Abstract
Flavodiiron proteins (FDPs, also called flavoproteins, Flvs) are modular enzymes widely present in Bacteria and Archaea. The evolution of cyanobacteria and oxygenic photosynthesis occurred in concert with the modulation of typical bacterial FDPs. Present cyanobacterial FDPs are composed of three domains, the β-lactamase-like, flavodoxin-like and flavin-reductase like domains. Cyanobacterial FDPs function as hetero- and homodimers and are involved in the regulation of photosynthetic electron transport. Whilst Flv2 and Flv4 proteins are limited to specific cyanobacterial species (β-cyanobacteria) and function in photoprotection of Photosystem II, Flv1 and Flv3 proteins, functioning in the “Mehler-like” reaction and safeguarding Photosystem I under fluctuating light conditions, occur in nearly all cyanobacteria and additionally in green algae, mosses and lycophytes. Filamentous cyanobacteria have additional FDPs in heterocyst cells, ensuring a microaerobic environment for the function of the nitrogenase enzyme under the light. Here, the evolution, occurrence and functional mechanisms of various FDPs in oxygenic photosynthetic organisms are discussed.
1. Introduction
Flavodiiron proteins (FDPs), previously called A-type flavoproteins (Flv) [1], are a large family of enzymes sharing sequence similarity. FDPs have been found mainly in anaerobic and some aerobic prokaryotes (Bacteria including cyanobacteria, and Archaea), and in Protozoa. Data mining of sequenced genomes has also led to the discovery of FDP homologs in some photosynthetic eukaryotes [2,3].
All FDPs share two conserved structural domains: the N-terminal metallo-β-lactamase-like domain, harboring a non-heme diiron center where O2 and/or NO reduction take place; and the C-terminal flavodoxin-like domain, containing a flavin mononucleotide (FMN) moiety [4,5]. X-ray crystallography of FDPs from different organisms has provided valuable data for the elucidation of the electron transfer properties in the active site during O2 and/or NO reduction [6,7,8,9]. The functional form of FDPs in anaerobic prokaryotes and eukaryotic protozoa has been resolved as a homodimer or homotetramer arranged in a “head to tail” configuration so that the diiron center of one monomer and the FMN in the other monomer closely contact each other, which ensures fast electron transfer between the cofactors.
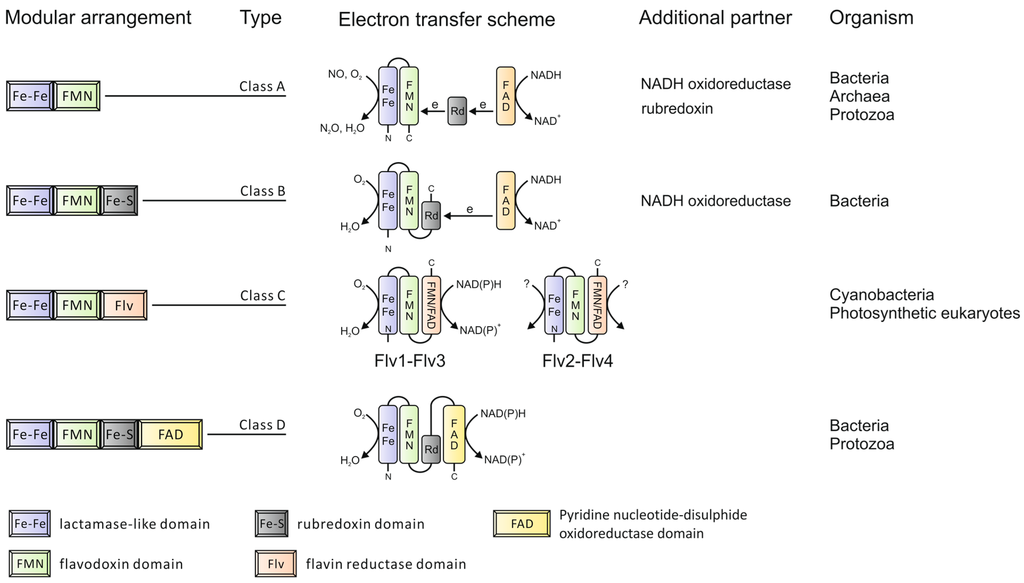
Figure 1.
Modular organization of the Flavodiiron protein (FDP) family.
Figure 1.
Modular organization of the Flavodiiron protein (FDP) family.
