Artificial Intelligence in the Image-Guided Care of Atrial Fibrillation
Abstract
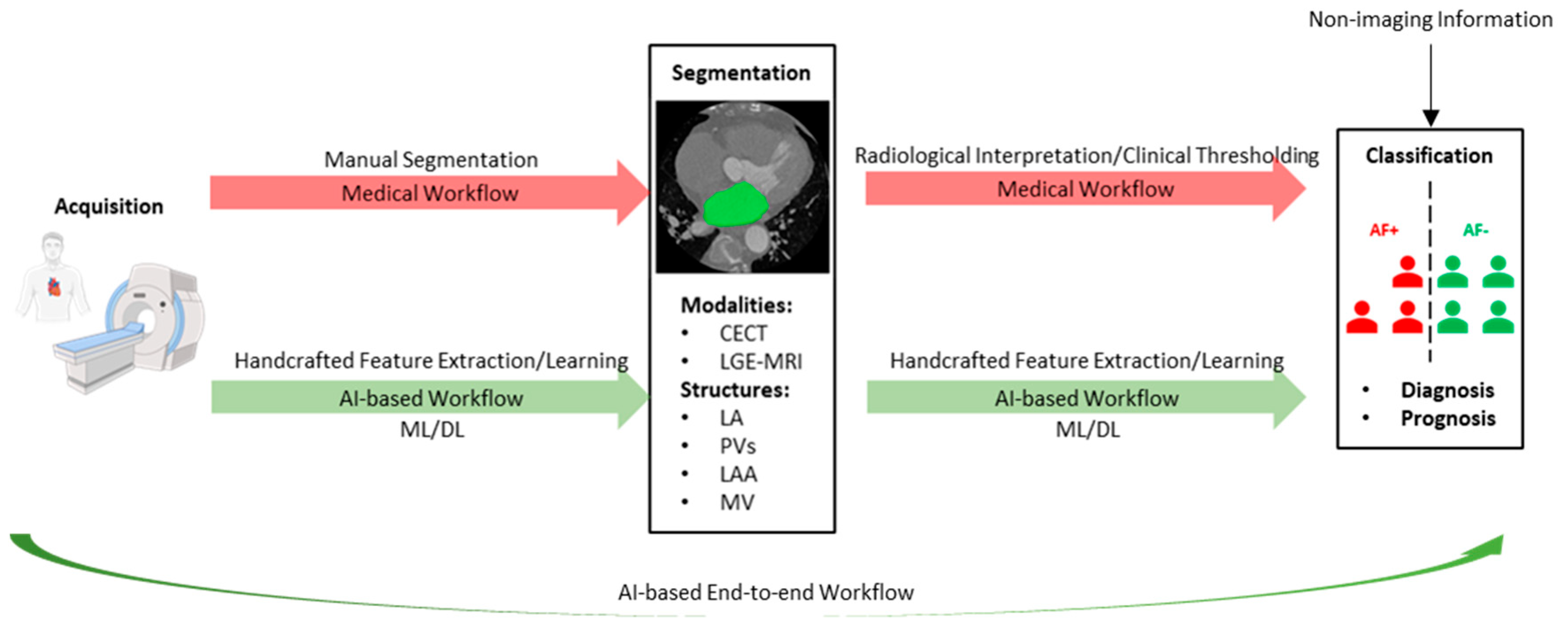
:1. Introduction
2. Artificial Intelligence for Segmentation
2.1. Methodologies
2.1.1. Architectures and Building Blocks
2.1.2. Training Segmentation Models
2.2. Performance of Segmentation Models
3. Artificial Intelligence for Classification
3.1. Feature Engineering
| Publication (Year) | Classification Task 1 | Imaging Modality | Evaluation Metrics | AUC 2 | Highlights 3 |
|---|---|---|---|---|---|
| Shade et al. (2020) [49] | Recurrent AF prediction AF+ (n = 12) AF− (n = 20) | LGE-MRI | AUC, sensitivity, specificity | 0.82 | Quadratic discriminant analysis with radiomic and biophysical modeling features. Contribution of biophysical modeling features is significantly greater than radiomic features. Using biophysical modeling features enables accurate recurrent AF prediction even with a small dataset. |
| Vinter et al. (2020) [50] | Electrical cardioversion success prediction | TTE | AUC | 0.60 (0.54–0.67) | Logistic regression with imaging biomarkers and non-imaging features. Sex-specific classification models achieved suboptimal performance in electrical cardioversion success prediction. |
| Women Success (n = 149) Failure (n = 183) | |||||
| Men Success (n = 394) Failure (n = 396) | 0.59 (0.55–0.63) | ||||
| Liu et al. (2020) [51] | AF Trigger origin stratification 4 Only PV trigger (n = 298) With non-PV trigger (n = 60) | CECT | AUC, accuracy, sensitivity, specificity | 0.88 ± 0.07 | ResNet34-based model identifies patients with non-PV triggers of AF from axial CECT slices. Decision making of the model is based on morphology of the LA, right atrium (RA), and PVs. |
| Zhou et al. (2020) [52] | Incident AF prediction AF+ (n = 653) AF− (n = 3656) | TTE | AUC, area under the precision-recall curve | 0.787 (0.782–0.792) | Logistic regression with imaging biomarkers and non-imaging features. Age is the sole predictive variable for incident AF prediction in oncology patients. Time-split data ensures model generalizability. |
| Hwang et al. (2020) [53] | Recurrent AF prediction AF+ (n = 163) AF− (n = 163) | TTE | AUC, accuracy, sensitivity, specificity | 0.861 | CNN-based model outperforms ML model in prediction of post-ablation AF recurrence when using curved M-mode images of global strain and global strain rate generated from TTE. |
| Firouznia et al. (2021) [54] | Recurrent AF prediction AF+ (n = 88) AF− (n = 115) | CECT | AUC | 0.87 (0.82–0.93) | Random forest with radiomic and non-imaging features. AF induced anatomical remodeling of the LA and PVs is associated with increased roughness in the morphology of these structures. |
| Matsumoto et al. (2022) [55] | AF detection 5 AF+ (n = 1724) AF− (n = 12144) | Radiography | AUC, accuracy, precision, negative predictive value, sensitivity, specificity | 0.80 (0.76–0.84) | Classification model based on EfficientNet identifies patients with AF from chest radiography. Regions that received more attention are the LA (the most) and the RA (the 2nd most) regions. |
| Zhang et al. (2022) [56] | AF detection 6 | CECT | AUC, accuracy, sensitivity, specificity | 0.92 (0.84–1.00) | Random forest with radiomic features. ML classification models identify patients with AF from EAT on chest CECT and CT. |
| n = 200 | |||||
| n = 300 | CT | 0.85 (0.77–0.92) | |||
| Roney et al. (2022) [57] | Recurrent AF prediction AF+ (n = 34) AF− (n = 65) | LGE-MRI | AUC, accuracy, precision, sensitivity | 0.85 ± 0.09 | SVM with PCA model, with imaging biomarker, biophysical modeling, and non-imaging features. ML classification model enables personalized prognosis of AF after catheter ablation |
| Yang et al. (2022) [58] | AF subtype stratification PAF (n = 207) PeAF (n = 107) | CECT | AUC, accuracy, sensitivity, specificity | 0.853 (0.755–0.951) | A nomogram integrating imaging biomarkers and radiomic features. |
| Recurrent AF prediction AF+ (n = 79)AF− (n = 235) | 0.793 (0.654–0.931) | Random forest with radiomic features. Radiomic features based on first order and texture correlate with the inflammatory tissue in the atria. | |||
| Dykstra et al. (2022) [59] | Incident AF prediction AF+ (n = 314) AF− (n = 7325) | LGE-MRI | AUC | 0.80/0.79/0.78 7 | Random survival forests with imaging biomarkers and non-imaging features. Time-dependent risk prediction of incident AF in patients with cardiovascular diseases. |
| Hamatani et al. (2022) [60] | Incident HF prediction HF+ (n = 606) HF− (n = 3790) | TTE Radiography | AUC, accuracy, sensitivity, specificity | 0.75 ± 0.01 | Random forest with imaging biomarkers and non-imaging features. Importance of imaging biomarkers extracted from TTE for incident HF in patients with AF. |
