Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping
Abstract
1. Introduction
2. Material and Methods
2.1. Specimen Collection
2.2. DNA Extraction
2.3. Pleo Qplex DNA Quantitation System
2.4. qPCR Reaction Set-Up
2.5. Pleo STRplex DNA Profiling System
2.6. PCR Set-Up
2.7. Fragment Analysis Set-Up
2.8. Databasing
3. Results
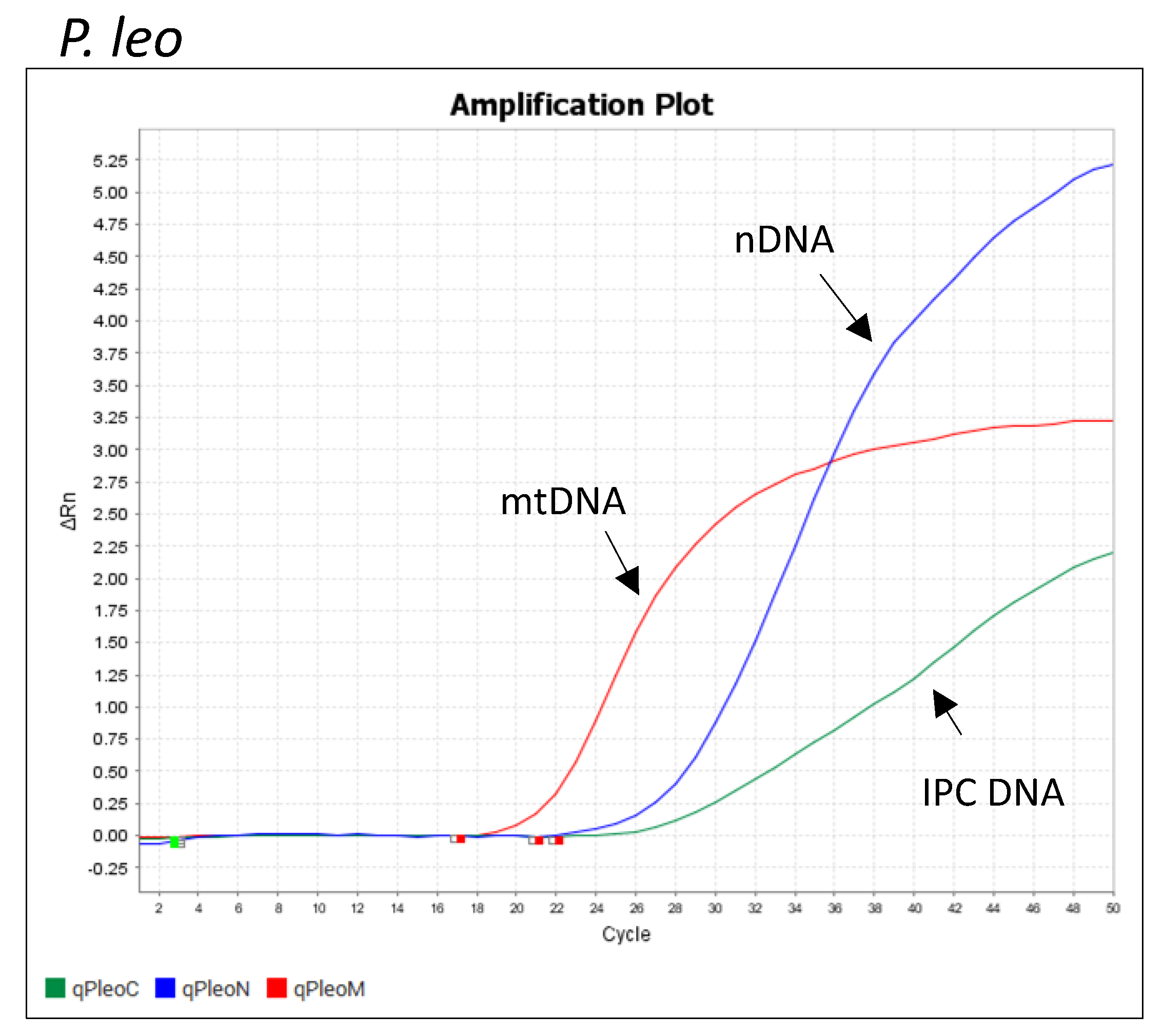
3.1. qPCR Assay Pleo Qplex
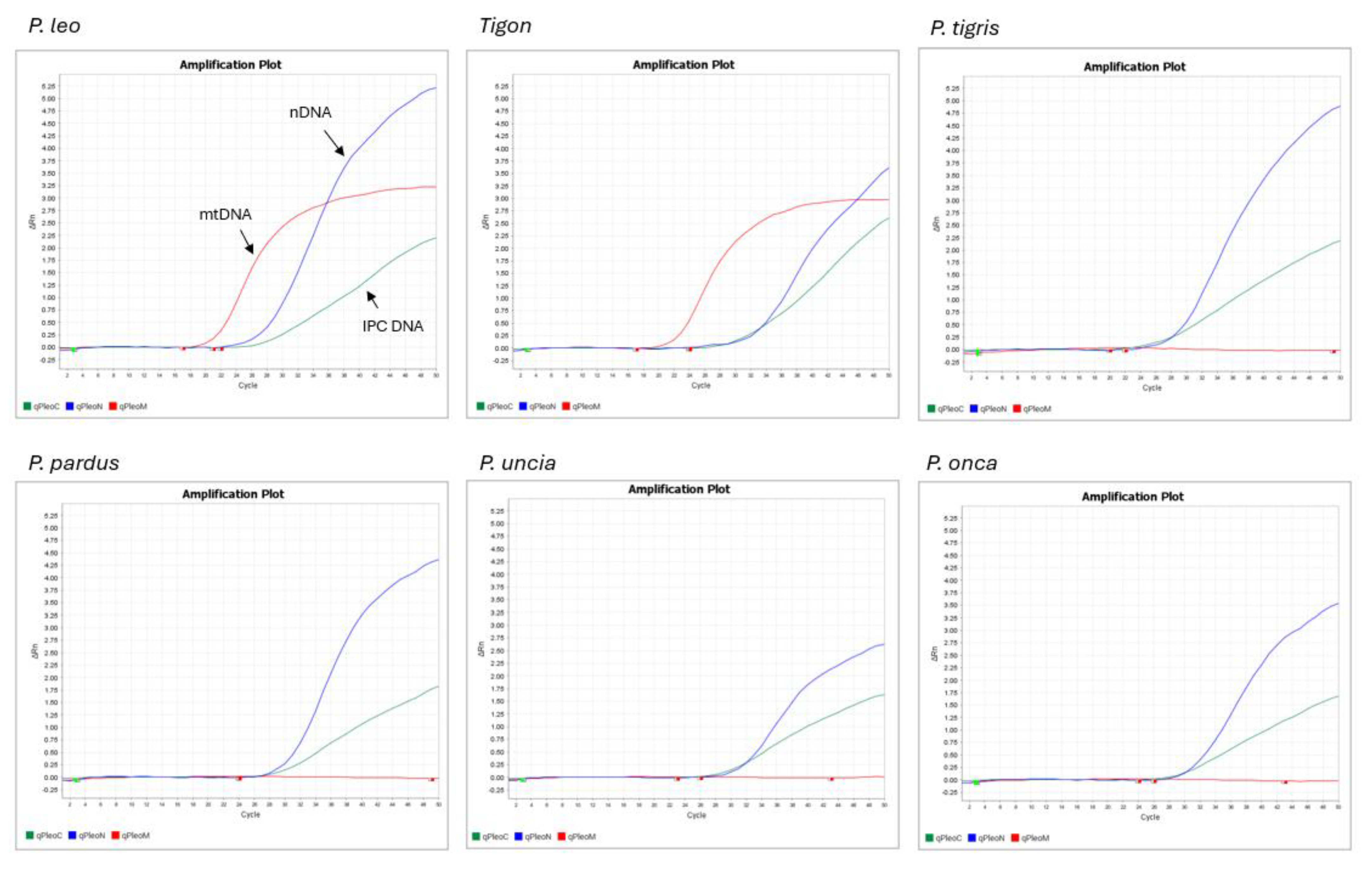
3.2. Pleo Qplex Specificity
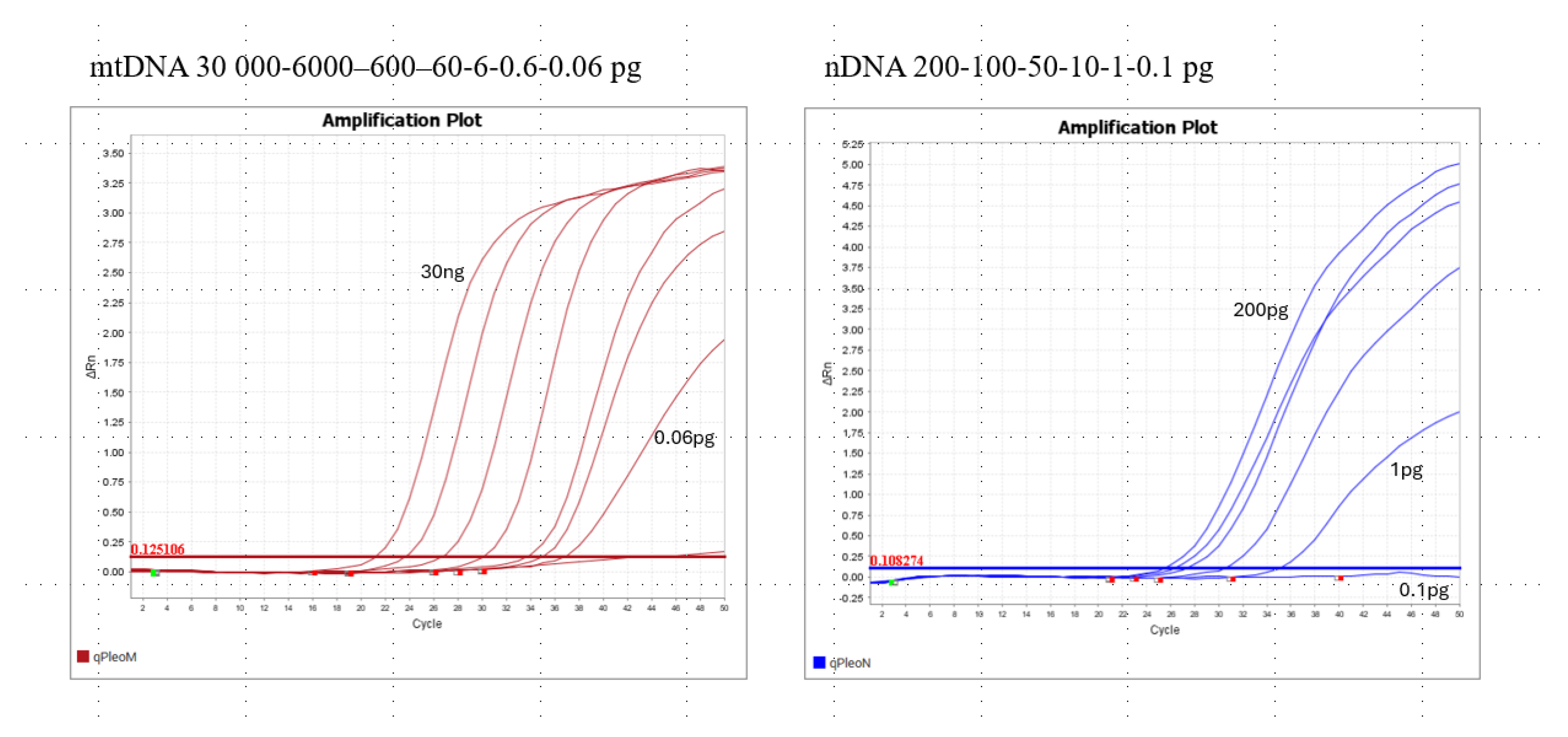
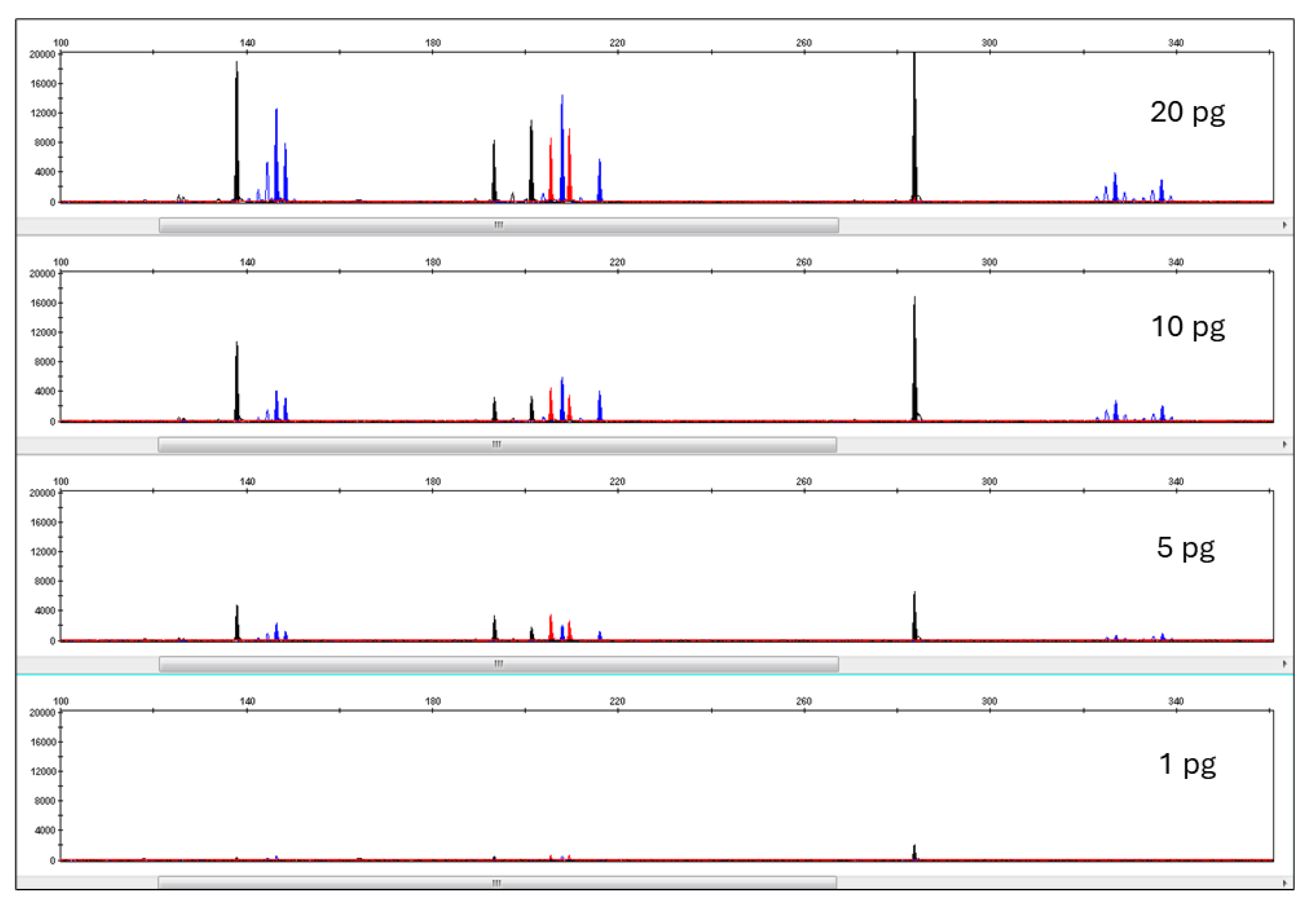
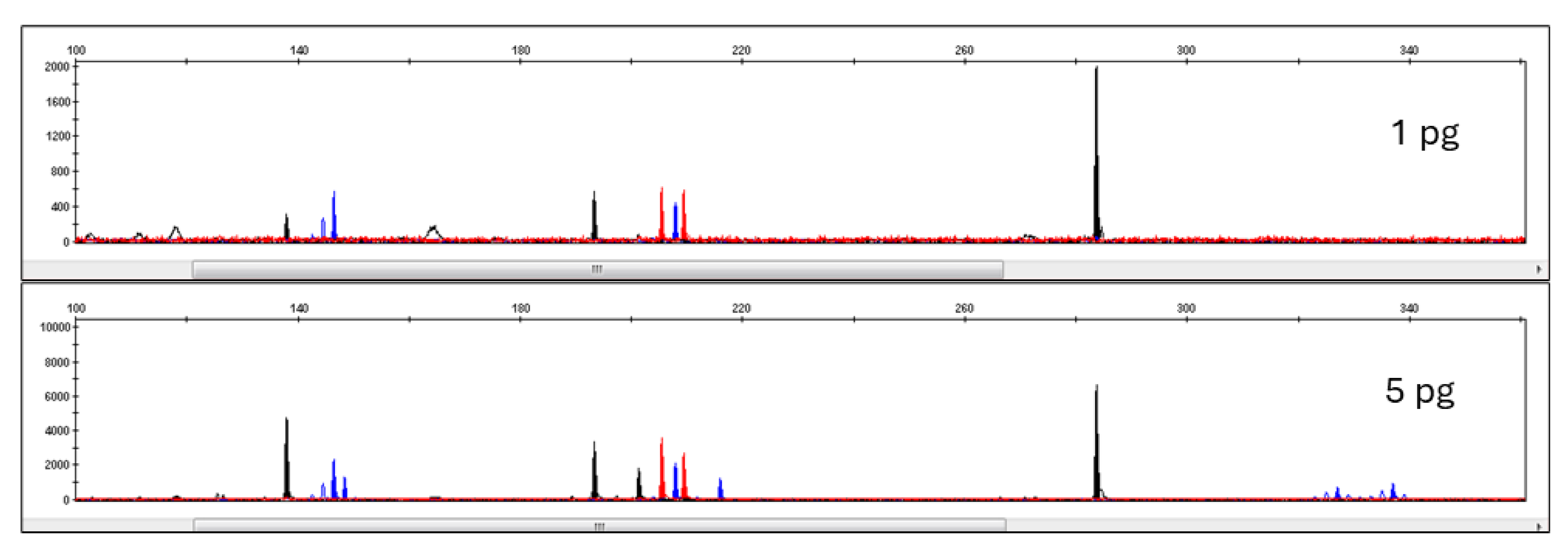
3.3. Pleo Qplex Sensitivity
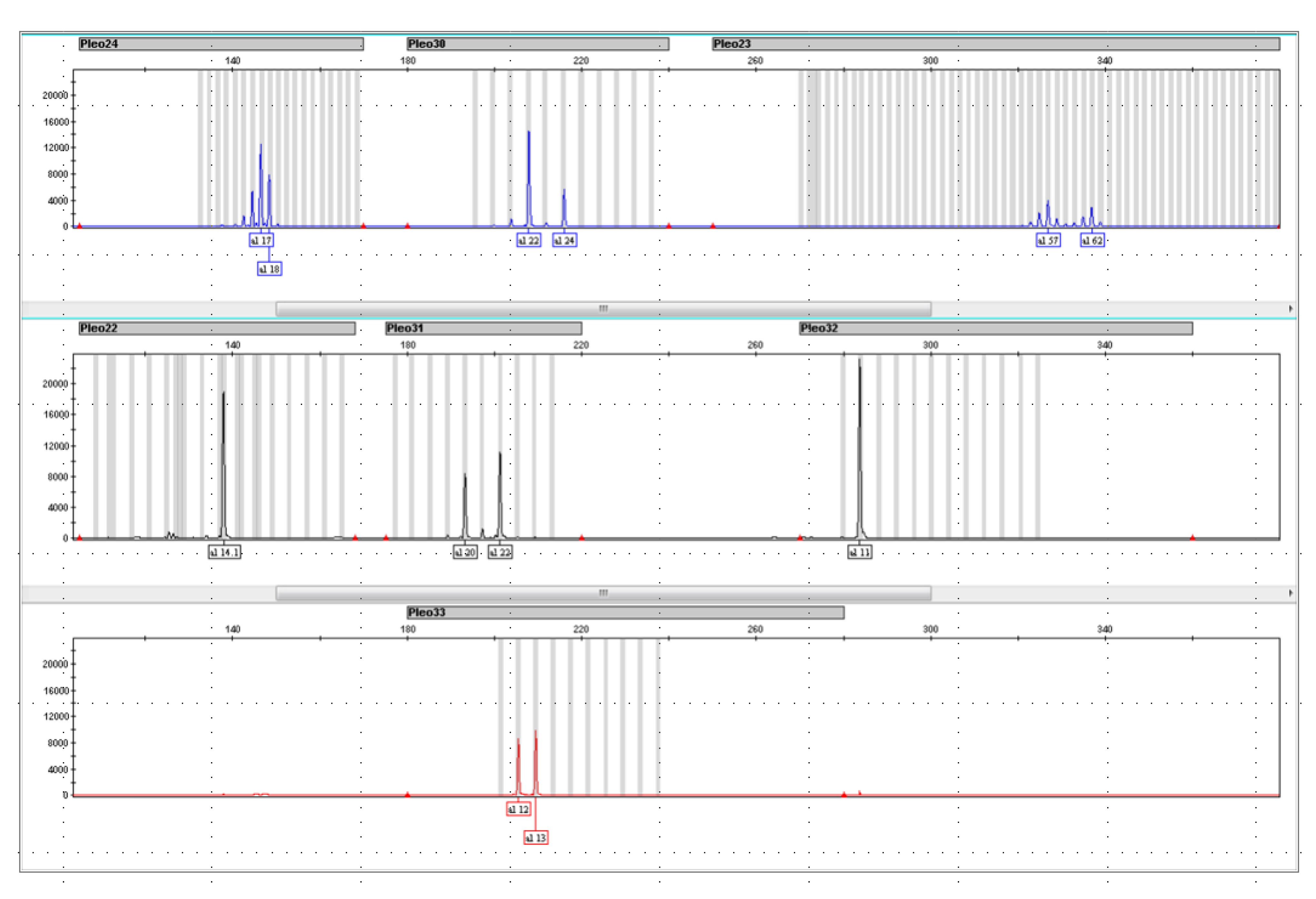
3.4. DNA Typing Pleo STRplex
Pleo 32 Locus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brugière, D.; Chardonnet, B.; Scholte, P. Large-scale extinction of large carnivores (lion Panthera leo, cheetah Acinonyx jubatus and wild dog Lycaon pictus) in protected areas of West and Central Africa. Trop. Conserv. Sci. 2015, 8, 513–527. [Google Scholar] [CrossRef]
- Cazalis, V.; Santini, L.; Lucas, P.M.; González-Suárez, M.; Hoffmann, M.; Benítez-López, A.; Pacifici, M.; Schipper, A.M.; Böhm, M.; Zizka, A.; et al. Prioritizing the reassessment of data-deficient species on the IUCN Red List. Conserv. Biol. 2023, 37, e14139. [Google Scholar] [CrossRef] [PubMed]
- Whitman, K.; Starfield, A.M.; Quadling, H.S.; Packer, C. Sustainable trophy hunting of African lions. Nature 2004, 428, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Johanisová, L.; Mauerhofer, V. Assessing trophy hunting in South Africa by comparing hunting and exporting databases. J. Nat. Conserv. 2023, 72, 126363. [Google Scholar] [CrossRef]
- Everatt, K.; Kokes, R.; Lopez Pereira, C. Evidence of a further emerging threat to lion conservation; targeted poaching for body parts. Biodivers. Conserv. 2019, 28, 4099–4114. [Google Scholar] [CrossRef]
- Coals, P.G.; Mbongwa, N.S.; Naude, V.N.; Williams, V.L. Contemporary cultural trade of lion body parts. Animals 2022, 12, 3169. [Google Scholar] [CrossRef]
- Bauer, H.; Nowell, K.; Sillero-Zubiri, C.; Macdonald, D.W. Lions in the modern arena of CITES. Conserv. Lett. 2018, 11, e12444. [Google Scholar] [CrossRef]
- Coals, P.; Moorhouse, T.P.; D’Cruze, N.C.; Macdonald, D.W.; Loveridge, A.J. Preferences for lion and tiger bone wines amongst the urban public in China and Vietnam. J. Nat. Conserv. 2020, 57, 125874. [Google Scholar] [CrossRef]
- Sibanda, L.; van der Meer, E.; Johnson, P.J.; Hughes, C.; Dlodlo, B.; Parry, R.H.; Mathe, L.J.; Hunt, J.E.; Macdonald, D.W.; Loveridge, A.J. Evaluating the effects of a conservation intervention on rural farmers’ attitudes toward lions. Hum. Dimens. Wildl. 2021, 26, 445–460. [Google Scholar] [CrossRef]