In addition to the common sequence core, some FDPs also have C-terminal extensions. Based on these C-terminal extensions, FDPs can be grouped into four classes [5] as depicted in Figure 1. The majority of FDPs belong to Class A, which is the simplest type with the shortest extension sequences, representing the minimal core structure. These can be found in Bacteria, Archaea and Protozoa. Class B FDPs are found in Enterobacteria, whilst Class D FDPs are present in some Bacteria and Protozoa. Class C FDPs seem to be specific to oxygenic photosynthetic organisms. The additional flavin reductase-like domain in this specific class makes it possible for nicotinamide adenine dinucleotide (phosphate), reduced form (NAD(P)H) to be directly used as an electron donor. The extension component is also coupled with extra cofactor(s), which bring additional features to FDPs during electron transfer. The number of redox partners required during electron transfer depends on the modular arrangement of FDPs. The more complex the FDPs, the fewer partners are involved. For example, rubredoxin donates electrons to many FDPs from Class A but is not needed for FDPs from Class B, which have a rubredoxin domain fused in the polypeptide.
The gene organization of FDPs in different organisms indicates a complex evolution. For example, a rubredoxin electron donor of Desulfovibrio gigas rubredoxin:oxygen oxidoreductase, Dg_ROO (Class A FDP), is encoded in the same operon as ROO [10] and the Escherichia coli flavorubredoxin (Class B FDP) and its partner nicotinamide adenine dinucleotide, reduced form (NADH): flavorubredoxin oxidoreductase are encoded by the same operon. Similar gene organizations have also been found in other Bacteria and Archaea. More interestingly, proteins homologous to the flavin-reductase domain of Class C FDPs are detected in some Bacteria and Archaea, but they are not in the same cistronic unit as FDPs. An example of the latter can be found in Nodularia spumigena CCY 9414, whose flavin-reductase-like protein is encoded directly downstream of a Class A FDP gene within the same operon. Thus, the complexity of the FDPs may result from multiple genome rearrangements and gene fusions during evolution.
The FDPs from anaerobic species have been proposed to protect against both O2 and/or NO toxicity by catalyzing the final step of O2 and/or NO reduction. It is worth mentioning that some FDPs act preferably as NO-reductases [11,12], others as O2-reductase [6,8,13], whereas some FDPs can catalyze both reactions [14]. Additionally, there seems to be functional relationship between respiratory terminal oxidases and the catalytic activity of FDPs. In Giardia, which lacks both the respiratory oxidases and reactive oxygen species (ROS) scavenging enzymes, FDP shows a high O2-reductase activity (>40 s−1), but very low NO-reductase activity (~0.2 s−1) [6]. In organisms containing respiratory oxidases (Escherichia coli, Desulfovibrio gigas, Desulfovibrio vulgaris, Moorella), FDPs are known to have either strict NO-reductase function or dual function, thus cooperating with respiratory oxidases in protection against O2 toxicity.
2. flv Genes in Oxygenic Photosynthetic Organisms
2.1. The flv Gene Family and its Organization in Cyanobacterial Genomes
Genes encoding FDPs (flv) can be found in most sequenced cyanobacteria, including the obligatory photoautotrophic species. Different cyanobacterial strains may possess several copies (2–6) of flv genes, thus comprising a small family encoding different FDPs. We have earlier shown that FDPs in oxygenic photosynthetic organisms can be grouped into almost symmetrical clusters (cluster A, including Flv1 and Flv2, and cluster B, including Flv3 and Flv4) and appear in pair(s) (flv1-flv3 or flv2-flv4) [2]. The distribution of the clusters is depicted in Figure 2 and the number of flv genes and their organization in the genomes of oxygenic photosynthetic organisms are summarized in Table 1. Cyanobacteria can be divided into two groups depending on RubBisCO and carboxysome types: α- and β-cyanobacteria [15,16,17]. The α-cyanobacteria have only one pair of flv genes (flv1-flv3). Some unicellular and filamentous, non-heterocystous β-cyanobacteria, including the model unicellular cyanobacterium Synechocystis sp. PCC 6803, possess the flv2-flv4 pair in addition to flv1-flv3. All heterocystous filamentous β-cyanobacteria contain two pairs of flv1-flv3, designated as flv1a-flv3a and flv1b-flv3b, thus having 4 or 6 flv genes depending on the presence of the flv2-flv4 pair. It seems that the flv1-flv3 pair is largely present in all organisms containing C Class FDPs, whereas the flv2-flv4 pair is present only in some β-cyanobacteria.