| Pujadas et al. (2022) [61] | Incident AF prediction AF+ (n = 193) AF− (n = 193) | MRI | AUC, accuracy, sensitivity, specificity | 0.76 ± 0.07 | SVM with radiomic and non-imaging features. Radiomic features based on shape and texture correlate with chamber enlargement and hypertrophy predispose AF, adverse changes in tissue composition of the myocardium, as well as LV diastolic dysfunction. |
3.2. Artificial Intelligence for Diagnosis
3.3. Artificial Intelligence for Prognosis
4. Future Directions
4.1. Unlabeled Datasets and Generalizability
4.2. Cutting-Edge Methods and Modalities
4.3. Clinical Applicability
- Consistently achieving the stated level of performance for every new sample.
- Providing outputs that clinicians can comprehend and interpret.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| AF | Atrial fibrillation |
| AI | Artificial intelligence |
| ASD | Average surface distance |
| ASPP | Atrous spatial pyramidal pooling |
| AUC | Area under the curve |
| CECT | Contrast-enhanced computed tomography |
| CI | Confidence interval |
| CNN | Convolutional neural network |
| ConvLSTM | Convolutional long short-term memory |
| CRF | Conditional random field |
| CT | Computed tomography |
| DL | Deep learning |
| DPM | Dual-path module |
| DSC | Dice similarity coefficient |
| EAM | Electroanatomic mapping |
| EAT | Epicardial adipose tissue |
| ECG | Electrocardiography |
| EP | Electrophysiology |
| GBMPM | Gated bidirectional message passing module |
| Grad-CAM | Gradient-weighted class activation mapping |
| HD | Hausdorff distance |
| HF | Heart failure |
| JSC | Jaccard similarity coefficient |
| LA | Left atrium |
| LAA | Left atrial appendage |
| LASC | Left atrium segmentation challenge |
| LGE-MRI | Late gadolinium-enhanced magnetic resonance imaging |
| LV | Left ventricle |
| LVEF | Left ventricular ejection fraction |
| ML | Machine learning |
| MRI | Magnetic resonance imaging |
| MSCM | Multiscale context-aware module |
| MV | Mitral valve |
| PAF | Paroxysmal atrial fibrillation |
| PCA | Principal component analysis |
| PeAF | Persistent atrial fibrillation |
| PET | Positron emission tomography |
| PV | Pulmonary vein |
| QC | Quality control |
| RA | Right atrium |
| SML | Symmetric multilevel supervision |
| SVM | Support vector machine |
| TTE | Transthoracic echocardiography |
| ViT | Vision transformer |
References
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.-H.; McAnulty, J.H.; Zheng, Z.-J.; et al. Worldwide Epidemiology of Atrial Fibrillation. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Staerk, L.; Sherer, J.A.; Ko, D.; Benjamin, E.J.; Helm, R.H. Atrial Fibrillation. Circ. Res. 2017, 120, 1501–1517. [Google Scholar] [CrossRef] [PubMed]
- Charitos, E.I.; Stierle, U.; Ziegler, P.D.; Baldewig, M.; Robinson, D.R.; Sievers, H.-H.; Hanke, T. A Comprehensive Evaluation of Rhythm Monitoring Strategies for the Detection of Atrial Fibrillation Recurrence. Circulation 2012, 126, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Wazni, O.M.; Tsao, H.-M.; Chen, S.-A.; Chuang, H.-H.; Saliba, W.; Natale, A.; Klein, A.L. Cardiovascular Imaging in the Management of Atrial Fibrillation. Focus Issue Card. Imaging 2006, 48, 2077–2084. [Google Scholar] [CrossRef]
- Burstein, B.; Nattel, S. Atrial Fibrosis: Mechanisms and Clinical Relevance in Atrial Fibrillation. J. Am. Coll. Cardiol. 2008, 51, 802–809. [Google Scholar] [CrossRef]
- Abecasis, J.; Dourado, R.; Ferreira, A.; Saraiva, C.; Cavaco, D.; Santos, K.R.; Morgado, F.B.; Adragão, P.; Silva, A. Left atrial volume calculated by multi-detector computed tomography may predict successful pulmonary vein isolation in catheter ablation of atrial fibrillation. EP Eur. 2009, 11, 1289–1294. [Google Scholar] [CrossRef]
- Njoku, A.; Kannabhiran, M.; Arora, R.; Reddy, P.; Gopinathannair, R.; Lakkireddy, D.; Dominic, P. Left atrial volume predicts atrial fibrillation recurrence after radiofrequency ablation: A meta-analysis. EP Eur. 2018, 20, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, R.; Al-Kaisey, A.M.; Kalman, J.M. Catheter ablation for atrial fibrillation: Current indications and evolving technologies. Nat. Rev. Cardiol. 2021, 18, 210–225. [Google Scholar] [CrossRef]
- Lip, G.Y.H.; Nieuwlaat, R.; Pisters, R.; Lane, D.A.; Crijns, H.J.G.M. Refining Clinical Risk Stratification for Predicting Stroke and Thromboembolism in Atrial Fibrillation Using a Novel Risk Factor-Based Approach: The Euro Heart Survey on Atrial Fibrillation. Chest 2010, 137, 263–272. [Google Scholar] [CrossRef]
