Towards Improved XAI-Based Epidemiological Research into the Next Potential Pandemic
Abstract
1. Introduction
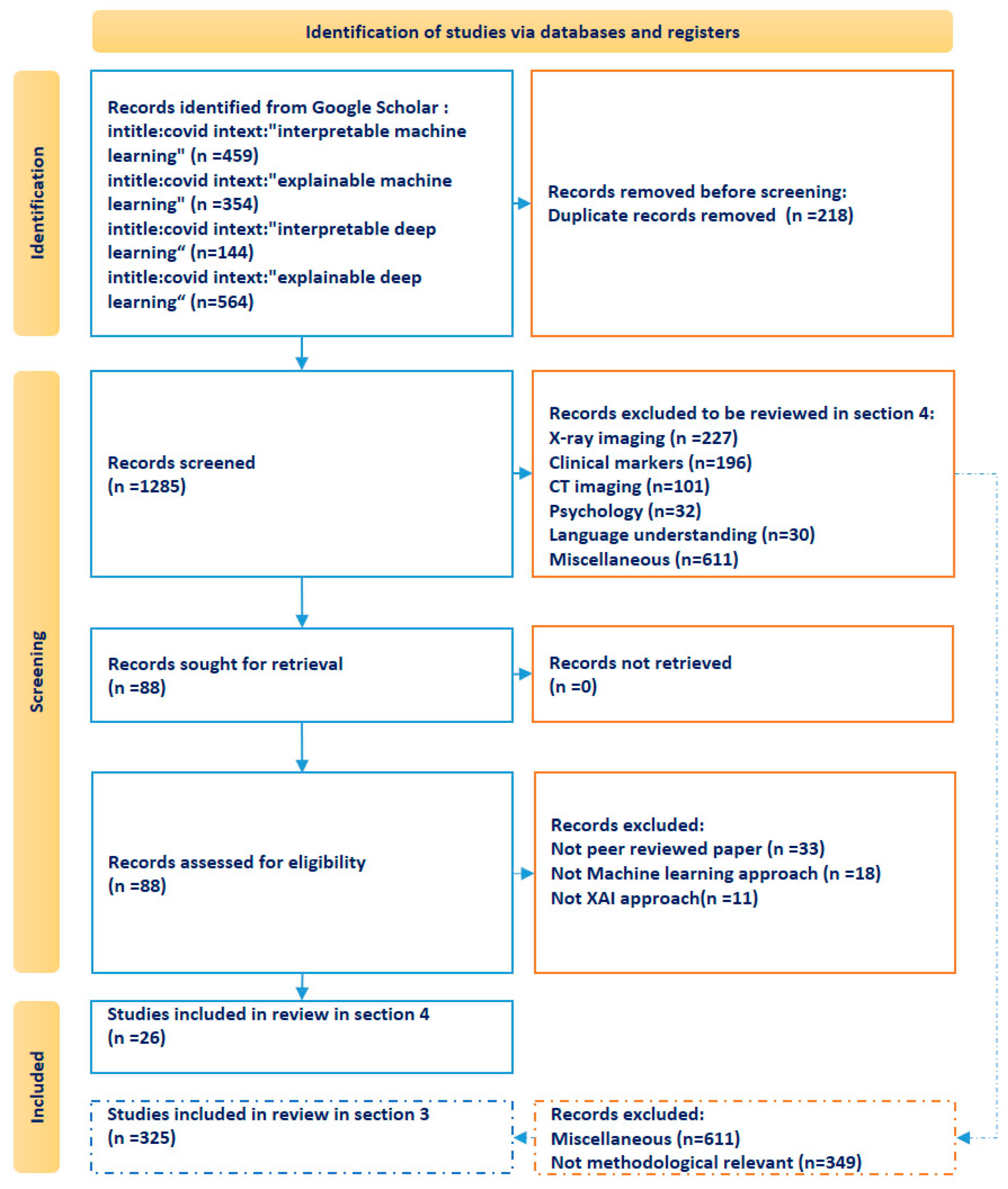
2. Research Methodology
3. Literature Review on XAI Pipelines of SARS-CoV-2 and Reference Framework for the Subsequent Epidemiological Study
3.1. Data Preprocessing
3.1.1. Missing Data
3.1.2. Unbalanced Data
3.1.3. Sparse Data
3.2. Feature Engineering
3.2.1. Pre-Trained Feature Extracting
3.2.2. Reduced Dimensional Feature Extraction
3.2.3. Feature Selection
3.3. Model Parameter Tuning
3.4. Model Training
3.5. Model Evaluation
3.6. Model Interpretation
3.6.1. Intrinsically Interpretable Models
3.6.2. Non-Intrinsically Interpretable Models
- -
- using formal concept analysis (FCA) to create a set of association rules with different confidence intervals [120];
- -
- applying a Bayesian network to visualize the effect of the potential influencers on decision making [24];
- -
- proposing a single associated decision tree (DT) to represent a random forest (RF) model [68];
- -
- applying the anchors method to help explain predictions by decision rules [23];
- -
- utilizing a probabilistic graphical model (PGM-Explainer) as a simpler interpretable Bayesian network in order to interpret GNNs [121];
- -
- applying the symbolic meta modeling approach, which integrates various simple parameterized functions to obtain a closed-form and interpretable expression for the meta model [122].
4. Literature Analysis of Epidemiological AI Research
- -
- the internal connected neurons’ weights in [72];
- -
- attention weights (to determine which input features should be given more attention over others, and the weight of importance for each historical temporal step) in [127]
- -
- integrated gradient in [98];
- -
- the XGB feature is important in [138], representing the percentage of trees that use a variable in the ensemble tree model.
