L.; Drummond, B.J.; Chamberlin, M.A.; Williams, R.W.; Lafitte, H.R.; Weers, B.P. Overexpression of ARGOS Genes Modifies Plant Sensitivity to Ethylene, Leading to Improved Drought Tolerance in Both Arabidopsis and Maize. Plant Physiol. 2015, 169, 266–282. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield Under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Illouz-Eliaz, N.; Nissan, I.; Nir, I.; Ramon, U.; Shohat, H.; Weiss, D. Mutations in the Tomato Gibberellin Receptors Suppress Xylem Proliferation and Reduce Water Loss Under Water-Deficit Conditions. J. Exp. Bot. 2020, 71, 3603–3612. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhang, N. CRISPR/Cas9 Targeted Mutagenesis of SlLBD40, a Lateral Organ Boundaries Domain Transcription Factor, Enhances Drought Tolerance in Tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced Drought Tolerance by CRISPR/Cas9-Mediated SlMAPK3 Mutagenesis in Tomato Plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SlNPR1 Mutagenesis Reduces Tomato Plant Drought Tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar] [CrossRef]
- Abdallah, N.A.; Elsharawy, H.; Abulela, H.A.; Thilmony, R.; Abdelhadi, A.A.; Elarabi, N.I. Multiplex CRISPR/Cas9-Mediated Genome Editing to Address Drought Tolerance in Wheat. GM Crops Food 2022, 6, 1–17. [Google Scholar] [CrossRef]
- He, X.; Luo, X.; Wang, T.; Liu, S.; Zhang, X.; Zhu, L. GhHB12 Negatively Regulates Abiotic Stress Tolerance in Arabidopsis and Cotton. Environ. Exp. Bot. 2020, 176, 104087. [Google Scholar] [CrossRef]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B. Roles of the Brassica napus Della Protein BnaA6. RGA, in modulating drought tolerance by interacting with the ABA signalling component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising Temperatures Reduce Global Wheat Production. Nat. Clim. Chang. 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Yang, H.; Huang, T.; Ding, M.; Lu, D.; Lu, W. High Temperature During Grain Filling Impacts on Leaf Senescence in Waxy Maize. Agron. J. 2017, 109, 906–916. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Zhou, J.; Hu, S.; Chen, H.; Xiang, J.; Zhang, Y.; Zeng, Y.; Shi, Q.; Zhu, D.; et al. Research Progress on Heat Stress of Rice at Flowering Stage. Rice Sci. 2019, 26, 1–10. [Google Scholar] [CrossRef]
- Ali, M.G.M.; Ahmed, M.; Ibrahim, M.M.; El Baroudy, A.A.; Ali, E.F.; Shokr, M.S.; Aldosari, A.A.; Majrashi, A.; Kheir, A.M.S. Optimizing Sowing Window, Cultivar Choice, and Plant Density to Boost Maize Yield Under RCP8. 5 Climate Scenario of CMIP5. Int. J. Biometeorol. 2022, 66, 971–985. [Google Scholar] [CrossRef]
- Ejaz, M.; Abbas, G.; Fatima, Z.; Iqbal, P.; Raza, M.A.; Kheir, A.M.S.; Ahmed, M.; Kakar, K.M.; Ahmad, S. Modelling Climate Uncertainty and Adaptations for Soybean-Based Cropping System. Int. J. Plant Prod. 2022, 16, 235–250. [Google Scholar] [CrossRef]
- Haider, S.; Raza, A.; Iqbal, J.; Shaukat, M.; Mahmood, T. Analyzing the Regulatory Role of Heat Shock Transcription Factors in Plant Heat Stress Tolerance: A Brief Appraisal. Mol. Biol. Rep. 2022, 49, 5771–5785. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Mathivanan, S. Abiotic Stress-Induced Molecular and Physiological Changes and Adaptive Mechanisms in Plants. In Abiotic Stress in Plants; Fahad, S., Saud, S., Chen, Y., Wu, C., Wang, D., Eds.; Intech Open: Rijeka, Croatia, 2020. [Google Scholar] [CrossRef]