- Sermesant, M.; Delingette, H.; Cochet, H.; Jaïs, P.; Ayache, N. Applications of artificial intelligence in cardiovascular imaging. Nat. Rev. Cardiol. 2021, 18, 600–609. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Haïssaguerre, M.; Jaïs, P.; Shah, D.C.; Takahashi, A.; Hocini, M.; Quiniou, G.; Garrigue, S.; Le Mouroux, A.; Le Métayer, P.; Clémenty, J. Spontaneous Initiation of Atrial Fibrillation by Ectopic Beats Originating in the Pulmonary Veins. N. Engl. J. Med. 1998, 339, 659–666. [Google Scholar] [CrossRef]
- Xiong, Z.; Xia, Q.; Hu, Z.; Huang, N.; Bian, C.; Zheng, Y.; Vesal, S.; Ravikumar, N.; Maier, A.; Yang, X.; et al. A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Med. Image Anal. 2021, 67, 101832. [Google Scholar] [CrossRef]
- Cho, Y.; Cho, H.; Shim, J.; Choi, J.-I.; Kim, Y.-H.; Kim, N.; Oh, Y.-W.; Hwang, S.H. Efficient Segmentation for Left Atrium With Convolution Neural Network Based on Active Learning in Late Gadolinium Enhancement Magnetic Resonance Imaging. J. Korean Med. Sci. 2022, 37, e271. [Google Scholar] [CrossRef]
- Abdulkareem, M.; Brahier, M.S.; Zou, F.; Taylor, A.; Thomaides, A.; Bergquist, P.J.; Srichai, M.B.; Lee, A.M.; Vargas, J.D.; Petersen, S.E. Generalizable Framework for Atrial Volume Estimation for Cardiac CT Images Using Deep Learning With Quality Control Assessment. Front. Cardiovasc. Med. 2022, 9, 822269. [Google Scholar] [CrossRef]
- Yang, G.; Chen, J.; Gao, Z.; Zhang, H.; Ni, H.; Angelini, E.; Mohiaddin, R.; Wong, T.; Keegan, J.; Firmin, D. Multiview Sequential Learning and Dilated Residual Learning for a Fully Automatic Delineation of the Left Atrium and Pulmonary Veins from Late Gadolinium-Enhanced Cardiac MRI Images. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; Volume 2018, pp. 1123–1127. [Google Scholar] [CrossRef]
- Razeghi, O.; Sim, I.; Roney, C.H.; Karim, R.; Chubb, H.; Whitaker, J.; O’Neill, L.; Mukherjee, R.; Wright, M.; O’Neill, M.; et al. Fully Automatic Atrial Fibrosis Assessment Using a Multilabel Convolutional Neural Network. Circ. Cardiovasc. Imaging 2020, 13, e011512. [Google Scholar] [CrossRef]
- Grigoriadis, G.I.; Zaridis, D.; Pezoulas, V.C.; Nikopoulos, S.; Sakellarios, A.I.; Tachos, N.S.; Naka, K.K.; Michalis, L.K.; Fotiadis, D.I. Segmentation of left atrium using CT images and a deep learning model. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, Scotland, UK, 11–15 July 2022; Volume 2022, pp. 3839–3842. [Google Scholar] [CrossRef]
- Jin, C.; Feng, J.; Wang, L.; Yu, H.; Liu, J.; Lu, J.; Zhou, J. Left Atrial Appendage Segmentation Using Fully Convolutional Neural Networks and Modified Three-Dimensional Conditional Random Fields. IEEE J. Biomed. Health Inform. 2018, 22, 1906–1916. [Google Scholar] [CrossRef]
- Wang, Y.; di Biase, L.; Horton, R.P.; Nguyen, T.; Morhanty, P.; Natale, A. Left Atrial Appendage Studied by Computed Tomography to Help Planning for Appendage Closure Device Placement. J. Cardiovasc. Electrophysiol. 2010, 21, 973–982. [Google Scholar] [CrossRef]
- Zhuang, X. Multivariate Mixture Model for Myocardial Segmentation Combining Multi-Source Images. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 2933–2946. [Google Scholar] [CrossRef]
- Zhuang, X.; Shen, J. Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Med. Image Anal. 2016, 31, 77–87. [Google Scholar] [CrossRef]
- Luo, X.; Zhuang, X. Χ-Metric: An N-Dimensional Information-Theoretic Framework for Groupwise Registration and Deep Combined Computing. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 9206–9224. [Google Scholar] [CrossRef]
- Xiong, Z.; Fedorov, V.V.; Fu, X.; Cheng, E.; Macleod, R.; Zhao, J. Fully Automatic Left Atrium Segmentation From Late Gadolinium Enhanced Magnetic Resonance Imaging Using a Dual Fully Convolutional Neural Network. IEEE Trans. Med. Imaging 2019, 38, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Yin, S.; Tang, R.; Liu, Y.; Song, Y.; Zhang, Y.; Liu, H.; Li, S. Segmentation and visualization of left atrium through a unified deep learning framework. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 589–600. [Google Scholar] [CrossRef]
- Borra, D.; Andalo, A.; Paci, M.; Fabbri, C.; Corsi, C. A fully automated left atrium segmentation approach from late gadolinium enhanced magnetic resonance imaging based on a convolutional neural network. Quant. Imaging Med. Surg. 2020, 10, 1894–1907. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, W.; Luo, G.; Wang, K.; Liang, D.; Li, S. Uncertainty-guided symmetric multilevel supervision network for 3D left atrium segmentation in late gadolinium-enhanced MRI. Med. Phys. 2022, 49, 4554–4565. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Available online: https://papers.nips.cc/paper/2012/hash/c399862d3b9d6b76c8436e924a68c45b-Abstract.html (accessed on 21 February 2023).