- Vieira Da Silva, C.; Afonso Costa, H.; Costa Santos, J.; Espinheira, R. Forensic Genetics as a Tool for Peace and Justice: An Overview on DNA Quantification. J. Forensic Res. 2012, 4. [Google Scholar] [CrossRef]
- Vajpayee, K.; Dash, H.R.; Parekh, P.B.; Shukla, R.K. PCR Inhibitors and Facilitators-Their Role in Forensic DNA Analysis. Forensic Sci. Int. 2023, 349, 111773. [Google Scholar] [CrossRef] [PubMed]
- Kuffel, A.; Gray, A.; Daeid, N.N. Impact of metal ions on PCR inhibition and RT-PCR efficiency. Int. J. Leg. Med. 2021, 135, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Hedman, J.; Rådström, P. Overcoming inhibition in real-time diagnostic PCR. In PCR Detection of Microbial Pathogens; Springer Science & Business Media: Berlin, Germany, 2013; pp. 17–48. [Google Scholar]
- Andréasson, H.; Gyllensten, U.; Allen, M. Real-time DNA quantification of nuclear and mitochondrial DNA in forensic analysis. Biotechniques 2002, 33, 402–411. [Google Scholar] [CrossRef]
- Fregeau, C.J.; Fourney, R.M. DNA typing with fluorescently tagged short tandem repeats: A sensitive and accurate approach to human identification. Biotechniques 1993, 15, 100–119. [Google Scholar]
- Puri, A. An international DNA database: Balancing hope, privacy, and scientific error. BC Int’l Comp. L. Rev. 2000, 24, 341. [Google Scholar]
- Khan, R. Is the FBI’s Criminal Justice Database, CODIS, Approaching Its Expiration Date? Forensic Genom. 2021, 1, 39–40. [Google Scholar] [CrossRef]
- Bataille, M.; Crainic, K.; Leterreux, M.; Durigon, M.; de Mazancourt, P. Multiplex amplification of mitochondrial DNA for human and species identification in forensic evaluation. Forensic Sci. Int. 1999, 99, 165–170. [Google Scholar] [CrossRef]
- Hellmann, A.P.; Rohleder, U.; Eichmann, C.; Pfeiffer, I.; Parson, W.; Schleenbecker, U. A proposal for standardization in forensic canine DNA typing: Allele nomenclature of six canine-specific STR loci. J. Forensic Sci. 2006, 51, 274–281. [Google Scholar] [CrossRef]
- Kanthaswamy, S. domestic animal forensic genetics–biological evidence, genetic markers, analytical approaches and challenges. Anim. Genet. 2015, 46, 473–484. [Google Scholar] [CrossRef]
- Menotti-Raymond, M.; Stephens, J.; Lyons, L.; O’Brien, S.; David, V. Genetic individualization of domestic cats using feline STR loci for forensic applications. J. Forensic Sci. 1997, 42, 1039–1051. [Google Scholar] [CrossRef]