- Boyer, J.S.; Westgate, M.E. Grain Yields with Limited Water. J. Exp. Bot. 2004, 55, 2385–2394. [Google Scholar] [CrossRef]
- Hazel, J.R. Thermal Adaptation in Biological Membranes: Is Homeoviscous Adaptation the Explanation? Annu. Rev. Physiol. 1995, 57, 19–42. [Google Scholar] [CrossRef]
- Awasthi, R.; Bhandari, K.; Nayyar, H. Temperature Stress and Redox Homeostasis in Agricultural Crops. Front. Environ. Sci. 2015, 3, 11. [Google Scholar] [CrossRef]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic Engineering Strategies for Biotic and Abiotic Stress Tolerance and Quality Enhancement in Horticultural Crops: A Comprehensive Review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a Regulatory Element Responsible for Salt Induction of Rice OsRAV2 Through Ex Situ and In Situ Promoter Analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Nandy, S.; Pathak, B.; Zhao, S.; Srivastava, V. Heat-Shock-Inducible CRISPR/Cas9 System Generates Heritable Mutations in Rice. Plant Direct 2019, 3, e00145. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down Regulation and Loss of Auxin Response Factor 4 Function Using CRISPR/Cas9 Alters Plant Growth, Stomatal Function and Improves Tomato Tolerance to Salinity and Osmotic Stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance Through FERONIA Receptor-Like Kinase-Mediated Reactive Oxygen Species Signalling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Qiu, Z.; Kang, S.; He, L.; Zhao, J.; Zhang, S.; Hu, J.; Zeng, D.; Zhang, G.; Dong, G.; Gao, Z.; et al. The Newly Identified Heat-Stress Sensitive Albino 1 Gene Affects Chloroplast Development in Rice. Plant Sci. 2018, 267, 168–179. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Si, X.; Tian, Y.; Chen, K.; Liu, J.; Chen, H.; Gao, C. Generation of Thermosensitive Male-Sterile Maize by Targeted Knockout of the ZmTMS5 Gene. J. Genet. Genom. 2017, 44, 465–468. [Google Scholar] [CrossRef]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce (Lactuca sativa). G3 Genes Genomes Genet. 2018, 8, 1513–1521. [Google Scholar] [CrossRef]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene Response Factor (ERF) Family Proteins in Abiotic Stresses and CRISPR–Cas9 Genome Editing of ERFs for Multiple Abiotic Stress Tolerance in Crop Plants: A Review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Chang, Y.; Nguyen, B.H.; Xie, Y.; Xiao, B.; Tang, N.; Zhu, W.; Mou, T.; Xiong, L. Co-overexpression of the Constitutively Active Form of OsbZIP46 and ABA-Activated Protein Kinase SAPK6 Improves Drought and Temperature Stress Resistance in Rice. Front. Plant Sci. 2017, 8, 1102. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K. Cold Stress Tolerance Mechanisms in Plants. A Review. Agron. Sustain. Dev. 2010, 30, 515–527. [Google Scholar] [CrossRef]
- Muller, O.; Stewart, J.J.; Cohu, C.M.; Polutchko, S.K.; Demmig-Adams, B.; Adams, W.W., III. Leaf Architectural, Vascular and Photosynthetic Acclimation to Temperature in Two Biennials. Physiol. Plant. 2014, 152, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Adams, W.W., III; Stewart, J.J.; Cohu, C.M.; Muller, O.; Demmig-Adams, B. Habitat Temperature and Precipitation of Arabidopsis thaliana Ecotypes Determine the Response of Foliar Vasculature, Photosynthesis, and Transpiration to Growth Temperature. Front. Plant Sci. 2016, 7, 1026. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Xiang, C.; Farooq, M.; Muhammad, N.; Yan, Z.; Hui, X.; Yuanyuan, K.; Bruno, A.K.; Lele, Z.; Jincai, L. Cold Stress in Wheat: Plant Acclimation Responses and Management Strategies. Front. Plant Sci. 2021, 12, 676884. [Google Scholar] [CrossRef] [PubMed]