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; Lange, T.D.; Halvorsen, P.; Johansen, H.D. ResUNet++: An Advanced Architecture for Medical Image Segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–2255. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, H.; Yeung, D.-Y.; Wong, W.; Woo, W. Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2015; Available online: https://proceedings.neurips.cc/paper/2015/hash/07563a3fe3bbe7e3ba84431ad9d055af-Abstract.html (accessed on 23 February 2023).
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015; Bach, F., Blei, D., Eds.; PMLR: London, UK, 2015; Volume 37, pp. 448–456. Available online: https://proceedings.mlr.press/v37/ioffe15.html (accessed on 20 February 2023).
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is All you Need. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30, Available online: https://proceedings.neurips.cc/paper/2017/hash/3f5ee243547dee91fbd053c1c4a845aa-Abstract.html (accessed on 22 February 2023).
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Xia, Q.; Yao, Y.; Hu, Z.; Hao, A. Automatic 3D Atrial Segmentation from GE-MRIs Using Volumetric Fully Convolutional Networks. In Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges; Pop, M., Sermesant, M., Zhao, J., Li, S., McLeod, K., Young, A., Rhode, K., Mansi, T., Eds.; Springer International Publishing: Cham, Germany, 2019; pp. 211–220. [Google Scholar]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. Available online: https://www.frontiersin.org/articles/10.3389/fcvm.2020.00025 (accessed on 4 May 2023). [CrossRef] [PubMed]
- Rahman, Z.; Jobson, D.J.; Woodell, G.A. Multi-scale retinex for color image enhancement. In Proceedings of the 3rd IEEE International Conference on Image Processing, Lausanne, Switzerland, 19 September 1996; Volume 3, pp. 1003–1006. [Google Scholar] [CrossRef]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best practices for convolutional neural networks applied to visual document analysis. In Proceedings of the Seventh International Conference on Document Analysis and Recognition, Edinburgh, Scotland, 3–6 August 2003; pp. 958–963. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y.; Chen, J.; Zhang, S.; Chen, D.Z. Suggestive Annotation: A Deep Active Learning Framework for Biomedical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2017; Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S., Eds.; Springer International Publishing: Cham, Germany, 2017; pp. 399–407. [Google Scholar]
- Christ, P.F.; Elshaer, M.E.A.; Ettlinger, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; Rempfler, M.; Armbruster, M.; Hofmann, F.; D’Anastasi, M.; et al. Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Germany, 2016; pp. 415–423. [Google Scholar]
- Paulus, W.J.; Tschöpe, C.; Sanderson, J.E.; Rusconi, C.; Flachskampf, F.A.; Rademakers, F.E.; Marino, P.; Smiseth, O.A.; De Keulenaer, G.; Leite-Moreira, A.F.; et al. How to diagnose diastolic heart failure: A consensus statement on the diagnosis of heart failure with normal left ventricular ejection fraction by the Heart Failure and Echocardiography Associations of the European Society of Cardiology. Eur. Heart J. 2007, 28, 2539–2550. [Google Scholar] [CrossRef]
- Shade, J.K.; Ali, R.L.; Basile, D.; Popescu, D.; Akhtar, T.; Marine, J.E.; Spragg, D.D.; Calkins, H.; Trayanova, N.A. Preprocedure Application of Machine Learning and Mechanistic Simulations Predicts Likelihood of Paroxysmal Atrial Fibrillation Recurrence Following Pulmonary Vein Isolation. Circ. Arrhythm. Electrophysiol. 2020, 13, e008213. [Google Scholar] [CrossRef] [PubMed]
- Vinter, N.; Frederiksen, A.S.; Albertsen, A.E.; Lip, G.Y.H.; Fenger-Gron, M.; Trinquart, L.; Frost, L.; Moller, D.S. Role for machine learning in sex-specific prediction of successful electrical cardioversion in atrial fibrillation? Open Heart 2020, 7, e001297. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-M.; Chang, S.-L.; Chen, H.-H.; Chen, W.-S.; Lin, Y.-J.; Lo, L.-W.; Hu, Y.-F.; Chung, F.-P.; Chao, T.-F.; Tuan, T.-C.; et al. The Clinical Application of the Deep Learning Technique for Predicting Trigger Origins in Patients With Paroxysmal Atrial Fibrillation With Catheter Ablation. Circ. Arrhythm. Electrophysiol. 2020, 13, e008518. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Hussain, M.; Brown, S.; Budd, T.; Tang, W.H.W.; Abraham, J.; Xu, B.; Shah, C.; Moudgil, R.; et al. Machine Learning–Based Risk Assessment for Cancer Therapy–Related Cardiac Dysfunction in 4300 Longitudinal Oncology Patients. J. Am. Heart Assoc. 2020, 9, e019628. [Google Scholar] [CrossRef]