- Halverson, J.L.; Basten, C. Forensic DNA identification of animal-derived trace evidence: Tools for linking victims and suspects. Croat. Med. J. 2005, 46, 598. [Google Scholar] [PubMed]
- Lorenzini, R.; Cabras, P.; Fanelli, R.; Carboni, G.L. Wildlife molecular forensics: Identification of the Sardinian mouflon using STR profiling and the Bayesian assignment test. Forensic Sci. Int. Genet. 2011, 5, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Caratti, S.; Rossi, L.; Sona, B.; Origlia, S.; Viara, S.; Martano, G.; Torre, C.; Robino, C. Analysis of 11 tetrameric STRs in wild boars for forensic purposes. Forensic Sci. Int. Genet. 2010, 4, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.K. RhODIS®(The Rhinoceros DNA Index System): The Application of Simple Forensic and Genetic Tools Help Conserve African Rhinoceros. In Wildlife Biodiversity Conservation: Multidisciplinary and Forensic Approaches; Springer: Berlin/Heidelberg, Germany, 2021; pp. 463–485. [Google Scholar]
- Harper, C.; Ludwig, A.; Clarke, A.; Makgopela, K.; Yurchenko, A.; Guthrie, A.; Dobrynin, P.; Tamazian, G.; Emslie, R.; van Heerden, M.; et al. Robust forensic matching of confiscated horns to individual poached African rhinoceros. Curr. Biol. 2018, 28, R13–R14. [Google Scholar] [CrossRef]
- Singh, A.; Priyambada, P.; Jabin, G.; Singh, S.K.; Joshi, B.D.; Venkatraman, C.; Chandra, K.; Sharma, L.K.; Thakur, M. Pangolin Indexing System: Implications in forensic surveillance of large seizures. Int. J. Leg. Med. 2020, 134, 1613–1618. [Google Scholar] [CrossRef]
- Roberto, B.; Mauro, Z.; Claudia, C.; Gianluca, D.; Marta, B.; Michel, D.; Luciano, D.T.; Luigi, L.F.; Oliviero, O.; Francesco, P.; et al. Who’s who in the western Hermann’s tortoise conservation: A STR toolkit and reference database for wildlife forensic genetic analyses. bioRxiv 2018, 484030. [Google Scholar] [CrossRef]
- Biello, R.; Zampiglia, M.; Corti, C.; Deli, G.; Biaggini, M.; Crestanello, B.; Delaugerre, M.; Di Tizio, L.; Leonetti, F.L.; Casari, S.; et al. Mapping the geographic origin of captive and confiscated Hermann’s tortoises: A genetic toolkit for conservation and forensic analyses. Forensic Sci. Int. Genet. 2021, 51, 102447. [Google Scholar] [CrossRef]
- Jan, C.; Fumagalli, L. Polymorphic DNA microsatellite markers for forensic individual identification and parentage analyses of seven threatened species of parrots (family Psittacidae). PeerJ 2016, 4, e2416. [Google Scholar] [CrossRef]
- Willows-Munro, S.; Kleinhans, C. Testing microsatellite loci for individual identification of captive African grey parrots (Psittacus erithacus): A molecular tool for parentage analysis that will aid in monitoring legal trade. Conserv. Genet. Resour. 2020, 12, 489–495. [Google Scholar] [CrossRef]