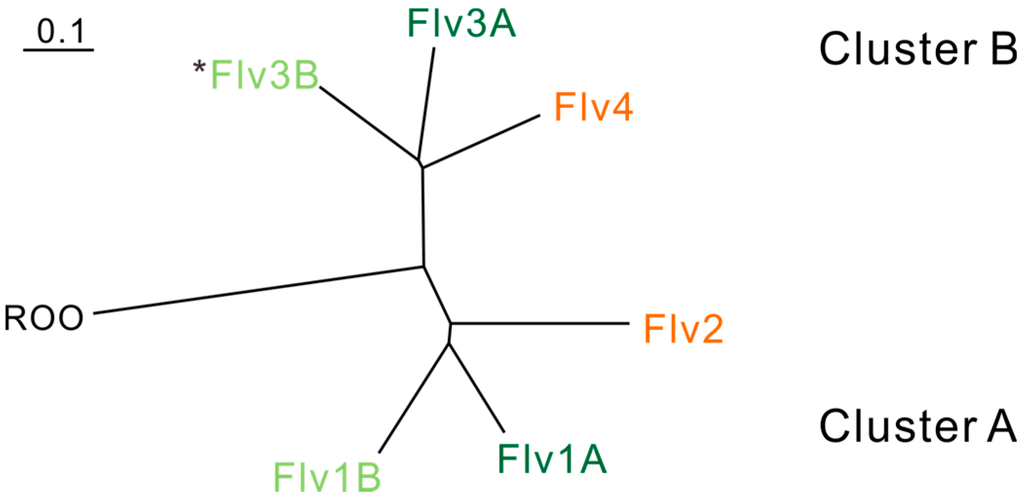
Figure 2.
Relationship between cyanobacterial FDPs based on sequence similarity and physiological functions. Phylogenic analysis is based on FDPs in Anabaena sp. PCC 7120. ROO (rubredoxin:oxygen oxidoreductase) from Desulfovibrio gigas is used as an outgroup. The FDPs functioning in the photoprotection of Photosystem II are indicated in orange. The FDPs functioning in the Mehler-like reaction and protection of Photosystem I are indicated in green. The FDPs indicated in dark green function in vegetative cells, and those in light green function in heterocysts. *Flv3B can be arranged and function as a homodimer [18].
Figure 2.
Relationship between cyanobacterial FDPs based on sequence similarity and physiological functions. Phylogenic analysis is based on FDPs in Anabaena sp. PCC 7120. ROO (rubredoxin:oxygen oxidoreductase) from Desulfovibrio gigas is used as an outgroup. The FDPs functioning in the photoprotection of Photosystem II are indicated in orange. The FDPs functioning in the Mehler-like reaction and protection of Photosystem I are indicated in green. The FDPs indicated in dark green function in vegetative cells, and those in light green function in heterocysts. *Flv3B can be arranged and function as a homodimer [18].
A further survey of the flv gene loci indicated that the genes encoding many of the flv paralog pairs are localized sequentially in genomes. Usually, the flv2 and flv4 genes are organized as an operon consisting of three genes arranged as in tandem orientation in the same direction of transcription (flv4-ORF-flv2). The structure of the flv4-ORF-flv2 operon is highly conserved, except for Microcoleus sp. PCC 7113, which possesses five ORFs between flv2 and flv4 genes (Table 1). In some cases, flv4 and flv2 are also located far away from each other and probably do not organize an operon.
The organization of the flv1 and flv3 genes is not conserved. The majority of α-cyanobacteria arrange flv1 and flv3 in the flv3-flv1 operons, whilst in β-cyanobacteria, the genes might be organized as flv3a-flv1a operons, or separated by 1–5 ORF(s) (designated as flv3a-ORF(s)-flv1a in Table 1), or otherwise be spread out in the genome. In contrast, the flv1b and flv3b genes are always arranged together in the flv3b-flv1b operon and are likely co-transcribed, as demonstrated in Anabaena sp. PCC 7120 [19]. In the genomes of eukaryotes, flv3-flv1 are not usually clustered together. However, some eukaryotes retain the clustering. For instance, Paulinella chromatophora, the photosynthetic protozoa bearing photosynthetic entities (chromatophores), derived from cyanobacteria [20]. In some green algae, flv1-flv3 also retain clustering, even though ORF(s) exist in between.

Table 1.