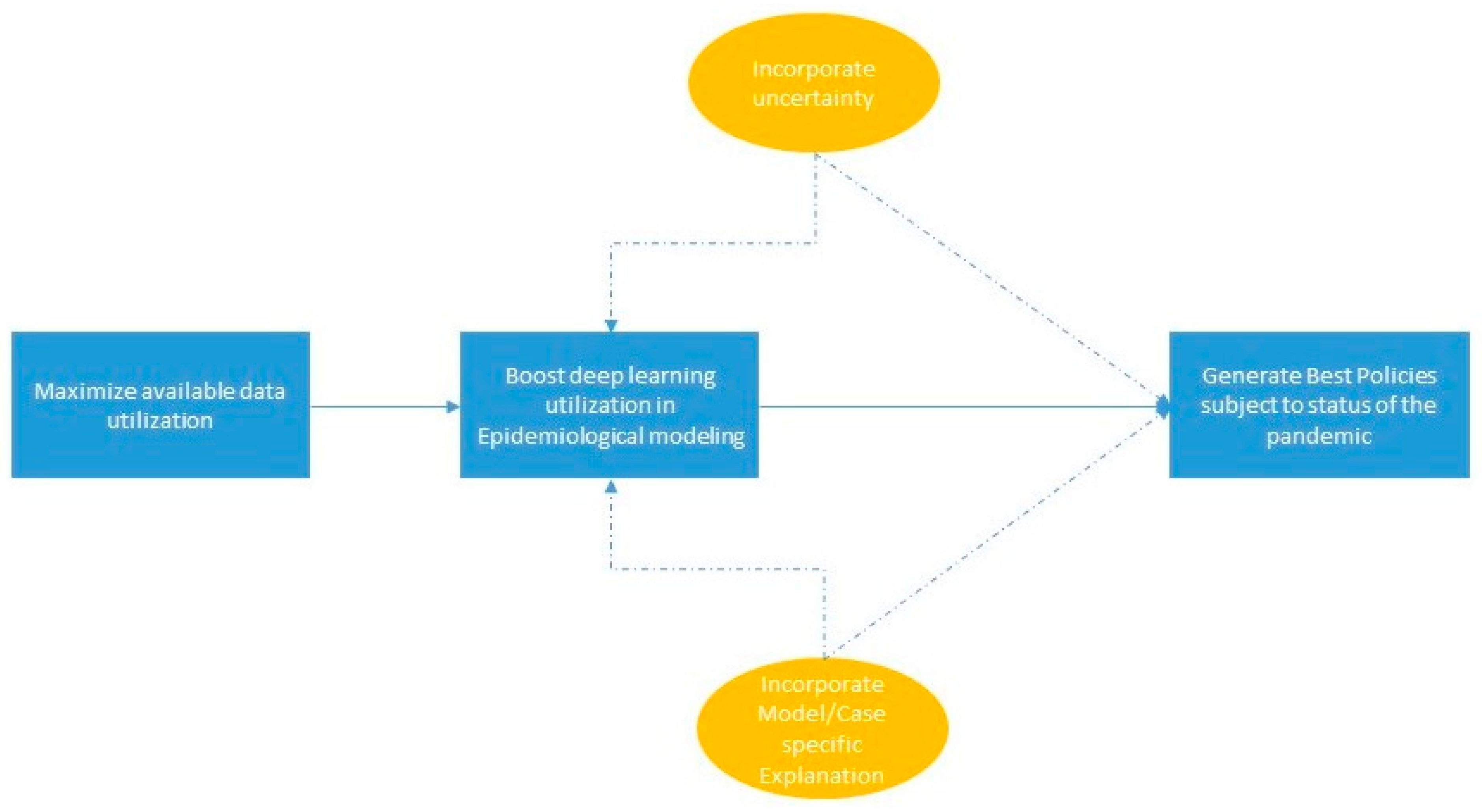
5. Discussion and Conclusions
5.1. Data
5.2. Modelling
5.3. Explanation
5.4. Uncertainty
5.5. Generation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ML abbreviations | |
| ADAB | adaptive boosting model |
| ANN | artificial neural network |
| AR | autoregressive model |
| BI-LSTM | bidirectional long short-term memory network |
| BN | Bayesian network model |
| CATB | categorical boosting model |
| CBR | case-based reasoning |
| CONV-LSTM | convolutional long short term memory network |
| DT | decision tree |
| EBM | explainable boosting machine |
| ENR | elastic net regularization regression |
| GAMS | generalized additive model |
| GAN | generative network |
| GB | gradient boosting model |
| KNN | k-nearest neighbors model |
| LGB | light gradient boosting model |
| LOG-R | logistic regression |
| LR | linear regression |
| LSTM | long short-term memory network |
| MLP | multi-layer perception network |
| NN | neural network |
| ODEs | ordinary differential equations |
| PINNs | physics-informed neural networks |
| RF | random forest |
| RNN | recurrent neural network |
| SEIR | susceptible exposed infected recovered model |
| SVM | support vector machine |
| XGB | extreme gradient boosting model |
| XAI abbreviations | |
| ALE | accumulated local effects |
| CW | connection weights |
| FCA | formal concept analysis |
| ICE | individual conditional expectation |
| IG | integrated gradients |
| LIME | local interpretable model-agnostic explanation |
| MCW | modified connection weights |
| MS | most squares |
| SHAP | Shapley value-based explanation |
| PDP | partial dependence plot |
| PFI | permutation feature importance |
| PGM | probabilistic graphical model |
| VB | Variance-based model |
References
- Vinod, D.N.; Prabaharan, S.R.S. COVID-19-The Role of Artificial Intelligence, Machine Learning, and Deep Learning: A Newfangled. Arch. Comput. Methods Eng. 2023, 30, 2667–2682. [Google Scholar] [CrossRef] [PubMed]
- Miller, T. Explanation in artificial intelligence: Insights from the social sciences. Artif. Intell. 2019, 267, 1–38. [Google Scholar] [CrossRef]
- Allgaier, J.; Mulansky, L.; Draelos, R.L.; Pryss, R. How does the model make predictions? A systematic literature review on the explainability power of machine learning in healthcare. Artif. Intell. Med. 2023, 143, 102616. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Swisher, C.L.; Chung, C.; Jaffray, D.; Sidey-Gibbons, C. On the importance of interpretable machine learning predictions to inform clinical decision making in oncology. Front. Oncol. 2023, 13, 1129380. [Google Scholar] [CrossRef] [PubMed]
- Confalonieri, R.; Coba, L.; Wagner, B.; Besold, T.R. A historical perspective of explainable Artificial Intelligence. WIREs Data Min. Knowl. Discov. 2021, 11, e1391. [Google Scholar] [CrossRef]
- Angelov, P.P.; Soares, E.A.; Jiang, R.; Arnold, N.I.; Atkinson, P.M. Explainable artificial intelligence: An analytical review. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2021, 11, e1424. [Google Scholar] [CrossRef]
- Vilone, G.; Longo, L. Notions of explainability and evaluation approaches for explainable artificial intelligence. Inf. Fusion 2021, 76, 89–106. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4793–4813. [Google Scholar] [CrossRef] [PubMed]
- Abiodun, K.M.; Awotunde, J.B.; Aremu, D.R.; Adeniyi, E.A. Explainable AI for Fighting COVID-19 Pandemic: Opportunities, Challenges, and Future Prospects. In Computational Intelligence for COVID-19 and Future Pandemics. Disruptive Technologies and Digital Transformations for Society 5.0; Kose, U., Watada, J., Deperlioglu, O., Marmolejo Saucedo, J.A., Eds.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Janssen, M.; Hartog, M.; Matheus, R.; Yi Ding, A.; Kuk, G. Will Algorithms Blind People? The Effect of Explainable AI and Decision-Makers’ Experience on AI-supported Decision-Making in Government. Soc. Sci. Comput. Rev. 2022, 40, 478–493. [Google Scholar] [CrossRef]
- Sun, J.; Shi, W.; Giuste, F.O. Improving explainable AI with patch perturbation-based evaluation pipeline: A COVID-19 X-ray image analysis case study. Sci. Rep. 2023, 13, 19488. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. The PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Severn, C.; Suresh, K.; Görg, C.; Choi, Y.S.; Jain, R.; Ghosh, D. A Pipeline for the Implementation and Visualization of Explainable Machine Learning for Medical Imaging Using Radiomics Features. Sensors 2022, 22, 5205. [Google Scholar] [CrossRef] [PubMed]
- Zoabi, Y.; Deri-Rozov, S.; Shomron, N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. NPJ Digit. Med. 2021, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, P.; MuraliKrishna, I. ARIMA and Predicted Geospatial Distribution of COVID-19 in India. In Interpretable Cognitive Internet of Things for Healthcare. Internet of Things; Kose, U., Gupta, D., Khanna, A., Rodrigues, J.J.P.C., Eds.; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Aldhahi, W.; Sull, S. Uncertain-CAM: Uncertainty-Based Ensemble Machine Voting for Improved COVID-19 CXR Classification and Explainability. Diagnostics 2023, 13, 441. [Google Scholar] [CrossRef] [PubMed]
- Docquier, F.; Golenvaux, N.; Nijssen, S. Cross-border mobility responses to COVID-19 in Europe: New evidence from facebook data. Glob. Health 2022, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Hinns, J.; Fan, X.; Liu, S.; Raghava Reddy Kovvuri, V.; Yalcin, M.O.; Roggenbach, M. An Initial Study of Machine Learning Underspecification Using Feature Attribution Explainable AI Algorithms: A COVID-19 Virus Transmission Case Study. In PRICAI 2021: Trends in Artificial Intelligence; Pham, D.N., Theeramunkong, T., Governatori, G., Liu, F., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 13031. [Google Scholar] [CrossRef]
- Andonov, D.I.; Ulm, B.; Graessner, M. Impact of the COVID-19 pandemic on the performance of machine learning algorithms for predicting perioperative mortality. BMC Med. Inf. Decis. Mak. 2023, 23, 67. [Google Scholar] [CrossRef] [PubMed]