- Hwang, Y.-T.; Lee, H.-L.; Lu, C.-H.; Chang, P.-C.; Wo, H.-T.; Liu, H.-T.; Wen, M.-S.; Lin, F.-C.; Chou, C.-C. A Novel Approach for Predicting Atrial Fibrillation Recurrence After Ablation Using Deep Convolutional Neural Networks by Assessing Left Atrial Curved M-Mode Speckle-Tracking Images. Front. Cardiovasc. Med. 2020, 7, 605642. [Google Scholar] [CrossRef]
- Firouznia, M.; Feeny, A.K.; LaBarbera, M.A.; McHale, M.; Cantlay, C.; Kalfas, N.; Schoenhagen, P.; Saliba, W.; Tchou, P.; Barnard, J.; et al. Machine Learning–Derived Fractal Features of Shape and Texture of the Left Atrium and Pulmonary Veins From Cardiac Computed Tomography Scans Are Associated With Risk of Recurrence of Atrial Fibrillation Postablation. Circ. Arrhythm. Electrophysiol. 2021, 14, e009265. [Google Scholar] [CrossRef]
- Matsumoto, T.; Ehara, S.; Walston, S.L.; Mitsuyama, Y.; Miki, Y.; Ueda, D. Artificial intelligence-based detection of atrial fibrillation from chest radiographs. Eur. Radiol. 2022, 32, 5890–5897. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Z.; Jiang, B.; Zhang, Y.; Wang, L.; de Bock, G.H.; Vliegenthart, R.; Xie, X. Machine-learning-based radiomics identifies atrial fibrillation on the epicardial fat in contrast-enhanced and non-enhanced chest CT. Br. J. Radiol. 2022, 95, 20211274. [Google Scholar] [CrossRef]
- Roney, C.H.; Sim, I.; Yu, J.; Beach, M.; Mehta, A.; Alonso Solis-Lemus, J.; Kotadia, I.; Whitaker, J.; Corrado, C.; Razeghi, O.; et al. Predicting Atrial Fibrillation Recurrence by Combining Population Data and Virtual Cohorts of Patient-Specific Left Atrial Models. Circ. Arrhythm. Electrophysiol. 2022, 15, e010253. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Cao, Q.; Xu, Z.; Ge, Y.; Li, S.; Yan, F.; Yang, W. Development and Validation of a Machine Learning-Based Radiomics Model on Cardiac Computed Tomography of Epicardial Adipose Tissue in Predicting Characteristics and Recurrence of Atrial Fibrillation. Front. Cardiovasc. Med. 2022, 9, 813085. [Google Scholar] [CrossRef]
- Dykstra, S.; Satriano, A.; Cornhill, A.K.; Lei, L.Y.; Labib, D.; Mikami, Y.; Flewitt, J.; Rivest, S.; Sandonato, R.; Feuchter, P.; et al. Machine learning prediction of atrial fibrillation in cardiovascular patients using cardiac magnetic resonance and electronic health information. Front. Cardiovasc. Med. 2022, 9, 998558. Available online: https://www.frontiersin.org/articles/10.3389/fcvm.2022.998558 (accessed on 15 March 2023). [CrossRef]
- Hamatani, Y.; Nishi, H.; Iguchi, M.; Esato, M.; Tsuji, H.; Wada, H.; Hasegawa, K.; Ogawa, H.; Abe, M.; Fukuda, S.; et al. Machine Learning Risk Prediction for Incident Heart Failure in Patients With Atrial Fibrillation. JACC Asia 2022, 2, 706–716. [Google Scholar] [CrossRef]
- Pujadas, E.R.; Raisi-Estabragh, Z.; Szabo, L.; McCracken, C.; Morcillo, C.I.; Campello, V.M.; Martin-Isla, C.; Atehortua, A.M.; Vago, H.; Merkely, B.; et al. Prediction of incident cardiovascular events using machine learning and CMR radiomics. Eur. Radiol. 2022, 33, 3488–3500. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Szczypiński, P.M.; Klepaczko, A.; Kociołek, M. QMaZda—Software tools for image analysis and pattern recognition. In Proceedings of the 2017 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA), Poznan, Poland, 20–22 September 2017; pp. 217–221. [Google Scholar] [CrossRef]
- Rodríguez, J.O.; Prieto, S.E.; Correa, C.; Bernal, P.A.; Puerta, G.E.; Vitery, S.; Soracipa, Y.; Muñoz, D. Theoretical generalization of normal and sick coronary arteries with fractal dimensions and the arterial intrinsic mathematical harmony. BMC Med. Phys. 2010, 10, 1. [Google Scholar] [CrossRef]
- Ali, R.L.; Hakim, J.B.; Boyle, P.M.; Zahid, S.; Sivasambu, B.; Marine, J.E.; Calkins, H.; Trayanova, N.A.; Spragg, D.D. Arrhythmogenic propensity of the fibrotic substrate after atrial fibrillation ablation: A longitudinal study using magnetic resonance imaging-based atrial models. Cardiovasc. Res. 2019, 115, 1757–1765. [Google Scholar] [CrossRef]
- Boyle, P.M.; Zghaib, T.; Zahid, S.; Ali, R.L.; Deng, D.; Franceschi, W.H.; Hakim, J.B.; Murphy, M.J.; Prakosa, A.; Zimmerman, S.L.; et al. Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat. Biomed. Eng. 2019, 3, 870–879. [Google Scholar] [CrossRef]