- Eom, S.H.; Ahn, M.A.; Kim, E.; Lee, H.J.; Lee, J.H.; Wi, S.H.; Kim, S.K.; Lim, H.B.; Hyun, T.K. Plant Response to Cold Stress: Cold Stress Changes Antioxidant Metabolism in Heading Type Kimchi Cabbage (Brassica rapa L. ssp. pekinensis). Antioxidants 2022, 11, 700. [Google Scholar] [CrossRef]
- Karavolias, N.G.; Horner, W.; Abugu, M.N.; Evanega, S.N. Application of Gene Editing for Climate Change in Agriculture. Front. Sustain. Food Syst. 2021, 5, 685801. [Google Scholar] [CrossRef]
- Ahmad, M. Plant Breeding Advancements with “CRISPR-Cas” Genome Editing Technologies Will Assist Future Food Security. Front. Plant Sci. 2023, 14, 1133036. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L. The OsMYb30 Transcription Factor Suppresses Cold Tolerance by Interacting with a JAZ Protein and Suppressing β-Amylase Expression1. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a Subfamily of Abscisic Acid Receptor Genes Promote Rice Growth and Productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Molla, K.A.; Shih, J.; Yang, Y. Single-Nucleotide Editing for zebra3 and wsl5 Phenotypes in Rice Using CRISPR/Cas9-Mediated Adenine Base Editors. aBIOTECH 2020, 1, 106–118. [Google Scholar] [CrossRef]
- Liu, X.; Lan, J.; Huang, Y.; Cao, P.; Zhou, C.; Ren, Y.; He, N.; Liu, S.; Tian, Y.; Nguyen, T.; et al. WSL5, a Pentatricopeptide Repeat Protein, Is Essential for Chloroplast Biogenesis in Rice Under Cold Stress. J. Exp. Bot. 2018, 69, 3949–3961. [Google Scholar] [CrossRef]
- Shakiba, E.; Edwards, J.D.; Jodari, F.; Duke, S.E.; Baldo, A.M.; Korniliev, P.; McCouch, S.R.; Eizenga, G.C. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 2017, 12, e0172133. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 System. Front. Plant Sci. 2019, 10, 1663. [Google Scholar] [CrossRef]
- Cui, Y.; Jiang, N.; Xu, Z.; Xu, Q. Heterotrimeric G Protein Are Involved in the Regulation of Multiple Agronomic Traits and Stress Tolerance in Rice. BMC Plant Biol. 2020, 20, 90. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of Tomato-Plant Chilling Tolerance by CRISPR–Cas9- Mediated SlCBF1 Mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, C.; Wang, G.; Mao, G.; Habben, J.E.; Chen, G.; Liu, M.; Shi, Y.; Wang, W.; Wang, X.; et al. Knockouts of Drought Sensitive Genes Improve Rice Grain Yield Under Both Drought and Well-Watered Field Conditions. Adv. Crop Sci. Technol. 2020, 8, 444. [Google Scholar]
- Wang, B.; Zhong, Z.; Wang, X.; Han, X.; Yu, D.; Wang, C.; Song, W.; Zheng, X.; Chen, C.; Zhang, Y. Knockout of the OsNAC006 Transcription Factor Causes Drought and Heat Sensitivity in Rice. Int. J. Mol. Sci. 2020, 21, 2288. [Google Scholar] [CrossRef]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, D.; Chen, M.; Liang, W.; Wei, J.; Qi, Y.; Yuan, Z. Development of Japonica Photo- Sensitive Genic Male Sterile Rice Lines by Editing Carbon Starved Anther Using CRISPR/Cas9. J. Genet. Genom. 