- Cabitza, F.; Campagner, A.; Ferrari, D.; Di Resta, C.; Ceriotti, D.; Sabetta, E.; Colombini, A.; De Vecchi, E.; Banfi, G.; Locatelli, M.; et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin. Chem. Lab. Med. (CCLM) 2021, 59, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Chadaga, K.; Chakraborty, C.; Prabhu, S. Clinical and Laboratory Approach to Diagnose COVID-19 Using Machine Learning. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 452–470. [Google Scholar] [CrossRef]
- Casiraghi, E.; Malchiodi, D.; Trucco, G.; Frasca, M.; Cappelletti, L.; Fontana, T.; Esposito, A.A.; Avola, E.; Jachetti, A.; Reese, J.; et al. Explainable Machine Learning for Early Assessment of COVID-19 Risk Prediction in Emergency Departments. IEEE Access 2020, 8, 196299–196325. [Google Scholar] [CrossRef]
- Alle, S.; Kanakan, A.; Siddiqui, S.; Garg, A.; Karthikeyan, A. COVID-19 Risk Stratification and Mortality Prediction in Hospitalized Indian Patients: Harnessing clinical data for public health benefits. PLoS ONE 2022, 17, e0264785. [Google Scholar] [CrossRef] [PubMed]
- Khanna, V.V.; Chadaga, K.; Sampathila, N.; Prabhu, S.; Chadaga, R. A machine learning and explainable artificial intelligence triage-prediction system for COVID-19. Decis. Anal. J. 2023, 7, 100246. [Google Scholar] [CrossRef]
- Adeoye, E.A.; Rozenfeld, Y.; Beam, J. Who was at risk for COVID-19 late in the US pandemic? Insights from a population health machine learning model. Med. Biol. Eng. Comput. 2022, 60, 2039–2049. [Google Scholar] [CrossRef] [PubMed]
- Rezapour, M.; Elmshaeuser, S.K. Artificial intelligence-based analytics for impacts of COVID-19 and online learning on college students’ mental health. PLoS ONE 2022, 17, e0276767. [Google Scholar] [CrossRef] [PubMed]
- Wendland, P.; Schmitt, V.; Zimmermann, J.; Häger, L.; Göpel, S.; Schenkel-Häger, C.; Kschischo, M. Machine learning models for predicting severe COVID-19 outcomes in hospitals. Inform. Med. Unlocked 2023, 37, 101188. [Google Scholar] [CrossRef] [PubMed]
- Croci, S.; Venneri, M.A.; Mantovani, S.; Fallerini, C.; Benetti, E.; Picchiotti, N.; Campolo, F.; Imperatore, F.; Palmieri, M.; Daga, S.; et al. The polymorphism L412F in TLR3 inhibits autophagy and is a marker of severe COVID-19 in males. Autophagy 2022, 18, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Huang, Y.; Huang, W.; Tan, H.; Chen, Z.; Zhong, Z.; Li, X.; Zhang, Y.; Gao, X. Deep supervised learning using self-adaptive auxiliary loss for COVID-19 diagnosis from imbalanced CT images. Neurocomputing 2021, 458, 232–245. [Google Scholar] [CrossRef] [PubMed]
- AlJame, M.; Ahmad, I.; Imtiaz, A.; Mohammed, A. Ensemble learning model for diagnosing COVID-19 from routine blood tests. Inform. Med. Unlocked 2020, 21, 100449. [Google Scholar] [CrossRef] [PubMed]
- Cortes, M.P.; Schultz, C.S.; Sinclair, S.I.J.E.; Bhakta, S.; Kunze, K.L.; Johnson, P.W.; Cowart, J.B.; Carter, R.E.; Franco, P.M.; Sanghavi, D.K.; et al. The Pitfalls of Mining for QuantiFERON Gold in Severely Ill Patients with COVID-19. Mayo Clinic Proceedings: Innovations. Qual. Outcomes 2022, 6, 409–419. [Google Scholar] [CrossRef]
- Esposito, A.; Casiraghi, E.; Chiaraviglio, F.; Scarabelli, A.; Stellato, E.; Plensich, G.; Lastella, G.; Di Meglio, L.; Fusco, S.; Avola, E.; et al. Artificial Intelligence in Predicting Clinical Outcome in COVID-19 Patients from Clinical, Biochemical and a Qualitative Chest X-Ray Scoring System. Rep. Med. Imaging 2021, 14, 27–39. [Google Scholar] [CrossRef]
- Liang, W.; Yao, J.; Chen, A. Early triage of critically ill COVID-19 patients using deep learning. Nat. Commun. 2020, 11, 3543. [Google Scholar] [CrossRef] [PubMed]
- Abbasimehr, H.; Paki, R.; Bahrini, A. A novel approach based on combining deep learning models with statistical methods for COVID-19 time series forecasting. Neural Comput. Applic. 2022, 34, 3135–3149. [Google Scholar] [CrossRef] [PubMed]
- Amin, J.; Sharif, M.; Gul, N. Quantum Machine Learning Architecture for COVID-19 Classification Based on Synthetic Data Generation Using Conditional Adversarial Neural Network. Cogn. Comput. 2022, 14, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Pandey, R.; Babu, R.N. COVIDScreen: Explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput. Applic. 2021, 33, 8871–8892. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, A.K.; Ramachandran, H.K. A preliminary analysis of AI based smartphone application for diagnosis of COVID-19 using chest X-ray images. Expert Syst. Appl. 2021, 183, 115401. [Google Scholar] [CrossRef] [PubMed]
- Laatifi, M.; Douzi, S.; Ezzine, H. Explanatory predictive model for COVID-19 severity risk employing machine learning, SHAPley addition, and LIME. Sci. Rep. 2023, 13, 5481. [Google Scholar] [CrossRef] [PubMed]
- Khozeimeh, F.; Sharifrazi, D.; Izadi, N.H. Combining a convolutional neural network with autoencoders to predict the survival chance of COVID-19 patients. Sci. Rep. 2021, 11, 15343. [Google Scholar] [CrossRef]
- Khobahi, S.; Agarwal, C.; Soltanalian, M. CoroNet: A Deep Network Architecture for Semi-Supervised Task-Based Identification of COVID-19 from Chest X-ray Images; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Ntakolia, C.; Priftis, D.; Charakopoulou-Travlou, M.; Rannou, I.; Magklara, K.; Giannopoulou, I.; Kotsis, K.; Serdari, A.; Tsalamanios, E.; Grigoriadou, A. An Explainable Machine Learning Approach for COVID-19’s Impact on Mood States of Children and Adolescents during the First Lockdown in Greece. Healthcare 2022, 10, 149. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, S.; Salam, N.; Nisha, U.B.; Abdullah, R.Y. COVID-19 Risk Prediction with Regularized Discriminant Analysis and Lasso Regression Using Booster Tree. In Information and Communication Technology for Competitive Strategies (ICTCS 2021); Kaiser, M.S., Xie, J., Rathore, V.S., Eds.; Lecture Notes in Networks and Systems; Springer: Singapore, 2023; Volume 401. [Google Scholar] [CrossRef]
- Beranová, L.; Joachimiak, M.P.; Kliegr, T. Why was this cited? Explainable machine learning applied to COVID-19 research literature. Scientometrics 2022, 127, 2313–2349. [Google Scholar] [CrossRef]
- De Falco, I.; De Pietro, G.; Sannino, G. Classification of Covid-19 chest X-ray images by means of an interpretable evolutionary rule-based approach. Neural Comput. Applic. 2023, 35, 16061–16071. [Google Scholar] [CrossRef]
- Gomes, D.P.S.; Horry, M.J.; Ulhaq, A.; Paul, M.; Chakraborty, S.; Saha, M.; Debnath, T.; Motiur Rahaman, D.M. MAVIDH Score: A COVID-19 Severity Scoring using Chest X-ray Pathology Features. arXiv 2021, arXiv:2011.14983. [Google Scholar]