- De Bruyn, M.; Dalton, D.L.; Mwale, M.; Ehlers, K.; Kotze, A. Development and Validation of a Novel Forensic STR Multiplex Assay for Blue (Anthropoides paradiseus), Wattled (Bugeranus carunculatus), and Grey-Crowned Crane (Balearica regulorum). Forensic Sci. Int. Genet. 2024, 73, 103100. [Google Scholar] [CrossRef]
- Potoczniak, M.J.; Chermak, M.; Quarino, L.; Tobe, S.S.; Conte, J. Development of a multiplex, PCR-based genotyping assay for African and Asian elephants for forensic purposes. Int. J. Leg. Med. 2020, 134, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Kinuthia, J.; Harper, C.; Muya, S.; Kimwele, C.; Alakonya, A.; Muigai, A.; Gakuya, F.; Mwaniki, M.; Gatebe, E. The selection of a standard STR panel for DNA profiling of the African elephant (Loxodonta africana) in Kenya. Conserv. Genet. Resour. 2015, 7, 305–307. [Google Scholar] [CrossRef]
- Vaněk, D.; Ehler, E.; Vaňková, L. Development of DNA quantitation and STR typing systems for Panthera tigris species determination and individual identification in forensic casework. Eur. J. Environ. Sci. 2021, 11, 113–118. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Chen, M.; Wang, C.; Li, S. A unified STR profiling system across multiple species with whole genome sequencing data. BMC Bioinform. 2019, 20, 671. [Google Scholar] [CrossRef]
- Dawnay, N.; Ogden, R.; Wetton, J.H.; Thorpe, R.S.; McEwing, R. Genetic data from 28 STR loci for forensic individual identification and parentage analyses in 6 bird of prey species. Forensic Sci. Int. Genet. 2009, 3, e63–e69. [Google Scholar] [CrossRef]
- Olsson, I.A.S.; Silva, S.P.d.; Townend, D.; Sandøe, P. Protecting animals and enabling research in the European Union: An overview of development and implementation of directive 2010/63/EU. ILAR J. 2017, 57, 347–357. [Google Scholar] [CrossRef]
- Hebenstreitova, K.; Salaba, O.; Trubac, J.; Kufnerova, J.; Vanek, D. The Influence of Tanning Chemical Agents on DNA Degradation: A Robust Procedure for the Analysis of Tanned Animal Hide—A Pilot Study. Life 2024, 14, 147. [Google Scholar] [CrossRef]
- Wu, J.-H.; Lei, Y.-L.; Fang, S.-G.; Wan, Q.-H. Twenty-one novel tri-and tetranucleotide microsatellite loci for the Amur tiger (Panthera tigris altaica). Conserv. Genet. 2009, 10, 567–570. [Google Scholar] [CrossRef]
- Swango, K.L.; Hudlow, W.R.; Timken, M.D.; Buoncristiani, M.R. Developmental validation of a multiplex qPCR assay for assessing the quantity and quality of nuclear DNA in forensic samples. Forensic Sci. Int. 2007, 170, 35–45. [Google Scholar] [CrossRef]