2016, 43, 415–419. [Google Scholar] [CrossRef]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B.; et al. Development of Commercial Thermo-Sensitive Genic Male Sterile Rice Accelerates Hybrid Rice Breeding Using the CRISPR/Cas9-Mediated TMS5 Editing System. Sci. Rep. 2016, 6, 37395. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 Improves Drought Tolerance in Rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, Y.; Huang, L.; Du, F.; Zhao, X.; Li, Z.; Wang, W.; Fu, B. A U-Box E3 Ubiquitin Ligase OsPUB67 Is Positively Involved in Drought Tolerance in Rice. Plant Mol. Biol. 2020, 102, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. Crispr/cas9 and talens generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and Characterization of GmMYB118 Responses to Drought and Salt Stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef] [PubMed]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato Facultative Parthenocarpy Results from SlAGAMOUS-LIKE 6 Loss of Function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock Out of the Annexin Gene OsAnn3 via CRISPR/Cas9-Mediated Genome Editing Decreased Cold Tolerance in Rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Nawaz, G.; Han, Y.; Usman, B.; Liu, F.; Qin, B.; Li, R. Knockout of OsPRP1, a Gene Encoding Proline-Rich Protein, Confers Enhanced Cold Sensitivity in Rice (Oryza sativa L.) at the Seedling Stage. 3 Biotech 2019, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Su, C.T.; Lin, C.H.; Huang, G.J.; Lin, Y.H. Expression of Sweet Potato Cysteine Protease SPCP2 Altered Developmental Characteristics and Stress Responses in Transgenic Arabidopsis Plants. J. Plant Physiol. 2010, 167, 838–847. [Google Scholar] [CrossRef]
- Shahid, S.A.; Zaman, M.; Heng, L. Introduction to Soil Salinity, Sodicity and Diagnostics Techniques. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–42. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F. Plant Abiotic Stress Response and Nutrient Use Efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt Stress Under the Scalpel–Dissecting the Genetics of Salt Tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene Expression Profiling of Plants Under Salt Stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil Salinity: A Serious Environmental Issue and Plant Growth Promoting Bacteria as One of the Tools for Its Alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of Salt Stress on Ion Concentration, Proline Content, Antioxidant Enzyme Activities and Gene Expression in Tomato Cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef] [PubMed]
- Rahneshan, Z.; Nasibi, F.; Moghadam, A.A. Effects of Salinity Stress on Some Growth, Physiological, Biochemical Parameters and Nutrients in Two Pistachio (Pistacia vera L.) Rootstocks. J. Plant Interact. 2018, 13, 73–82. [Google Scholar] [CrossRef]
- Ali, Q.; Daud, M.K.; Haider, M.Z.; Ali, S.; Rizwan, M.; Aslam, N.; Noman, A.; Iqbal, N.; Shahzad, F.; Deeba, F.; et al. Seed Priming by Sodium Nitroprusside Improves Salt Tolerance in Wheat (Triticum aestivum L.) by Enhancing Physiological and Biochemical Parameters. Plant Physiol. Biochem. 2017, 119, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Ekinci, M.; Ors, S.; Turan, M.; Yildiz, S.; Yildirim, E. Effects of Individual and Combined Effects of Salinity and Drought on Physiological, Nutritional and Biochemical Properties of Cabbage (Brassica oleracea var. capitata). Sci. Hortic. 2018, 240, 196–204. [Google Scholar] [CrossRef]