- Plank, G.; Loewe, A.; Neic, A.; Augustin, C.; Huang, Y.-L.; Gsell, M.A.F.; Karabelas, E.; Nothstein, M.; Prassl, A.J.; Sánchez, J.; et al. The openCARP simulation environment for cardiac electrophysiology. Comput. Methods Programs Biomed. 2021, 208, 106223. [Google Scholar] [CrossRef]
- Shafiq-ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef]
- Raisi-Estabragh, Z.; Gkontra, P.; Jaggi, A.; Cooper, J.; Augusto, J.; Bhuva, A.N.; Davies, R.H.; Manisty, C.H.; Moon, J.C.; Munroe, P.B.; et al. Repeatability of Cardiac Magnetic Resonance Radiomics: A Multi-Centre Multi-Vendor Test-Retest Study. Front. Cardiovasc. Med. 2020, 7, 586236. Available online: https://www.frontiersin.org/articles/10.3389/fcvm.2020.586236 (accessed on 7 February 2023). [CrossRef]
- Kudo, M.; Sklansky, J. Comparison of algorithms that select features for pattern classifiers. Pattern Recognit. 2000, 33, 25–41. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Available online: https://proceedings.neurips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html (accessed on 7 February 2023).
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Beach, CA, USA, 9–15 June 2019; PMLR: London, UK, 2019; pp. 6105–6114. Available online: https://proceedings.mlr.press/v97/tan19a.html (accessed on 8 March 2023).
- Wang, T.J.; Parise, H.; Levy, D.; D’Agostino, R.B.; Wolf, P.A.; Vasan, R.S.; Benjamin, E.J. Obesity and the Risk of New-Onset Atrial Fibrillation. JAMA 2004, 292, 2471–2477. [Google Scholar] [CrossRef]
- Mahajan, R.; Nelson, A.; Pathak, R.K.; Middeldorp, M.E.; Wong, C.X.; Twomey, D.J.; Carbone, A.; Teo, K.; Agbaedeng, T.; Linz, D.; et al. Electroanatomical Remodeling of the Atria in Obesity: Impact of Adjacent Epicardial Fat. JACC Clin. Electrophysiol. 2018, 4, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- US Preventive Services Task Force. Screening for Atrial Fibrillation: US Preventive Services Task Force Recommendation Statement. JAMA 2022, 327, 360–367. [Google Scholar] [CrossRef]
- Kahwati, L.C.; Asher, G.N.; Kadro, Z.O.; Keen, S.; Ali, R.; Coker-Schwimmer, E.; Jonas, D.E. Screening for Atrial Fibrillation: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2022, 327, 368–383. [Google Scholar] [CrossRef]
- Du, Y.; Li, Q.; Sidorenkov, G.; Vonder, M.; Cai, J.; de Bock, G.H.; Guan, Y.; Xia, Y.; Zhou, X.; Zhang, D.; et al. Computed Tomography Screening for Early Lung Cancer, COPD and Cardiovascular Disease in Shanghai: Rationale and Design of a Population-based Comparative Study. Acad. Radiol. 2021, 28, 36–45. [Google Scholar] [CrossRef]
- Hahn, V.S.; Lenihan, D.J.; Ky, B. Cancer Therapy–Induced Cardiotoxicity: Basic Mechanisms and Potential Cardioprotective Therapies. J. Am. Heart Assoc. 2014, 3, e000665. [Google Scholar] [CrossRef]
- Farmakis, D.; Parissis, J.; Filippatos, G. Insights Into Onco-Cardiology: Atrial Fibrillation in Cancer. J. Am. Coll. Cardiol. 2014, 63, 945–953. [Google Scholar] [CrossRef]
- Littlejohns, T.J.; Holliday, J.; Gibson, L.M.; Garratt, S.; Oesingmann, N.; Alfaro-Almagro, F.; Bell, J.D.; Boultwood, C.; Collins, R.; Conroy, M.C.; et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat. Commun. 2020, 11, 2624. [Google Scholar] [CrossRef] [PubMed]
- Akao, M.; Chun, Y.-H.; Wada, H.; Esato, M.; Hashimoto, T.; Abe, M.; Hasegawa, K.; Tsuji, H.; Furuke, K. Current status of clinical background of patients with atrial fibrillation in a community-based survey: The Fushimi AF Registry. J. Cardiol. 2013, 61, 260–266. [Google Scholar] [CrossRef]
- Kannel, W.B.; D’Agostino, R.B.; Silbershatz, H.; Belanger, A.J.; Wilson, P.W.F.; Levy, D. Profile for Estimating Risk of Heart Failure. Arch. Intern. Med. 1999, 159, 1197–1204. [Google Scholar] [CrossRef]
- Hussein, A.A.; Lindsay, B.; Madden, R.; Martin, D.; Saliba, W.I.; Tarakji, K.G.; Saqi, B.; Rausch, D.J.; Dresing, T.; Callahan, T.; et al. New Model of Automated Patient-Reported Outcomes Applied in Atrial Fibrillation. Circ. Arrhythm. Electrophysiol. 2019, 12, e006986. [Google Scholar] [CrossRef]
- Krishnan, R.; Rajpurkar, P.; Topol, E.J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Chen, C.; Tarroni, G.; Duan, J.; Guitton, F.; Petersen, S.E.; Guo, Y.; Matthews, P.M.; Rueckert, D. Self-Supervised Learning for Cardiac MR Image Segmentation by Anatomical Position Prediction. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Cham, Germany, 2019; pp. 541–549. [Google Scholar]
- Kervadec, H.; Dolz, J.; Tang, M.; Granger, E.; Boykov, Y.; Ben Ayed, I. Constrained-CNN losses for weakly supervised segmentation. Med. Image Anal. 2019, 54, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yuan, Q.; Gao, Y.; He, K.; Wang, S.; Tang, X.; Tang, J.; Shen, D. Weakly Supervised Segmentation of COVID19 Infection with Scribble Annotation on CT Images. Pattern Recognit. 2022, 122, 108341. [Google Scholar] [CrossRef]