- Holt, A.; Wootton, S.C.; Mulero, J.J.; Brzoska, P.M.; Langit, E.; Green, R.L. Developmental validation of the Quantifiler® HP and Trio Kits for human DNA quantification in forensic samples. Forensic Sci. Int. Genet. 2016, 21, 145–157. [Google Scholar] [CrossRef]
- Holmes, A.S.; Houston, R.; Elwick, K.; Gangitano, D.; Hughes-Stamm, S. Evaluation of four commercial quantitative real-time PCR kits with inhibited and degraded samples. Int. J. Leg. Med. 2018, 132, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Ewing, M.M.; Thompson, J.M.; McLaren, R.S.; Purpero, V.M.; Thomas, K.J.; Dobrowski, P.A.; DeGroot, G.A.; Romsos, E.L.; Storts, D.R. Human DNA quantification and sample quality assessment: Developmental validation of the PowerQuantĘr) system. Forensic Sci. Int. Genet. 2016, 23, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Simoes Dutra Correa, H.; Brescia, G.; Cortellini, V.; Cerri, N.; Verzeletti, A. DNA quantitation and degradation assessment: A quantitative PCR protocol designed for small forensic genetics laboratories. Electrophoresis 2020, 41, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Pineda, G.M.; Montgomery, A.H.; Thompson, R.; Indest, B.; Carroll, M.; Sinha, S.K. Development and validation of InnoQuant™, a sensitive human DNA quantitation and degradation assessment method for forensic samples using high copy number mobile elements Alu and SVA. Forensic Sci. Int. Genet. 2014, 13, 224–235. [Google Scholar] [CrossRef]
- Dawnay, N.; Ogden, R.; McEwing, R.; Carvalho, G.R.; Thorpe, R.S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. 2007, 173, 1–6. [Google Scholar] [CrossRef]
- Galan, M.; Pagès, M.; Cosson, J.-F. Next-generation sequencing for rodent barcoding: Species identification from fresh, degraded and environmental samples. PLoS ONE 2012, 7, e48374. [Google Scholar] [CrossRef]
- Vankova, L.; Vanek, D. Capillary-Electrophoresis-Based Species Barcoding of Big Cats: CR-mtDNA-Length Polymorphism. Life 2024, 14, 497. [Google Scholar] [CrossRef]
- Alves, C.; Pereira, R.; Prieto, L.; Aler, M.; Amaral, C.R.; Arévalo, C.; Berardi, G.; Di Rocco, F.; Caputo, M.; Carmona, C.H.; et al. Species identification in forensic samples using the SPInDel approach: A GHEP-ISFG inter-laboratory collaborative exercise. Forensic Sci. Int. Genet. 2017, 28, 219–224. [Google Scholar] [CrossRef]
- Dule, E.J.; Kinimi, E.; Bakari, G.G.; Max, R.A.; Lyimo, C.M.; Mushi, J.R. Species authentication in meat products sold in Kilosa District in Tanzania using HRM-enhanced DNA barcoding. J. Consum. Prot. Food Saf. 2024, 20, 41–52. [Google Scholar] [CrossRef]