- Nguyen, X.V.; Dikici, E.; Candemir, S.; Ball, R.L.; Prevedello, L.M. Mortality Prediction Analysis among COVID-19 Inpatients Using Clinical Variables and Deep Learning Chest Radiography Imaging Features. Tomography 2022, 8, 1791–1803. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, J.H.; Aquino, G.; Nance, S. Automated diagnosis and prognosis of COVID-19 pneumonia from initial ER chest X-rays using deep learning. BMC Infect. Dis. 2022, 22, 637. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Kou, Z.; Zhang, Y.; Wang, D. A Duo-Generative Approach to Explainable Multimodal COVID-19 Misinformation Detection; Association for Computing Machinery: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Thenata, A.; Suryadi, M. Machine Learning Prediction of Anxiety Levels in the Society of Academicians during the COVID-19 Pandemic. J. Varian 2022, 6, 81–88. [Google Scholar] [CrossRef]
- Janko, V.; Slapničar, G.; Dovgan, E.; Reščič, N.; Kolenik, T.; Gjoreski, M.; Smerkol, M.; Gams, M.; Luštrek, M. Machine Learning for Analyzing Non-Countermeasure Factors Affecting Early Spread of COVID-19. Int. J. Environ. Res. Public Health 2021, 18, 6750. [Google Scholar] [CrossRef] [PubMed]
- Chieregato, M.; Frangiamore, F.; Morassi, M. A hybrid machine learning/deep learning COVID-19 severity predictive model from CT images and clinical data. Sci. Rep. 2022, 12, 4329. [Google Scholar] [CrossRef] [PubMed]
- Hueniken, K.; Somé, N.H.; Abdelhack, M.; Taylor, G.; Elton Marshall, T.; Wickens, C.M.; Hamilton, H.A.; Wells, S.; Felsky, D. Machine Learning–Based Predictive Modeling of Anxiety and Depressive Symptoms during 8 Months of the COVID-19 Global Pandemic: Repeated Cross-sectional Survey Study. JMIR Ment. Health 2021, 8, e32876. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Sohail, A.; Nofal, T.; Tavares, J.O.; Manuel, R.S. Explainability of neural network clustering in interpreting the COVID-19 emergency data. Fractals 2022, 30, 2240122. [Google Scholar] [CrossRef]
- Souza, A.A.d.; Almeida, D.C.d.; Barcelos, T.S. Simple hemogram to support the decision-making of COVID-19 diagnosis using clusters analysis with self-organizing maps neural network. Soft Comput. 2023, 27, 3295–3306. [Google Scholar] [CrossRef]
- Lannou, E.L.; Post, B.; Haar, S.; Brett, S.J.; Kardirvelu, B.; Faisal, A.A. Clustering of Patient Comorbidities within Electronic Medical Records Enables High-Precision COVID-19 Mortality Prediction; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Higaki, A.; Okayama, H.; Homma, Y. Predictive value of neutrophil-to-lymphocyte ratio for the fatality of COVID-19 patients complicated with cardiovascular diseases and/or risk factors. Sci. Rep. 2022, 12, 13606. [Google Scholar] [CrossRef]
- Cooper, A.; Doyle, O.; Bourke, A. Supervised Clustering for Subgroup Discovery: An Application to COVID-19 Symptomatology. In Machine Learning and Principles and Practice of Knowledge Discovery in Databases; Kamp, M., Koprinska, I., Bibal, A., Bouadi, T., Frenay, B., Galarraga, L., Oramas, J., Adilova, L., Krishnamurthy, Y., Kang, B., et al., Eds.; ECML PKDD 2021; Communications in Computer and Information Science; Springer: Cham, Switzerland, 2021; Volume 1525. [Google Scholar] [CrossRef]
- Lande, J.; Pillay, A.; Chandra, R. Deep learning for COVID-19 topic modelling via Twitter: Alpha, Delta and Omicron. arXiv 2023, arXiv:2303.00135. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Zhou, Y.; Patterson, M. Efficient analysis of COVID-19 clinical data using machine learning models. Med. Biol. Eng. Comput. 2022, 60, 1881–1896. [Google Scholar] [CrossRef] [PubMed]
- Trajanoska, M.; Trajanov, R.; Eftimov, T. Dietary, comorbidity, and geo-economic data fusion for explainable COVID-19 mortality prediction. Expert Syst. Appl. 2022, 209, 118377. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wong, K.W.; Zhu, D. Drug-CoV: A drug-origin knowledge graph discovering drug repurposing targeting COVID-19. Knowl. Inf. Syst. 2023, 65, 5289–5308. [Google Scholar] [CrossRef]
- Ma, J.; Li, B.; Li, Q.; Fan, C.; Mostafavi, A. Attributed Network Embedding Model for Exposing COVID-19 Spread Trajectory Archetypes. arXiv 2022, arXiv:2209.09448. [Google Scholar]
- Zhu, J.; Weng, F.; Zhuang, M.; Lu, X.; Tan, X.; Lin, S.; Zhang, R. Revealing Public Opinion towards the COVID-19 Vaccine with Weibo Data in China: BertFDA-Based Model. Int. J. Environ. Res. Public Health 2022, 19, 13248. [Google Scholar] [CrossRef] [PubMed]
- de Paiva, B.B.M.; Pereira, P.D.; de Andrade, C.M.V. Potential and limitations of machine meta-learning (ensemble) methods for predicting COVID-19 mortality in a large inhospital Brazilian dataset. Sci. Rep. 2023, 13, 3463. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Duan, X.; Hou, S.; Xing, W.; Yang, M.; Ma, Y.; Wang, Z.; Li, W.; Li, Q.; He, C.; et al. Intelligent Classification of B-Line and White Lung from COVID-19 Pneumonia Ultrasound Images Using Radiomics Analysis; Association for Computing Machinery: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Lingelbach, K.; Gado, S.; Janssen, D.; Piechnik, D.; Eichler, M.; Knopf, D.; Hentschel, L.; Schuler, M.; Sernatinger, D.; Peissner, M. Identifying the Effects of COVID-19 on Psychological Well-Being Through Unsupervised Clustering for Mixed Data. In Proceedings of Sixth International Congress on Information and Communication Technology; Yang, X.S., Sherratt, S., Dey, N., Joshi, A., Eds.; Lecture Notes in Networks and Systems; Springer: Singapore, 2022; Volume 235. [Google Scholar] [CrossRef]
- Rostami, M.; Oussalah, M.A. Novel explainable COVID-19 diagnosis method by integration of feature selection with random forest. Inform. Med. Unlocked 2022, 30, 100941. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Jiang, Y.; Shi, Q.Z.L.; Kong, D.; Qian, M.; Chu, J. Impact of COVID-19 on Anxiety, Stress, and Coping Styles in Nurses in Emergency Departments and Fever Clinics: A Cross-Sectional Survey. Risk Manag. Healthc. Policy 2021, 14, 585–594. [Google Scholar] [CrossRef]
- Hegde, S.J.; Ng, M.T.M.; Rios, M.; Mahmassani, H.S.; Chen, Y.; Smilowitz, K. Capacity Analysis and Determinants of the Global COVID-19 Vaccine Distribution Process. Preprint 2023. [CrossRef]