- Ghazali, E.G.E. SuaedavermiculataForssk. Ex JF Gmel: Structural Characteristics and Adaptations to Salinity and Drought: A Review. Int. J. Sci. 2020, 9, 28–33. [Google Scholar]
- Pan, T.; Liu, M.; Kreslavski, V.D.; Zharmukhamedov, S.K.; Nie, C.; Yu, M.; Kuznetsov, V.V.; Allakhverdiev, S.I.; Shabala, S. Non-stomatal Limitation of Photosynthesis by Soil Salinity. Crit. Rev. Environ. Sci. Technol. 2021, 51, 791–825. [Google Scholar] [CrossRef]
- Navada, S.; Vadstein, O.; Gaumet, F.; Tveten, A.K.; Spanu, C.; Mikkelsen, Ø.; Kolarevic, J. Biofilms Remember: Osmotic Stress Priming as a Microbial Management Strategy for Improving Salinity Acclimation in Nitrifying Biofilms. Water Res. 2020, 176, 115732. [Google Scholar] [CrossRef]
- Atieno, J.; Li, Y.; Langridge, P.; Dowling, K.; Brien, C.; Berger, B.; Varshney, R.K.; Sutton, T. Exploring Genetic Variation for Salinity Tolerance in Chickpea Using Image-Based Phenotyping. Sci. Rep. 2017, 7, 1300. [Google Scholar] [CrossRef]
- Huang, Y.; Guan, C.; Liu, Y.; Chen, B.; Yuan, S.; Cui, X.; Zhang, Y.; Yang, F. Enhanced Growth Performance and Salinity Tolerance in Transgenic Switchgrass via Overexpressing Vacuolar Na+ (K+)/H+ Antiporter Gene (PvNHX1). Front. Plant Sci. 2017, 8, 458. [Google Scholar] [CrossRef]
- Zhang, Q.; Dai, W. Plant Response to Salinity Stress. In Stress Physiology of Woody Plants; Dai, W., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 155–173. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap Accelerates Breeding of a Salt-Tolerant Rice Cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced Rice Salinity Tolerance via CRISPR/Cas9-Targeted Mutagenesis of the OsRR22 Gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [PubMed]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of Rice Paraquat TOLERANCE 3 Confers Enhanced Resistance to Abiotic Stresses and Increases Grain Yield in Field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.; Cao, H.; Liu, B. OsmiR535, a Potential Genetic Editing Target for Drought and Salinity Stress Tolerance in Oryza sativa. Plants 2020, 9, 1337. [Google Scholar] [CrossRef]
- Alam, M.S.; Kong, J.; Tao, R.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.H. CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Negm, M.; Wu, D.; Li, J. The Trihelix Transcription Factor OsGTc-2 Is Involved Adaption to Salt Stress in Rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Chen, Z.; Li, P.; Xu, H.; Liu, K.; Zha, W.; Li, S.; Chen, J.; Yang, G.; Huang, J.; et al. Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability 2022, 14, 2621. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Wang, W.C.; Lin, T.C.; Kieber, J.; Tsai, Y.C. Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef]
- Bo, W.; Zhaohui, Z.; Huanhuan, Z.; Xia, W.; Binglin, L.; Lijia, Y.; Xiangyan, H.; Deshui, Y.; Xuelian, Z.; Chunguo, W.; et al. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Sci. 2019, 26, 98–108. [Google Scholar] [CrossRef]