- Chang, M.; Kim, J.-Y.; Lee, H.; Lee, E.-J.; Lee, W.-H.; Moon, S.; Choe, S.; Choung, C.M. Development of diagnostic SNP markers and a novel SNP genotyping assay for distinguishing opium poppies. Forensic Sci. Int. 2022, 339, 111416. [Google Scholar] [CrossRef]
- Gouveia, N.; Brito, P.; Serra, A.; Balsa, F.; Andrade, L.; Bento, M.S.; Cunha, P.; Bogas, V.; Lopes, V.; Porto, M. Validation of Quantifiler® Trio DNA Quantification kit in forensic samples. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e24–e25. [Google Scholar] [CrossRef]
- Wang, D.Y.; Chang, C.W.; Lagacé, R.E.; Calandro, L.M.; Hennessy, L.K. Developmental validation of the AmpFℓSTR® Identifiler® Plus PCR Amplification Kit: An established multiplex assay with improved performance. J. Forensic Sci. 2012, 57, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Ogden, R.; Linacre, A. Wildlife forensic science: A review of genetic geographic origin assignment. Forensic Sci. Int. Genet. 2015, 18, 152–159. [Google Scholar] [CrossRef]
- Wheeldon, T.J.; Rutledge, L.Y.; Patterson, B.R.; White, B.N.; Wilson, P.J. Y-chromosome evidence supports asymmetric dog introgression into eastern coyotes. Ecol. Evol. 2013, 3, 3005–3020. [Google Scholar] [CrossRef]
- Verscheure, S.; Backeljau, T.; Desmyter, S. Reviewing population studies for forensic purposes: Dog mitochondrial DNA. ZooKeys 2013, 365, 381–411. [Google Scholar] [CrossRef]
- Berger, C.; Heinrich, J.; Berger, B.; Hecht, W.; Parson, W.; CaDNAP. Towards forensic DNA phenotyping for predicting visible traits in dogs. Genes 2021, 12, 908. [Google Scholar] [CrossRef]
- Enenkel, K.A. 2 The Species and Beyond: Classification and the Place of Hybrids in Early Modern Zoology. In Zoology in Early Modern Culture: Intersections of Science, Theology, Philology, and Political and Religious Education; Brill: Leiden, The Netherlands, 2014; pp. 55–148. [Google Scholar]
- Pérez-Espona, S.; Consortium, C. Conservation-focused biobanks: A valuable resource for wildlife DNA forensics. Forensic Sci. Int. Anim. Environ. 2021, 1, 100017. [Google Scholar] [CrossRef]
- Alaeddini, R. Forensic implications of PCR inhibition—A review. Forensic Sci. Int. Genet. 2012, 6, 297–305. [Google Scholar] [CrossRef]
- Ramón-Laca, A.; Soriano, L.; Gleeson, D.; Godoy, J.A. A simple and effective method for obtaining mammal DNA from faeces. Wildl. Biol. 2015, 21, 195–203. [Google Scholar] [CrossRef]
- Ruggieri, J.; Kemp, R.; Forman, S.; Van Eden, M.E. Techniques for nucleic acid purification from plant, animal, and microbial samples. In Sample Preparation Techniques for Soil, Plant, and Animal Samples; Humana Press: Totowa, NJ, USA, 2016; pp. 41–52. [Google Scholar]