- Youha, S.A.; Alkhamis, M.; Al Mazeedi, S. Using Machine Learning to Unveil Demographic and Clinical Features of COVID-19 Symptomatic and Asymptomatic Patients. Preprint 2020. [CrossRef]
- Kianfar, N.; Mesgari, M.S.; Mollalo, A.; Kaveh, M. Spatio-temporal modeling of COVID-19 prevalence and mortality using artificial neural network algorithms. Spat. Spatio-Temporal Epidemiol. 2022, 40, 100471. [Google Scholar] [CrossRef] [PubMed]
- Harmouche-Karaki, M.; Mahfouz, M.; Salameh, P.; El Helou, N. Physical Activity Levels and Predictors during COVID-19 Lockdown among Lebanese Adults: The Impacts of Sociodemographic Factors, Type of Physical Activity and Work Location. Healthcare 2023, 11, 2080. [Google Scholar] [CrossRef] [PubMed]
- Baqui, P.; Marra, V.; Alaa, A.M. Comparing COVID-19 risk factors in Brazil using machine learning: The importance of socioeconomic, demographic and structural factors. Sci. Rep. 2021, 11, 15591. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Annadi, R.R.; Chaudhri, I.; Munir, K.; Hajagos, J.; Saltz, J.; Hoai, M.; Mallipattu, S.K.; Moffitt, R.; Koraishy, F.M. Short- and Long-Term Recovery after Moderate/Severe AKI in Patients with and without COVID-19. Kidney360 2021, 3, 242–257. [Google Scholar] [CrossRef]
- Cao, Z.; Tang, F.; Chen, C.; Zhang, C.; Guo, Y.; Lin, R.; Huang, Z.; Teng, Y.; Xie, T.; Xu, Y.; et al. Impact of Systematic Factors on the Outbreak Outcomes of the Novel COVID-19 Disease in China: Factor Analysis Study. J. Med. Internet Res. 2020, 22, e23853. [Google Scholar] [CrossRef] [PubMed]
- Rinderknecht, M.D.; Klopfenstein, Y. Predicting critical state after COVID-19 diagnosis: Model development using a large US electronic health record dataset. NPJ Digit. Med. 2021, 4, 113. [Google Scholar] [CrossRef] [PubMed]
- Doblhammer, G.; Kreft, D.; Reinke, C. Regional Characteristics of the Second Wave of SARS-CoV-2 Infections and COVID-19 Deaths in Germany. Int. J. Environ. Res. Public Health 2021, 18, 10663. [Google Scholar] [CrossRef] [PubMed]
- Abbaspour, S.; Robbins, G.K.; Blumenthal, K.G.; Hashimoto, D.; Hopcia, K.; Mukerji, S.S.; Shenoy, E.S.; Wang, W.; Klerman, E.B. Identifying Modifiable Predictors of COVID-19 Vaccine Side Effects: A Machine Learning Approach. Vaccines 2022, 10, 1747. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Byeon, H. Predicting Depression during the COVID-19 Pandemic Using Interpretable TabNet: A Case Study in South Korea. Mathematics 2023, 11, 3145. [Google Scholar] [CrossRef]
- Jiang, Z.; Yin, J.; Han, P.; Chen, N.; Kang, Q.; Qiu, Y.; Li, Y.; Lao, Q.; Sun, M.; Yang, D.; et al. Wavelet transformation can enhance computed tomography texture features: A multicenter radiomics study for grade assessment of COVID-19 pulmonary lesions. Quant. Imaging Med. Surg. 2022, 12, 4758–4770. [Google Scholar] [CrossRef] [PubMed]
- Chadaga, K.; Prabhu, S.; Bhat, V.; Sampathila, N.; Umakanth, S.; Chadaga, R. Artificial intelligence for diagnosis of mild–moderate COVID-19 using haematological markers. Ann. Med. 2023, 55, 1. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Han, Z.; Heidari, A.A.; Shou, Y.; Ye, H.; Wang, L.; Huang, X.; Chen, H.; Chen, Y.; Wu, P. Detection of COVID-19 severity using blood gas analysis parameters and Harris hawks optimized extreme learning machine. Comput. Biol. Med. 2022, 142, 105166. [Google Scholar] [CrossRef] [PubMed]
- Kalustian, K.; Ruth, N. Evacuate the Dancefloor: Exploring and classifying Spotify music listening before and during the COVID-19 pandemic in DACH countries. Jahrb. Musikpsychol. 2021, 30, e95. [Google Scholar] [CrossRef]
- Vernikou, S.; Lyras, A.; Kanavos, A. Multiclass sentiment analysis on COVID-19-related tweets using deep learning models. Neural Comput. Applic. 2022, 34, 19615–19627. [Google Scholar] [CrossRef] [PubMed]
- Taye, A.D.; Borga, L.G.; Greiff, S. A machine learning approach to predict self-protecting behaviors during the early wave of the COVID-19 pandemic. Sci. Rep. 2023, 13, 6121. [Google Scholar] [CrossRef] [PubMed]
- Maitre, J.; Bergeron-Leclerc, C.; Maltais, D.; Gaboury, S. Exploring Anxiety of Qubec University Community during COVID-19 Pandemic via Machine Learning; Association for Computing Machinery: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Kareem, A.; Olufemi, A.; Lawal, N. Development of a COVID-19 Patients’ Fatality Prediction System Using Swarm Intelligent Convolution Neural Network. Asian J. Res. Comput. Sci. 2023, 16, 12–35. [Google Scholar] [CrossRef]
- Najaran, M.H.T. A probabilistic meta-heuristic optimisation algorithm for image multi-level thresholding. Genet. Program Evolvable Mach. 2023, 24, 14. [Google Scholar] [CrossRef]
- Mora-Garcia, R.T.; Cespedes-Lopez, M.F.; Perez-Sanchez, V.R. Housing Price Prediction Using Machine Learning Algorithms in COVID-19 Times. Land 2022, 11, 2100. [Google Scholar] [CrossRef]
- Shade, J.; Doshi, A.; Sung, E. Real-Time Prediction of Mortality, Cardiac Arrest, and Thromboembolic Complications in Hospitalized Patients with COVID-19. JACC Adv. 2022, 1, 100043. [Google Scholar] [CrossRef]
- Qiu, Z.; Cao, Z.; Zou, M. The effectiveness of governmental nonpharmaceutical interventions against COVID-19 at controlling seasonal influenza transmission: An ecological study. BMC Infect. Dis. 2022, 22, 331. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Bath, P.A.; Marincowitz, C.; Sutton, L.; Pilbery, R.; Hopfgartner, F.; Mazumdar, S.; Campbell, R.; Stone, T.; Thomas, B.; et al. Pre-hospital prediction of adverse outcomes in patients with suspected COVID-19: Development, application and comparison of machine learning and deep learning methods. Comput. Biol. Med. 2022, 151 (Part A), 106024. [Google Scholar] [CrossRef]
- Shiri, I.; Mostafaei, S.; Haddadi Avval, A. High-dimensional multinomial multiclass severity scoring of COVID-19 pneumonia using CT radiomics features and machine learning algorithms. Sci. Rep. 2022, 12, 14817. [Google Scholar] [CrossRef] [PubMed]
- Thimoteo, L.M.; Vellasco, M.M.; Amaral, J. Explainable Artificial Intelligence for COVID-19 Diagnosis Through Blood Test Variables. J. Control Autom. Electr. Syst. 2022, 33, 625–644. [Google Scholar] [CrossRef]