- Rajchl, M.; Lee, M.C.H.; Oktay, O.; Kamnitsas, K.; Passerat-Palmbach, J.; Bai, W.; Damodaram, M.; Rutherford, M.A.; Hajnal, J.V.; Kainz, B.; et al. DeepCut: Object Segmentation From Bounding Box Annotations Using Convolutional Neural Networks. IEEE Trans. Med. Imaging 2017, 36, 674–683. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
- Xu, L.; Bennamoun, M.; Boussaid, F.; An, S.; Sohel, F. An Improved Approach to Weakly Supervised Semantic Segmentation. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1897–1901. [Google Scholar] [CrossRef]
- Christopher, M.; Eugene, K.; Joshua, B.; Sathya, V.; Thomas, H.; Joshua, C.; Edward, D.; Nathan, B.; Brent, W.; Alton, A.; et al. Dark Regions of No-Reflow on Late Gadolinium Enhancement Magnetic Resonance Imaging Result in Scar Formation After Atrial Fibrillation Ablation. J. Am. Coll. Cardiol. 2011, 58, 177–185. [Google Scholar] [CrossRef]
- McGann, C.; Akoum, N.; Patel, A.; Kholmovski, E.; Revelo, P.; Damal, K.; Wilson, B.; Cates, J.; Harrison, A.; Ranjan, R.; et al. Atrial Fibrillation Ablation Outcome Is Predicted by Left Atrial Remodeling on MRI. Circ. Arrhythm. Electrophysiol. 2014, 7, 23–30. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Kigali, Rwanda, 1–5 May 2023; Available online: https://openreview.net/forum?id=YicbFdNTTy (accessed on 8 March 2023).
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A Hybrid Transformer Architecture for Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Germany, 2021; pp. 61–71. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. In Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 1748–1758. [Google Scholar] [CrossRef]
- Watanabe, E.; Miyagawa, M.; Uetani, T.; Kinoshita, M.; Kitazawa, R.; Kurata, M.; Ishimura, H.; Matsuda, T.; Tanabe, Y.; Kido, T.; et al. Positron emission tomography/computed tomography detection of increased 18F-fluorodeoxyglucose uptake in the cardiac atria of patients with atrial fibrillation. Int. J. Cardiol. 2019, 283, 171–177. [Google Scholar] [CrossRef]
- Xie, B.; Chen, B.-X.; Nanna, M.; Wu, J.-Y.; Zhou, Y.; Shi, L.; Wang, Y.; Zeng, L.; Wang, Y.; Yang, X.; et al. 18F-fluorodeoxyglucose positron emission tomography/computed tomography imaging in atrial fibrillation: A pilot prospective study. Eur. Heart J. Cardiovasc. Imaging 2022, 23, 102–112. [Google Scholar] [CrossRef]
- Pappone, C.; Oreto, G.; Lamberti, F.; Vicedomini, G.; Loricchio, M.L.; Shpun, S.; Rillo, M.; Calabrò, M.P.; Conversano, A.; Ben-Haim, S.A.; et al. Catheter Ablation of Paroxysmal Atrial Fibrillation Using a 3D Mapping System. Circulation 1999, 100, 1203–1208. [Google Scholar] [CrossRef]
- Zaharchuk, G. Next generation research applications for hybrid PET/MR and PET/CT imaging using deep learning. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2700–2707. [Google Scholar] [CrossRef]
- An, Q.; McBeth, R.; Zhou, H.; Lawlor, B.; Nguyen, D.; Jiang, S.; Link, M.S.; Zhu, Y. Prediction of Type and Recurrence of Atrial Fibrillation after Catheter Ablation via Left Atrial Electroanatomical Voltage Mapping Registration and Multilayer Perceptron Classification: A Retrospective Study. Sensors 2022, 22, 4058. [Google Scholar] [CrossRef] [PubMed]
- Valindria, V.V.; Lavdas, I.; Bai, W.; Kamnitsas, K.; Aboagye, E.O.; Rockall, A.G.; Rueckert, D.; Glocker, B. Reverse Classification Accuracy: Predicting Segmentation Performance in the Absence of Ground Truth. IEEE Trans. Med. Imaging 2017, 36, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Delingette, H.; Ayache, N. Explainable cardiac pathology classification on cine MRI with motion characterization by semi-supervised learning of apparent flow. Med. Image Anal. 2019, 56, 80–95. [Google Scholar] [CrossRef] [PubMed]







| Publication (Year) 1 | Dataset 2 | Framework | Evaluation Metrics | Highlights |
|---|---|---|---|---|
| Jin et al. (2018) [19] | 150 3 LAA | - | DSC, JSC | Transforming grayscale slices into pseudo color slices improves the spatial resolution of local feature learning. A 3D CRF for post-processing uses the volumetric information to improve 2D segmentation performance from the axial view. |
| Yang et al. (2018) [16] | 100 LA, PVs | TensorFlow | DSC, accuracy, sensitivity, specificity | Applying ConvLSTM to U-net learns the inter-slice correlation from the axial view. Integration of the sequential information with the complementary volumetric information from the coronal and the sagittal views improves 2D segmentation performance from the axial view. |