- Zhang, C.; Srivastava, A.K.; Sadanandom, A. Targeted Mutagenesis of the SUMO Protease, Overly Tolerant to Salt1 in Rice Through CRISPR/Cas9-Mediated Genome Editing Reveals a Major Role of This SUMO Protease In Salt Tolerance. bioRxiv 2019. bioRxiv:555706. [Google Scholar]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 Genes|Genomes|Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Lin, J.; Liu, X.; Chu, W.; Li, J.; Gao, Y.; An, K.; Song, W.; Xin, M.; Yao, Y.; et al. Histone Acetyltransferase TaHAG1 Acts as a Crucial Regulator to Strengthen Salt Tolerance of Hexaploid Wheat. Plant Physiol. 2021, 186, 1951–1969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xun, H.; Wang, W.; Ding, X.; Tian, H.; Hussain, S.; Dong, Q.; Li, Y.; Cheng, Y.; Wang, C.; et al. Mutation of GmAITR Genes by CRISPR/Cas9 Genome Editing Results in Enhanced Salinity Stress Tolerance in Soybean. Front. Plant Sci. 2021, 12, 779598. [Google Scholar] [CrossRef]
- Vlčko, T.; Ohnoutková, L. Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants 2020, 9, E195. [Google Scholar] [CrossRef]
- Tran, M.T.; Doan, D.T.H.; Kim, J.; Song, Y.J.; Sung, Y.W.; Das, S.; Kim, E.J.; Son, G.H.; Kim, S.H.; Van Vu, T.; et al. CRISPR/Cas9-Based Precise Excision of SlHyPRP1 Domain (s) to Obtain Salt Stress Tolerant Tomato. Plant Cell Rep. 2021, 40, 999–1011. [Google Scholar] [CrossRef]
- Guo, M.; Wang, X.S.; Guo, H.D.; Bai, S.Y.; Khan, A.; Wang, X.M.; Gao, Y.M.; Li, J.S. Tomato salt tolerance mechanisms and their potential applications for fighting salinity: A review. Front. Plant Sci. 2022, 13, 949541. [Google Scholar] [CrossRef]
- Alengebawy, A.; Abdelkhalek, S.T.; Qureshi, S.R.; Wang, M.Q. Heavy Metals and Pesticides Toxicity in Agricultural Soil and Plants: Ecological Risks and Human Health Implications. Toxics 2021, 9, 42. [Google Scholar] [CrossRef]
- Ghori, N.-H.; Ghori, T.; Hayat, M.Q.; Imadi, S.R.; Gul, A.; Altay, V.; Ozturk, M. Heavy Metal Stress and Responses in Plants. Int. J. Environ. Sci. Technol. 2019, 16, 1807–1828. [Google Scholar] [CrossRef]
- Aydinalp, C.; Marinova, S. The Effects of Heavy Metals on Seed Germination and Plant Growth on Alfalfa Plant (Medicago sativa). Bulg. J. Agric. Sci. 2009, 15, 347–350. [Google Scholar]
- Dağhan, H.; Öztürk, M.; Hakeem, K.; Sabir, M.; Mermut, A. Soil Pollution in Turkey and Remediation Methods. In Soil Remediation and Plants Prospects and Challenges; Hakeem, K.R., Sabir, M., Ozturk, M., Mermut, A.R., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 287–312. [Google Scholar]
- Rascio, N.; Navari-Izzo, F. Heavy Metal Hyperaccumulating Plants: How and Why Do They Do It? and What Makes Them so Interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Khalid, S.; Abbas, G.; Shahid, N.; Nadeem, M.; Sabir, M.; Aslam, M.; Dumat, C. Heavy Metal Stress and Crop Productivity. In Crop Production and Global Environmental Issues; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–25. [Google Scholar]
- Kaur, R.; Das, S.; Bansal, S.; Singh, G.; Sardar, S.; Dhar, H.; Ram, H. Heavy Metal Stress in Rice: Uptake, Transport, Signalling, and Tolerance Mechanisms. Physiol. Plant. 2021, 173, 430–448. [Google Scholar] [CrossRef] [PubMed]
- Baeg, G.J.; Kim, S.H.; Choi, D.M.; Tripathi, S.; Han, Y.J.; Kim, J. CRISPR/Cas9-Mediated Mutation of 5-Oxoprolinase Gene Confers Resistance to Sulfonamide Compounds in Arabidopsis. Plant Biotechnol. Rep. 2021, 15, 753–764. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Hakeem, K.R.; Nahar, K.; Alharby, H.F. Plant Abiotic Stress Tolerance: Agronomic, Molecular and Biotechnological Approaches; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar] [CrossRef]