- Xu, M.; Ouyang, L.; Han, L.; Sun, K.; Yu, T.; Li, Q.; Tian, H.; Safarnejad, L.; Zhang, H.; Gao, Y.; et al. Accurately Differentiating Between Patients with COVID-19, Patients with Other Viral Infections, and Healthy Individuals: Multimodal Late Fusion Learning Approach. J. Med. Internet Res. 2021, 23, e25535. [Google Scholar] [CrossRef] [PubMed]
- Yagin, F.H.; Cicek, I.B.; Alkhateeb, A.; Yagin, B.; Colak, C.; Azzeh, M.; Akbulut, S. Explainable artificial intelligence model for identifying COVID-19 gene biomarkers. Comput. Biol. Med. 2023, 154, 106619. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Dong, E.; Badr, H.S.; Petrone, M.E.; Grubaugh, N.D.; Gardner, L.M. Incorporating variant frequencies data into short-term forecasting for COVID-19 cases and deaths in the USA: A deep learning approach. eBioMedicine 2023, 89. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.; Cavalheiro, A.; Borges, A.; Duarte, A.F.; Soares, A.; Pereira, M.; Nunes, N.J.; Azevedo, L.; Oliveira, A. Modeling the Geospatial Evolution of COVID-19 Using Spatio-Temporal Convolutional Sequence-to-Sequence Neural Networks; Association for Computing Machinery: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Sengupta, S.; Loomba, J.; Sharma, S.; Brown, D.E.; Thorpe, L.; Haendel, M.A. Analyzing historical diagnosis code data from NIH N3C and RECOVER Programs using deep learning to determine risk factors for Long Covid. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 2797–2802. [Google Scholar] [CrossRef]
- Li, A.; Yadav, N. An Adaptable LSTM Network Predicting COVID-19 Occurrence Using Time Series Data. In Proceedings of the 2021 IEEE International Conference on Digital Health (ICDH), Chicago, IL, USA, 5–10 September 2021; pp. 172–177. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, S.; Yu, G. An interpretable hybrid predictive model of COVID-19 cases using autoregressive model and LSTM. Sci. Rep. 2023, 13, 6708. [Google Scholar] [CrossRef] [PubMed]
- Segovia-Dominguez, I.; Zhen, Z.; Wagh, R.; Lee, H.; Gel, Y.R. TLife-LSTM: Forecasting Future COVID-19 Progression with Topological Signatures of Atmospheric Conditions. In Advances in Knowledge Discovery and Data Mining; Karlapalem, K., Cheng, H., Ramakrishnan, N., Agrawal, R.K., Reddy, P.K., Srivastava, J., Chakraborty, T., Eds.; PAKDD 2021; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12712. [Google Scholar] [CrossRef]
- Alqaissi, E.; Alotaibi, F.; Ramzan, M.S. Graph data science and machine learning for the detection of COVID-19 infection from symptoms. PeerJ Comput. Sci. 2023, 9, e1333. [Google Scholar] [CrossRef]
- Fritz, C.; Dorigatti, E.; Rügamer, D. Combining Graph Neural Networks and Spatio-temporal Disease Models to Predict COVID-19 Cases in Germany. arXiv 2021, arXiv:2101.00661. [Google Scholar]
- Karthikeyan, A.; Garg, A.; Vinod, P.K.; Deva Priyakumar, U. Machine Learning Based Clinical Decision Support System for Early COVID-19 Mortality Prediction. Front. Public Health 2021, 9, 626697. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness & Correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Bottrighi, A.; Pennisi, M.; Roveta, A. A machine learning approach for predicting high risk hospitalized patients with COVID-19 SARS-Cov-2. BMC Med. Inform. Decis. Mak. 2022, 22, 340. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Zhou, X.; Jin, S.; Lu, Y.; Pan, J.; Lin, Q.; Yang, S.; Xu, T.; Basharat, Z.; Zippi, M.; et al. A Comparison of XGBoost, Random Forest, and Nomograph for the Prediction of Disease Severity in Patients with COVID-19 Pneumonia: Implications of Cytokine and Immune Cell Profile. Front. Cell. Infect. Microbiol. 2022, 12, 819267. [Google Scholar] [CrossRef] [PubMed]
- Nopour, R.; Kazemi-Arpanahi, H.; Shanbehzadeh, M.; Azizifar, A. Performance analysis of data mining algorithms for diagnosing COVID-19. J. Educ. Health Promot. 2021, 10, 405. [Google Scholar] [CrossRef] [PubMed]
- Onari, M.A.; Nobile, M.S.; Grau, I.; Fuchs, C.; Zhang, Y.; Boer, A.; Scharnhorst, V. Comparing Interpretable AI Approaches for the Clinical Environment: An Application to COVID-19. In Proceedings of the 2022 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Ottawa, ON, Canada, 15–17 August 2022; pp. 1–8. [Google Scholar] [CrossRef]
- Schiesser, L.; Witschel, H.F.; De La Harpe, A. Uncovering Cross-Platform Spreading Patterns of Fake News about COVID-19. In Proceedings of Society 5.0 Conference; EasyChair: Stockport, UK, 2023. [Google Scholar] [CrossRef]
- Seguel, R.J.; Gallardo, L.; Osses, M.; Rojas, N.Y.; Nogueira, T.; Menares, C.; Andrade, M.; Belalcázar, L.C.; Carrasco, P. Photochemical sensitivity to emissions and local meteorology in Bogotá, Santiago, and São Paulo: An analysis of the initial COVID-19 lockdowns. Elem. Sci. Anthr. 2022, 10, 00044. [Google Scholar] [CrossRef]
- Moslehi, S.; Mahjub, H.; Farhadian, M. Interpretable generalized neural additive models for mortality prediction of COVID-19 hospitalized patients in Hamadan, Iran. BMC Med. Res. Methodol. 2022, 22, 339. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tse, G.; Lee, S.; Liu, T.; Wu, W.K.K.; Cao, Z.; Zeng, D.D.; Chi Kei Wong, I.; Zhang, Q.; Yung Cheung, B.M. Identifying Main and Interaction Effects of Risk Factors to Predict Intensive Care Admission in Patients Hospitalized with COVID-19: A Retrospective Cohort Study in Hong Kong; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Derevitskii, I.V.; Mramorov, N.D.; Usoltsev, S.D.; Kovalchuk, S.V. Hybrid Bayesian Network-Based Modeling: COVID-19-Pneumonia Case. J. Pers. Med. 2022, 12, 1325. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, M.; Sun, G. An interpretable machine learning framework for diagnosis and prognosis of COVID-19. PLoS ONE 2023, 18, e0291961. [Google Scholar] [CrossRef]
- Rabby, G.; Berka, P. Multi-class classification of COVID-19 documents using machine learning algorithms. J. Intell. Inf. Syst. 2023, 60, 571–591. [Google Scholar] [CrossRef] [PubMed]
- Saleh, N.I.; Ghani, H.A.; Jilani, Z. Defining factors in hospital admissions during COVID-19 using LSTM-FCA explainable model. Artif. Intell. Med. 2022, 132, 102394. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, R.; Chan-Olmsted, S.; Chen, H.; Thai, M.T. Cultural-aware Machine Learning based Analysis of COVID-19 Vaccine Hesitancy. arXiv 2023, arXiv:2304.06953. [Google Scholar]