| Xiong et al. (2019) [24] | 2018 LASC 4 | TensorFlow | DSC, HD, sensitivity, specificity | Using the unique dual-path architecture with local and global encoders results in highly accurate segmentation of the LA. |
| Du et al. (2020) [25] | 2018 LASC | TensorFlow | DSC, HD | Gradual introduction of the DPM, MSCM, GBMPM, and the deep supervision module to the framework improves segmentation performance in each addition. |
| Razeghi et al. (2020) [17] | 207 5 Multilabel 6 | TensorFlow | DSC, accuracy, precision, sensitivity, specificity | Using a variant of U-net for automated segmentation of the LA enables reproducible assessment of atrial fibrosis in patients with AF. PV segmentation and MV segmentation result in lower accuracy and higher uncertainty than LA segmentation. |
| Borra et al. (2020) [26] 7 | 2018 LASC | Keras with TensorFlow backend | DSC, HD, sensitivity, specificity | LA segmentation using a 3D variant of U-net outperforms its 2D counterpart. Significant decline in local segmentation accuracy observed in the regions encompassing the PVs. |
| Liu et al. (2022) [27] | 2018 LASC | PyTorch | DSC, JSC, HD, ASD | SML structure and uncertainty-guided loss function improve local segmentation accuracy on the fuzzy surface of the LA. |
| Grigoriadis et al. (2022) [18] | 20 8 LA, PVs, LAA | TensorFlow-GPU and Keras library | DSC, HD, ASD, rand error index | Integration of attention blocks with variant of U-net for LA segmentation enhances feature learning. |
| Cho et al. (2022) [14] | 118 LA | PyTorch with TensorFlow backend | DSC, precision, sensitivity | Using active learning gradually improves the segmentation performance after each step of human intervention with an initially small, labeled dataset. |
| Abdulkareem et al. (2022) [15] | 337 LA | TensorFlow | DSC | Adoption of a QC mechanism for segmentation enables automated and reproducible estimation of the volume of LA. |
| Building Blocks | Usage and Significance for Segmentation |
|---|---|
| ConvLSTM | Integrated with U-net to connect the encoder and the decoder for learning the sequential information between adjacent slices from the axial view [16]. |
| Batch Normalization | Applied in each convolutional layer before the activation function so that the segmentation models are less sensitive to the initial parameters, therefore accelerating the training process [15,17,26]. |
| Squeeze and Excitation | An additional block included in each convolutional layer of ResUNet++ to adapt model response according to feature relevance [18]. |
| ASPP | Connects the encoder and the decoder in the ResUNet++ architecture to facilitate multiscale feature learning [18]. |
| Attention | Attention blocks in the decoder of the ResUNet++ architecture enhance focus on the essential region of the input slices [18]. |
| Dropout | Prevents model overfitting so that the developed models are more generalizable to unseen population [15,26]. |
| Publication (Year) | Architecture | DSC | HD (mm) |
|---|---|---|---|
| Du et al. (2020) [25] | 2D framework comprising DPM, MSCM, and GBMPM. | 0.94 | 11.89 |
| Borra et al. (2020) [26] | 3D variant of U-net. | 0.91 | 8.34 |
| Liu et al. (2022) [27] | 3D network based on V-net with integrated SML structure. | 0.92 | 11.68 |
| Publication (Year) | Routine Assessments (Post-Ablation) | Symptomatic Assessments |
|---|---|---|
| Shade et al. (2020) [49] | 3, 6, and 12 months | Yes |
| Vinter et al. (2020) [50] | 3 months | Yes |
| Hwang et al. (2020) [53] | 1 week; 1, 3, and 6 months; and every 3–6 months | Yes |
| Firouznia et al. (2021) [54] | 3, 6, and 12 months * | Not specified |
| Roney et al. (2022) [57] | 2–4 appointments over 1 year | Not specified |
| Yang et al. (2022) [58] | Not specified | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, Y.; Bennamoun, M.; Sharif, N.; Lip, G.Y.H.; Dwivedi, G. Artificial Intelligence in the Image-Guided Care of Atrial Fibrillation. Life 2023, 13, 1870. https://doi.org/10.3390/life13091870
Lyu Y, Bennamoun M, Sharif N, Lip GYH, Dwivedi G. Artificial Intelligence in the Image-Guided Care of Atrial Fibrillation. Life. 2023; 13(9):1870. https://doi.org/10.3390/life13091870
Chicago/Turabian StyleLyu, Yiheng, Mohammed Bennamoun, Naeha Sharif, Gregory Y. H. Lip, and Girish Dwivedi. 2023. "Artificial Intelligence in the Image-Guided Care of Atrial Fibrillation" Life 13, no. 9: 1870. https://doi.org/10.3390/life13091870
APA StyleLyu, Y., Bennamoun, M., Sharif, N., Lip, G. Y. H., & Dwivedi, G. (2023). Artificial Intelligence in the Image-Guided Care of Atrial Fibrillation. Life, 13(9), 1870. https://doi.org/10.3390/life13091870