Genes encoding FDPs in cyanobacteria and photosynthetic eukaryotes.
| No. of flv's | flv1(a) | flv3(a) | flv2 | flv4 | flv1b | flv3b | Gene organization (flv1(a), flv3(a)) | Gene organization (flv2, flv4) | Gene organization (flv1b, flv3b) | |
|---|---|---|---|---|---|---|---|---|---|---|
| α-Cyanobacteria (unicellular) | ||||||||||
| Cyanobium gracile PCC 6307 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus AS9601 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MED4 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MIT 9202 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MIT 9211 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MIT 9215 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MIT 9301 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MIT 9303 | 2 | * | ||||||||
| Prochlorococcus marinus MIT 9312 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus MIT9313 | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus NATL1A | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus NATL2A | 2 | →flv3-flv1→ | ||||||||
| Prochlorococcus marinus SS120 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus BL107 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus CB0101 | 2 | * | ||||||||
| Synechococcus CB0205 | 2 | →flv3-6 ORF's flv1→ | ||||||||
| Synechococcus CC9311 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus CC9605 | 2 | →flv3-WP_011365453-flv1→ | ||||||||
| Synechococcus CC9902 | 2 | →flv3-2 ORF's-flv1→ | ||||||||
| Synechococcus RCC307 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus RS9916 | 2 | →flv3-WP_007099263-flv1→ | ||||||||
| Synechococcus RS9917 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus WH 7805 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus WH 8102 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus WH 8109 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus WH 5701 | 2 | →flv-WP_00617255-flv1→ | ||||||||
| Synechococcus WH 7803 | 2 | →flv3-flv1→ | ||||||||
| β-Cyanobacteria (unicellular) | ||||||||||
| Acaryochloris marina MBIC11017 | 2 | →flv3-AM1_1385-flv1→ | ||||||||
| Chroococcidiopsis thermalis PCC 7203 | 2 | →flv3-2 ORF's-flv1→ | ||||||||
| Cyanobacterium aponinum PCC 10605 | 2 | * | ||||||||
| Cyanobacterium stanieri PCC 7202 | 2 | * | ||||||||
| Cyanobacterium UCYN-A | 0 | * | ||||||||
| Cyanothece CCY 0110 | 4 | * | →flv4-WP_008278275-flv2→ | |||||||
| Cyanothece PCC 7424 | 4 | * | →flv4-PCC7424_0480-flv2→ | |||||||
| Cyanothece PCC 7425 | 2 | →flv3-WP_012627311-flv1→ | ||||||||
| Cyanothece PCC 7822 | 4 | * | →flv4-Cyan7822_3509-flv2→ | |||||||
| Cyanothece PCC 8801 | 4 | * | →flv4-PCC8801_3605-flv2→ | |||||||
| Cyanothece PCC 8802 | 4 | * | →flv4-Cyan8802_2509-flv2→ | |||||||
| Crocosphaera watsonii WH 8501 | 2 | * | ||||||||
| Dactylococcopsis salina PCC 8305 | 2 | →flv3-flv1→ | ||||||||
| Gloeobacter kilaueensis JS1 | 2 | →flv3-flv1→ | ||||||||
| Gloeobacter violaceus PCC 7421 | 2 | →flv3-flv1→ | ||||||||
| Gloeocapsa PCC 7428 | 2 | →flv3-flv1→ | ||||||||
| Halothece PCC 7418 | 4 | * | →flv4-PCC7418_1461-flv2→ | |||||||
| Microcystis aeruginosa NIES-843 | 4 | * | →flv4-YP_001660097-flv2→ | |||||||
| Microcystis aeruginosa PCC 7806 | 4 | * | →flv4-IPF_2587-flv2→ | |||||||
| Pleurocapsa PCC 7327 | 4 | →flv3-Ple7327_0831-flv1→ | →flv4-Ple7327_3773-flv2→ | |||||||
| Stanieria cyanosphaera PCC 7437 | 4 | * | →flv4-Sta7437_3860-flv2→ | |||||||