- Yang, D.; Eng, B.; Dudar, J.; Saunders, S.; Waye, J. Removal of PCR inhibitors using silica-based spin columns: Application to ancient bones. Can. Soc. Forensic Sci. J. 1997, 30, 1–5. [Google Scholar] [CrossRef]
- Queiroz, A.P.S.; Santos, F.; Sassaroli, A.; Hársi, C.; Monezi, T.; Mehnert, D. Electropositive filter membrane as an alternative for the elimination of PCR inhibitors from sewage and water samples. Appl. Environ. Microbiol. 2001, 67, 4614–4618. [Google Scholar] [CrossRef] [PubMed]
- Matheson, C.D.; Marion, T.E.; Hayter, S.; Esau, N.; Fratpietro, R.; Vernon, K.K. Removal of metal ion inhibition encountered during DNA extraction and amplification of copper-preserved archaeological bone using size exclusion chromatography. Am. J. Phys. Anthropol. Off. Publ. Am. Assoc. Phys. Anthropol. 2009, 140, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Huang, D.; Yang, L.; Wu, S.; Shen, X.; Pedersen-Bjergaard, S.; Huang, C. Removal of polymerase chain reaction inhibitors by electromembrane extraction. Anal. Chem. 2021, 93, 11488–11496. [Google Scholar] [CrossRef] [PubMed]
- Votrubova-Dubska, J.; Vanek, D.; Zikmund, J.; Mestek, O.; Urbanova, V.; Brzobohata, H.; Brestovansky, P. Efficient removal of a PCR inhibitory agent (vivianite) found on excavated bones. Forensic Sci. Int. 2016, 261, 8–13. [Google Scholar] [CrossRef]
- Teng, F.; Guan, Y.; Zhu, W. A simple and effective method to overcome the inhibition of Fe to PCR. J. Microbiol. Methods 2008, 75, 362–364. [Google Scholar] [CrossRef]
- Geng, T.; Mathies, R.A. Minimizing inhibition of PCR-STR typing using digital agarose droplet microfluidics. Forensic Sci. Int. Genet. 2015, 14, 203–209. [Google Scholar] [CrossRef]
- Minaguchi, K.; Takenaka, O. Structural variations of the VWA locus in humans and comparison with non-human primates. Forensic Sci. Int. 2000, 113, 9–16. [Google Scholar] [CrossRef]
- Crouse, C.A.; Schumm, J. Investigation of species specificity using nine PCR-based human STR systems. J. Forensic Sci. 1995, 40, 952–956. [Google Scholar] [CrossRef]
- Ely, J.J.; Gonzalez, D.L.; Reeves-Daniel, A.; Stone, W.H. Individual identification and paternity determination in chimpanzees (Pan troglodytes) using human short tandem repeat (STR) markers. Int. J. Primatol. 1998, 19, 255–271. [Google Scholar] [CrossRef]
- ENFSI D. DNA Database Management. 2016. Available online: https://enfsi.eu/wp-content/uploads/2016/09/final_version_enfsi_2016_document_on_dna-database_management_0.pdf (accessed on 24 September 2024).