- Jana, A.; Minacapelli, C.D.; Rustgi, V.; Metaxas, D. Global and local interpretation of black-box machine learning models to determine prognostic factors from early COVID-19 data. In Proceedings of the SPIE 12088, 17th International Symposium on Medical Information Processing and Analysis, Campinas, Brazil, 17–19 November 2021; p. 120880A. [Google Scholar] [CrossRef]
- Gao, X.W.; Gao, A. COVID-CBR: A Deep Learning Architecture Featuring Case-Based Reasoning for Classification of COVID-19 from Chest X-Ray Images. In Proceedings of the 2021 20th IEEE International Conference on Machine Learning and Applications (ICMLA), Pasadena, CA, USA, 13–16 December 2021; pp. 1319–1324. [Google Scholar] [CrossRef]
- Mahmoudi, S.A.; Stassin, S.; Daho, M.E.H.; Lessage, X.; Mahmoudi, S. Explainable Deep Learning for Covid-19 Detection Using Chest X-ray and CT-Scan Images. In Healthcare Informatics for Fighting COVID-19 and Future Epidemics. EAI/Springer Innovations in Communication and Computing; Garg, L., Chakraborty, C., Mahmoudi, S., Sohmen, V.S., Eds.; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.C.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M.; et al. COVLIAS 2.0-cXAI: Cloud-Based Explainable Deep Learning System for COVID-19 Lesion Localization in Computed Tomography Scans. Diagnostics 2022, 12, 1482. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.K.; Zhu, D.; Liu, Y.; Erkelens, A.; Daniello, N.; Fox, J. Interpreting County Level COVID-19 Infection and Feature Sensitivity using Deep Learning Time Series Models. arXiv 2022, arXiv:2210.03258. [Google Scholar]
- Jing, N.; Shi, Z.; Hu, Y. Cross-sectional analysis and data-driven forecasting of confirmed COVID-19 cases. Appl. Intell. 2022, 52, 3303–3318. [Google Scholar] [CrossRef] [PubMed]
- Nazir, A.; Ampadu, H.K. Interpretable deep learning for the prediction of ICU admission likelihood and mortality of COVID-19 patients. PeerJ Comput. Sci. 2022, 8, e889. [Google Scholar] [CrossRef] [PubMed]
- Nader, I.W.; Zeilinger, E.L.; Jomar, D. Onset of effects of non-pharmaceutical interventions on COVID-19 infection rates in 176 countries. BMC Public Health 2021, 21, 1472. [Google Scholar] [CrossRef]
- Balogh, A.; Harman, A.; Kreuter, F. Real-Time Analysis of Predictors of COVID-19 Infection Spread in Countries in the European Union through a New Tool. Int. J. Public Health 2022, 67, 1604974. [Google Scholar] [CrossRef]
- Zhou, C.; Wheelock, A.M.; Zhang, C.; Ma, J.; Dong, K.; Pan, J.; Li, Z.; Liang, W.; Gao, J.; Xu, L. The role of booster vaccination in decreasing COVID-19 age-adjusted case fatality rate: Evidence from 32 countries. Front. Public Health 2023, 11, 1150095. [Google Scholar] [CrossRef]
- Zheng, H.L.; An, S.Y.; Qiao, B.J. A data-driven interpretable ensemble framework based on tree models for forecasting the occurrence of COVID-19 in the USA. Environ. Sci. Pollut. Res. 2023, 30, 13648–13659. [Google Scholar] [CrossRef] [PubMed]
- Bakkeli, N.Z. Predicting COVID-19 exposure risk perception using machine learning. BMC Public Health 2023, 23, 1377. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Englert, P.; Varga, M. Socio-economic disparities and COVID-19 in the USA. J. Phys. Complex. 2021, 2, 035017. [Google Scholar] [CrossRef]
- Arık, S.Ö.; Shor, J.; Sinha, R. A prospective evaluation of AI-augmented epidemiology to forecast COVID-19 in the USA and Japan. NPJ Digit. Med. 2021, 4, 146. [Google Scholar] [CrossRef] [PubMed]
- Dlamini, W.M.D.; Simelane, S.P.; Nhlabatsi, N.M. Bayesian network-based spatial predictive modelling reveals COVID-19 transmission dynamics in Eswatini. Spat. Inf. Res. 2022, 30, 183–194. [Google Scholar] [CrossRef]
- Ren, X.; Mi, Z.; Georgopoulos, P.G. Socioexposomics of COVID-19 across New Jersey: A comparison of geostatistical and machine learning approaches. J. Expo. Sci. Environ. Epidemiol. 2023, 34, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Gautam, A.; Huson, D.H. On the Application of Advanced Machine Learning Methods to Analyze Enhanced, Multimodal Data from Persons Infected with COVID-19. Computation 2021, 9, 4. [Google Scholar] [CrossRef]
- Flores, C.; Taramasco, C.; Lagos, M.E.; Rimassa, C.; Figueroa, R. A Feature-Based Analysis for Time-Series Classification of COVID-19 Incidence in Chile: A Case Study. Appl. Sci. 2021, 11, 7080. [Google Scholar] [CrossRef]
- Zopluoglu, C. Detecting Examinees With Item Preknowledge in Large-Scale Testing Using Extreme Gradient Boosting (XGBoost). Educ. Psychol. Meas. 2019, 79, 931–961. [Google Scholar] [CrossRef]
- Molnar, C. Interpretable Machine Learning. Leanpub: Victoria, BC, Canada, 2020; Available online: https://leanpub.com/interpretable-machine-learning (accessed on 18 June 2024).
- Vega, R.; Flores, L.; Greiner, R. SIMLR: Machine Learning inside the SIR Model for COVID-19 Forecasting. Forecasting 2022, 4, 72–94. [Google Scholar] [CrossRef]
- Banerjee, T.; Paul, A.; Srikanth, V. Causal connections between socioeconomic disparities and COVID-19 in the USA. Sci. Rep. 2022, 12, 15827. [Google Scholar] [CrossRef] [PubMed]
- Solayman, S.; Aumi, S.A.; Mery, C.S.; Mubassir, M.; Khan, R. Automatic COVID-19 prediction using explainable machine learning techniques. Int. J. Cogn. Comput. Eng. 2023, 4, 36–46. [Google Scholar] [CrossRef]
- Raissi, M.; Perdikaris, P.; Karniadakis, G.E. Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J. Comput. Phys. 2019, 378, 686–707. [Google Scholar] [CrossRef]
- Ricky, T.; Chen, Q.; Rubanova, Y.; Bettencourt, J.; Duvenaud, D. Neural Ordinary Differential Equations. arXiv 2019, arXiv:1806.07366. [Google Scholar]
- Schiassi, E.; De Florio, M.; D’Ambrosio, A.; Mortari, D.; Furfaro, R. Physics-Informed Neural Networks and Functional Interpolation for Data-Driven Parameters Discovery of Epidemiological Compartmental Models. Mathematics 2021, 9, 2069. [Google Scholar] [CrossRef]
- Hu, H.; Kennedy, C.M.; Kevrekidis, P.G.; Zhang, H.-K. A Modified PINN Approach for Identifiable Compartmental Models in Epidemiology with Application to COVID-19. Viruses 2022, 14, 2464. [Google Scholar] [CrossRef] [PubMed]
- Kosma, C.; Nikolentzos, G.; Panagopoulos, G.; Steyaert, J.-M.; Vazirgiannis, M. Neural Ordinary Differential Equations for Modeling Epidemic Spreading. Trans. Mach. Learn. Res. 2023. Available online: https://openreview.net/forum?id=yrkJGne0vN (accessed on 18 June 2024).