- Songmei, L.; Jie, J.; Yang, L.; Jun, M.; Shouling, X.; Yuanyuan, T.; Youfa, L.; Qingyao, S.; Jianzhong, H. Characterization and Evaluation of OsLCT1 and OsNramp5 Mutants Generated Through CRISPR/Cas9-Mediated Mutagenesis for Breeding Low Cd Rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef]
- Chang, J.D.; Huang, S.; Yamaji, N.; Zhang, W.; Ma, J.F.; Zhao, F.J. OsNRAMP1 Transporter Contributes to Cadmium and Manganese Uptake in Rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef]
- Chu, C.; Huang, R.; Liu, L.; Tang, G.; Xiao, J.; Yoo, H.; Yuan, M. The Rice Heavy-Metal Transporter OsNRAMP1 Regulates Disease Resistance by Modulating ROS Homoeostasis. Plant Cell Environ. 2022, 45, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Z.; Chen, M.X.; Yu, L.J.; Xie, L.J.; Yuan, L.B.; Qi, H.; Xiao, M.; Guo, W.; Chen, Z.; Yi, K.; et al. OsARM1, an R2R3 MYB Transcription Factor, Is Involved in Regulation of the Response to Arsenic Stress in Rice. Front. Plant Sci. 2017, 8, 1868. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A.A. Production of Low-Cs+ Rice Plants by Inactivation of the K+ Transporter Os HAK 1 with the CRISPR-Cas System. Plant J. 2017, 92, 43–56. [Google Scholar] [CrossRef]
- Derksen, D.A.; Anderson, R.L.; Blackshaw, R.E.; Maxwell, B. Weed Dynamics and Management Strategies for Cropping Systems in the Northern Great Plains. Agron. J. 2002, 94, 174–185. [Google Scholar] [CrossRef]
- Riaz, M.; Jamil, M.; Mahmood, T.Z. Yield and Yield Components of Maize as Affected by Various Weed Control Methods Under Rain-Fed Conditions of Pakistan. Int. J. Agric. Biol. 2007, 9, 152–155. [Google Scholar]
- Chipman, D.; Barak, Z.; Schloss, J.V. Biosynthesis of 2-aceto-2-hydroxy Acids: Acetolactate Synthases and Acetohydroxyacid Synthases. Biochim. Biophys. Acta 1998, 1385, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Townsend, J.; Tepperman, J.; Black, M.; Chui, C.F.; Mazur, B.; Dunsmuir, P.; Bedbrook, J. The Molecular Basis of Sulfonylurea Herbicide Resistance in Tobacco. EMBO J. 1988, 7, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering Herbicide-Resistant Rice Plants Through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering Herbicide-Resistant Watermelon Variety Through CRISPR/Cas9-Mediated Base-Editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef]
- Kuang, Y.; Li, S.; Ren, B.; Yan, F.; Spetz, C.; Li, X.; Zhou, X.; Zhou, H. Base-Editing-Mediated Artificial Evolution of OsALS1 In Planta to Develop Novel Herbicide-Tolerant Rice Germplasms. Mol. Plant 2020, 13, 565–572. [Google Scholar] [CrossRef]
- Wang, F.; Xu, Y.; Li, W.; Chen, Z.; Wang, J.; Fan, F.; Tao, Y.; Jiang, Y.; Zhu, Q.-H.; Yang, J. Creating a Novel Herbicide-Tolerance OsALS Allele Using CRISPR/Cas9-Mediated Gene Editing. Crop J. 2021, 9, 305–312. [Google Scholar] [CrossRef]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.; Kabsch, W. Interaction of the Herbicide Glyphosate with Its Target Enzyme 5-Enolpyruvylshikimate 3-Phosphate Synthase in Atomic Detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef]