| Primer/Probe Name | Final Concentration (µM) | Sequence (5′-3′) | PCR Product Size (* bp) | Specificity | TaqMan Probe Fluorescent Label |
|---|---|---|---|---|---|
| qPleoM_f | 0.75 | ACCTATTAGGAGATCCCGACAAC | 150 | cytB (mtDNA) | --- |
| qPleoM_r | 0.75 | CTGTTTGGAAGTGTGGAGGGCA | --- | ||
| qPleoM_p | 0.25 | TACCCCCGCCAATCCTCTAAGCACC | probe | VIC | |
| qPleoN_f | 0.5 | CGTTCTTGGAACGCTGCATA | ~215–260 | STR locus Pati01 ** (nDNA) | --- |
| qPleoN_r | 0.5 | ATGGGCAGCACTCGTATGAT | --- | ||
| qPleoN_p | 0.25 | ATGCTACAGAAATAGAAGCCAA | probe | 6-FAM | |
| qPleoC_f | 0.5 | GAGACGAATACCAACCGGCA | 366 | IPC (Internal Positive Control) | --- |
| qPleoC_r | 0.5 | GGACCATGCTTGCGTTTGAG | --- | ||
| qPleoC_p | 0.25 | TCGACGATTCAAGCACGAT | probe | NED |
| qPCR Reaction Composition | 10 µL Reaction | Final Concentration in qPCR |
|---|---|---|
| 2× TaqMan Multiplex Master Mix | 5 µL | 1× |
| 20× qPleo mtDNA Assay Mix (20× qPleoM) | 0.5 µL | 1× |
| 20× qPleo nDNA Assay Mix (20× qPleoN) | 0.5 µL | 1× |
| 20× qPleo IPC DNA Assay Mix (20× qPleoC) | 0.5 µL | 1× |
| IPC DNA (0.1 pg/μL) | 1 µL | 0.1 pg |
| Template DNA | 1 µL | different |
| H2O | 1.5 µL |
| Name | Repeat Structure | Repetition | Size (bp *) | 5’ Primer Fluorescent Label |
|---|---|---|---|---|
| Pleo24 | (CA)n | 2n | 105–160 | FAM |
| Pleo30 | (ATGG)n (GATA)n (TAGA)n | 4n | 180–240 | FAM |
| Pleo23 | (ATGT)n (GT)n (AC)n (AT)n | 2n | 250–380 | FAM |
| Pleo22 | (TAGA)n | 4n | 105–168 | ATTO550 |
| Pleo31 | (GATA)n (GA)n | 4n | 175–220 | ATTO550 |
| Pleo32 | (TCTG)n (TCTA)n | 4n | 270–360 | ATTO550 |
| Pleo33 | (CAGA)n (TAGA)n | 4n | 180–280 | ATTO565 |
| STR Marker | Primer Sequence 5′ → 3′ |
|---|---|
| Pleo 24 | F: GTGTAGTTATGTGTATTATGAATGTGTGTATGC |
| R: AATATCTTAGCAGATGGAGCTGGG | |
| Pleo 30 | F: GGCCTTCTAACTTCCTTGCAGA |
| R: CATTTAGTTAGCCCATTTTCATCA | |
| Pleo 23 | F: CTGTTTGACAGTACAAGTATTACTGGCC |
| R: GGTCTATGTGTCTCTGTTTCCTCTTATG | |
| Pleo 22 | F: TAAGAATTTATGGATTACTCGGCAAAT |
| R: TATTCATTGTAGTCCCTGGGATTG | |
| Pleo 31 | F: GAGTTAGGACAAGATTATCAAGGAACTTG |
| R: CAGTCTGAGCTTAGAGTCTGCTCAAG | |
| Pleo 32 | F: GGGCAAATACACTAACCA |
| R: CTCCTGCTAGAATCTCCAA | |
| Pleo 33 | F: TCTTTGTTTGGCTATAACCATTCACTAG |
| R: AACCCAGTGTCTCCTTGTACCAC |
| PCR | 12.5 µL Reaction | Final Concentration in PCR |
|---|---|---|
| Gold Star 10× buffer | 1.25 µL | 1× |
| 10× Pleo STRPlex Primer Mix | 1.25 µL | 1× |
| Template DNA | different | 10 pg nuclear DNA |
| AmpliTaq Gold DNA polymerase | 0.25 µL | 2.5 U/PCR |
| H2O | to 12.5 µL |
| STRs | Alelles |
|---|---|
| Pleo24 | 15, 16, 19, 20, 21, 22, 23 |
| Pleo30 | 20, 21, 22, 23, 24, 25, 26, 27 |
| Pleo23 | 50, 51, 52, 53, 5, 56, 62, 65, 66, 67 |
| Pleo22 | 14, 14.1, 15, 16.1, 17, 18, 20 |
| Pleo31 | 18, 19, 20, 21 |
| Pleo32 | 11, 14, 15, 16, 17, 18, 19, 20 |
| Pleo33 | 13, 15, 16, 17, 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vankova, L.; Alaverdyan, J.; Vanek, D. Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping. Life 2025, 15, 664. https://doi.org/10.3390/life15040664
Vankova L, Alaverdyan J, Vanek D. Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping. Life. 2025; 15(4):664. https://doi.org/10.3390/life15040664
Chicago/Turabian StyleVankova, Lenka, Johana Alaverdyan, and Daniel Vanek. 2025. "Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping" Life 15, no. 4: 664. https://doi.org/10.3390/life15040664
APA StyleVankova, L., Alaverdyan, J., & Vanek, D. (2025). Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping. Life, 15(4), 664. https://doi.org/10.3390/life15040664