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable Deep Learning for Pulmonary Disease and Coronavirus COVID-19 Detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef] [PubMed]
- Olaide, N.; Absalom, E. A case-based reasoning framework for early detection and diagnosis of novel coronavirus. Inform. Med. Unlocked 2020, 20, 100395. [Google Scholar] [CrossRef]
- Rawat, D.; Sharma, S.; Bhadula, S. Case based Reasoning Technique in Digital Diagnostic System for Lung Cancer Detection. In Proceedings of the 2023 8th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 1–3 June 2023; pp. 1355–1361. [Google Scholar] [CrossRef]
- Phongchit, N.; Taeprasartsit, P. Prediction Performance and Explainability of COVID-19 Classification Models. In Proceedings of the 2021 25th International Computer Science and Engineering Conference (ICSEC), Chiang Rai, Thailand, 18–20 November 2021; pp. 383–387. [Google Scholar] [CrossRef]
- Zhou, J.; Gandomi, A.H.; Chen, F. Evaluating the quality of machine learning explanations: A survey on methods and metrics. Electronics 2021, 10, 593. [Google Scholar] [CrossRef]
- MacKay, D.J. A practical Bayesian framework for backpropagation networks. Neural Comput. 1992, 4, 448–472. [Google Scholar] [CrossRef]
- Ali, H.; Shah, Z. Combating COVID-19 Using Generative Adversarial Networks and Artificial Intelligence for Medical Images: Scoping Review. JMIR Med. Inform. 2022, 10, e37365. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, J.; Li, B.; Gu, X.; Luo, X. COVID-19 diagnosis on CT scan images using a generative adversarial network and concatenated feature pyramid network with an attention mechanism. Med. Phys. 2021, 48, 4334–4349. [Google Scholar] [CrossRef] [PubMed]








| Method [Source] | Explanation |
|---|---|
| K-Means [50,51] | Clustering algorithms that can detect complex patterns based on a partition system to group data into several clusters |
| PCA—principal component analysis [26,51,52,53] | A statistical procedure, which relies on linear transformation for reducing the dimensionality of datasets while preserving crucial information |
| AE—auto encoders [40] | Perform dimensionality reduction similar to PCA. However, unlike PCA, which relies on linear transformation, AEs carry out non-linear transformation using deep neural networks |
| SOM—self-organizing maps [54,55] | Is an unsupervised machine learning technique to cluster the high-dimensional data into low-dimensional outputs consisting of a similar structure like artificial neural networks (ANNs), with the difference that the organizing maps in SOM use competitive learning whereas the ANNs use error correction learning such as back-propagation with gradient descent |
| LDA—Latent Dirichlet Allocation [56] | Is a Bayesian unsupervised clustering method that is often employed to cluster topics of a set of documents in each cluster |
| t-SNE—t-stochastic neighborhood embedding [57] | Is a kind of unsupervised non-linear embedding dimensionality reduction: It embeds the points from a higher dimension to a lower dimension trying to preserve the local structure of data |
| UMAP—uniform manifold approximation and projection [58,59] | Is a flexible non-linear dimension reduction algorithm based on Riemannian geometry and algebraic topology to learn the manifold structure of the data and find a low dimensional embedding that preserves the essential topological structure of that manifold |
| RFF—Random Fourier Features [60] | An approximate kernel method, which maps the given data to a low dimensional randomized feature space based on Euclidean inner product space |
| Method [Source] | Description of Method |
|---|---|
| J48 [111] | Machine learning decision tree classification algorithm based on Iterative Dichotomies. |
| RIPPER [112,113] | Rule-based ML algorithm, in which rules are learned from the data directly. |
| pyFUME [112] | Can create rules based on fuzzy logic. |
| GAMs [114] | Used as non-linear regression tools that allow for non-parametric fittings of complex dependencies of responses. |
| GNAMs [115] | A hybrid ML-DL, which belongs to the GAMs family and learns a linear combination of multi-layer perceptron models. |
| EBM [116] | Explainable boosting machine is constructed with multiple hierarchically organized simple classifiers consisting of sequences of binary decisions and tree-based decision system. |
| JRip [109] | Rule-based classifier, which creates propositional rules that can be used to classify elements. |
| Quantum Lattice [25] | Inspired by the Richard Feynman path, which creates multiple possible graphical models composed of different mathematical operations. On selection of the best model, a Q-graph is created to provide the rationale behind a prediction. Further, a simplified equation for the model is obtained that provides insights into the mapping of inputs to outputs. |
| Bayesian networks [117] | Probabilistic graphical model for representing knowledge about an uncertain domain. |
| Modelling Aspect | SEIR Based ML | Time Explicitness | Dynamic Time | Graphical Model | Rule Creating XAI | Model-Specific XAI |
|---|---|---|---|---|---|---|
| [Source] | [135,142] | [18,81,92,98,102,120,127,129,132,135,138,139,142] | [98,102,120,127,135,138,139,142] | [136,142,143] | [51,120] | [72,98,127,138] |
| Paper | Selected Key Data |
|---|---|
| [133] | Compliance with NPIs, mobility patterns, work–life conflicts. |
| [134] | Income per capita, Population density. |
| [142] | Regional government policies. |
| [76] | Travel data, population density, medical endowments, environmental policy. |
| [143] | Socioeconomic disparities. |
| [120] | New cases, seasons, national lockdown, population vaccination number. |
| [102] | Population data, positive and total tests, number of cases and deaths, population vaccination number. |
| [81] | Population density, educational data, income data, household and housing data. |
| [129] | NPIs and how long which NPI has been in place. |
| [18] | Data on international travel bans, stringency of countries containment policies, Facebook users’ mobility data. |
| [130] | NPIs, different virus variants, average daily temperature, population characteristics, health expenditures, cultural participation data. |
| [135] | Confirmed case and deaths, Google mobility reports, government restrictions, demographic data. |
| [136] | Geographic data, e.g., proximity to major health facilities, churches, shopping centers and supermarkets, Average annual traffic density. |
| [131] | Health services indexes, GDP, behavioral risk factors. |
| [78] | Demographic data, economic data, health care data, unemployment, education, emissions. |
| [137] | Air pollution, proximity to industrial facilities, neighborhood and housing characteristics, age, poverty rate. |
| [61] | Dietary habits, past comorbidity prevalence, environmental policy factors such as seasonally averaged temperature geolocation, development indices. |
| [74] | Sex, age, ethnicity, comorbidities, socioeconomic data. |
| [92] | NPIs, influenza virology surveillance. |
| [132] | Vaccination data, wearing masks, mobility, government interventions. |
| [51] | Weather, culture, travel, health, economical data, development data. |
| [72] | Unemployment data, population density, air and rail transportation, urban population, gross national income per capita. |
| [127] | Demographic, public health data, population density, transportation, pollution, sex ratio. |
| [98] | Mobility, climate data, demographic data, virus variant frequencies. |
| [138] | Weather situation in the location of the infected person, medRxiv, and bioRxiv SARS-CoV-2 literature databases. |
| [139] | Mobility data, death number, patients in ICU, hospitalization by region. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalili, H.; Wimmer, M.A. Towards Improved XAI-Based Epidemiological Research into the Next Potential Pandemic. Life 2024, 14, 783. https://doi.org/10.3390/life14070783
Khalili H, Wimmer MA. Towards Improved XAI-Based Epidemiological Research into the Next Potential Pandemic. Life. 2024; 14(7):783. https://doi.org/10.3390/life14070783
Chicago/Turabian StyleKhalili, Hamed, and Maria A. Wimmer. 2024. "Towards Improved XAI-Based Epidemiological Research into the Next Potential Pandemic" Life 14, no. 7: 783. https://doi.org/10.3390/life14070783
APA StyleKhalili, H., & Wimmer, M. A. (2024). Towards Improved XAI-Based Epidemiological Research into the Next Potential Pandemic. Life, 14(7), 783. https://doi.org/10.3390/life14070783