- Ortega, J.L.; Rajapakse, W.; Bagga, S.; Apodaca, K.; Lucero, Y.; Sengupta-Gopalan, C. An Intragenic Approach to Confer Glyphosate Resistance in Chile (Capsicum annuum) by Introducing an In Vitro Mutagenized Chile EPSPS Gene Encoding for a Glyphosate Resistant EPSPS Protein. PLoS ONE 2018, 13, e0194666. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted Base Editing in Rice and Tomato Using a CRISPR-Cas9 Cytidine Deaminase Fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, J.; Chai, Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.; Li, J.; Jiang, L.; Gao, C. Generation of Herbicide Tolerance Traits and a New Selectable Marker in Wheat Using Base Editing. Nat. Plants 2019, 5, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, S.; Meng, X.; Chai, Z.; Wang, D.; Yuan, Y.; Chen, K.; Jiang, L.; Li, J.; Gao, C. Generating Broad-Spectrum Tolerance to ALS-Inhibiting Herbicides in Rice by Base Editing. Sci. China Life Sci. 2021, 64, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Z.B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-Guide RNA Directed Genome Editing in Soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef]
- Liu, L.; Kuang, Y.; Yan, F.; Li, S.; Ren, B.; Gosavi, G.; Spetz, C.; Li, X.; Wang, X.; Zhou, X.; et al. Developing a Novel Artificial Rice Germplasm for Dinitroaniline Herbicide Resistance by Base Editing of OsTubA2. Plant Biotechnol. J. 2021, 19, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zheng, Y.; Xiao, K.; Wei, Y.; Zhu, Y.; Cai, Q.; Chen, L.; Xie, H.; Zhang, J. OsPRX2 Contributes to Stomatal Closure and Improves Potassium Deficiency Tolerance in Rice. Biochem. Biophys. Res. Commun. 2018, 495, 461–467. [Google Scholar] [CrossRef]

| Crops | Gene | Trait | Technique | References |
|---|---|---|---|---|
| Rice | OsbHLH024 | Salinity | CRISPR/Cas9 | [235] |
| Rice | OsRR22 | Salinity | CRISPR/Cas9 | [232,237] |
| Rice | OsRAV2, OsNAC041, OsmiR535 | Salinity | CRISPR/Cas9 | [234,236,238] |
| Rice | OsRR9, OsRR10 | Salinity | CRISPR/Cas9 | [239] |
| Rice | OsNAC041 | Salinity | CRISPR/Cas9 | [240] |
| Rice | OsOTS1 | Salinity | CRISPR/Cas9 | [241,242] |
| Rice | OsDST | Drought and salinity | CRISPR/Cas9 | [151] |
| Rice | SAPK2 | Tolerance to salinity | CRISPR/Cas9 | [131] |
| Wheat | TaHAG1 | Salt tolerance | CRISPR/Cas9 | [243] |
| Maize | ZmHKTI | Tolerance to salinity | CRISPR/Cas9 | [244] |
| Soybean | GmAITR | Salt tolerance | CRISPR/Cas9 | [245] |
| Soybean | Drb2a, Drb2b | Tolerance to droughtand salinity stress | CRISPR/Cas9 | [208] |
| Barley | HvITPK1 | salinity | CRISPR/Cas9 | [246] |
| Tomato | SlHyPRP1, SlARF4 | salinity | CRISPR/Cas9 | [247,248] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, R.K.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Asati, R.; Chauhan, S.; Tiwari, P.N.; Payasi, D.K. Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants. Life 2023, 13, 1456. https://doi.org/10.3390/life13071456
Yadav RK, Tripathi MK, Tiwari S, Tripathi N, Asati R, Chauhan S, Tiwari PN, Payasi DK. Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants. Life. 2023; 13(7):1456. https://doi.org/10.3390/life13071456
Chicago/Turabian StyleYadav, Rakesh Kumar, Manoj Kumar Tripathi, Sushma Tiwari, Niraj Tripathi, Ruchi Asati, Shailja Chauhan, Prakash Narayan Tiwari, and Devendra K. Payasi. 2023. "Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants" Life 13, no. 7: 1456. https://doi.org/10.3390/life13071456
APA StyleYadav, R. K., Tripathi, M. K., Tiwari, S., Tripathi, N., Asati, R., Chauhan, S., Tiwari, P. N., & Payasi, D. K. (2023). Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants. Life, 13(7), 1456. https://doi.org/10.3390/life13071456