| Synechococcus PCC 7335 | 4 | * | * | |||||||
| Synechococcus PCC 7002 | 2 | * | ||||||||
| Synechococcus PCC 6312 | 2 | * | ||||||||
| Synechococcus PCC 7502 | 2 | * | ||||||||
| Synechococcus JA-3-3Ab | 2 | * | ||||||||
| Synechococcus JA-2-3B'a(2-13) | 2 | * | ||||||||
| Synechococcus elongatus PCC 7942 | 2 | →flv3-flv1→ | ||||||||
| Synechococcus elongatus PCC 6301 | 2 | →flv3-flv1→ | ||||||||
| Synechocystis PCC 6803 | 4 | * | →flv4-sll0218-flv2→ | |||||||
| Synechocystis PCC 6714 | 2 | →flv3-2 ORF's-flv1→ | ||||||||
| Thermosynechococcus NK55a | 2 | * | ||||||||
| Thermosynechococcus elongatus BP-1 | 2 | * | ||||||||
| β-Cyanobacteria (filamentous) | ||||||||||
| Anabaena 90 | 4 | →flv3-WP_015078091-flv1→ | →flv3b-flv1b→ | |||||||
| Anabaena PCC 7120 | 6 | →flv3a-all3892,all3893,all3894-flv1a→ | →flv4-all4445-flv2→ | →flv3b-flv1b→ | ||||||
| Anabaena cylindrica PCC 7122 | 4 | →flv3-flv1→ | →flv3b-flv1b→ | |||||||
| Anabaena variabilis ATCC 29413 | 6 | →flv3a-flv1a→ | →flv4-Ava_1370-flv2→ | →flv3b-flv1b→ | ||||||
| Arthrospira platensis NIES-39 | 2 | →flv3-flv1→ | ||||||||
| Calothrix 336/3 | 4 | →flv3-Cal336_3958-flv1→ | →flv3b–flv1b→ | |||||||
| Calothrix PCC 6303 | 4 | →flv3-flv1→ | →flv3b-flv1b→ | |||||||
| Calothrix PCC 7507 | 6 | →flv3a-flv1a→ | →flv4-Cal7507_5629-flv2→ | →flv3b-flv1b→ | ||||||
| Chamaesiphon minutus PCC 6605 | 2 | →flv3-2 ORF's-flv1→ | ||||||||
| Chloroflexus aurantiacus J-10-fl | 0 | |||||||||
| Cylindrospermopsis raciborskii CS-505 | 4 | →flv3-flv1→ | →flv3b-flv1b→ | |||||||
| Cylindrospermum stagnale PCC 7417 | 4 | →flv3-flv1→ | →flv3b-flv1b→ | |||||||
| Crinalium epipsammum PCC 9333 | 2 | →flv3-YP_007142380-flv1→ | ||||||||
| Geitlerinema PCC 7407 | 2 | →flv3-flv1→ | ||||||||
| Geminocystis herdmanii PCC 6308 | 0 | |||||||||
| Leptolyngbya PCC 7376 | 4 | * | →flv4-Lepto7376_3457-flv2→ | |||||||
| Lyngbya majuscula 3L | 2 | →flv3-flv1→ | ||||||||
| Lyngbya PCC 8106 | 2 | →flv3-WP_009783639-flv1→ | ||||||||
| Microcoleus chthonoplastes PCC 7420 | 4 | * | * | |||||||
| Microcoleus PCC 7113 | 4 | →flv3-3 ORF's-flv1→ | →flv4-5 ORF's-flv2→ | |||||||
| Microcoleus vaginatus FGP-2 | 2 | →flv3-EGK88546-flv1→ | ||||||||
| Nodularia spumigena CCY 9414 ** | 6 | * | →flv4-flv2→ | →flv3b-flv1b→ | ||||||
| Nostoc PCC 7107 | 6 | →flv3a-10 ORF's-flv1a→ | →flv4-WP_015113616-flv2→ | →flv3b-flv1b→ | ||||||
| Nostoc PCC 7524 | 6 | →flv3a-6 ORF's-flv1a→ | →flv4-Nos7524_2687-flv2→ | →flv3b-flv1b→ | ||||||
| Nostoc punctiforme ATCC 29133 | 5 | →flv3a-flv1a→ | →Npun_R0592-flv2→ | →flv3b-flv1b→ | ||||||
| Nostoc azollae 0708 | 4 | →flv3-flv1→ | →flv3b-flv1b→ | |||||||
| Oscillatoria PCC 6506 | 2 | * | ||||||||
| Oscillatoria acuminata PCC 6304 | 2 | →flv3-flv1→ | ||||||||
| Oscillatoria nigroviridis PCC 7112 | 2 | →flv3-Osc7112_2977-flv1→ | ||||||||
| Oscillatoriales JSC-1 | 2 | * | ||||||||
| Planktothrix agardhii NIVA-CYA 126/8 | 2 | →flv3-flv1→ | ||||||||
| Pseudanabaena sp. PCC 7367 | 4 | →flv3-flv1→ | →flv4-Pse7367_3922-flv2→ | |||||||
| Raphidiopsis brookii D9 | 2 | →flv3-flv1→ | ||||||||
| Rivularia PCC 7116 | 6 | →flv3a-flv1a→ | →flv4-Riv7116_6032-flv2→ | →flv3b-flv1b→ | ||||||
| Trichodesmium erythraeum IMS101 | 2 | * | ||||||||
| Photosynthetic protozoa | ||||||||||
| Paulinella chromatophora | 2 | →flv3-flv1→ | ||||||||
| Green algae | ||||||||||
| Chlamydomonas reinhardtii | 2 | * | ||||||||
| Chlorella variabilis | 2 | * | ||||||||
| Micromonas pusilla CCMP1545 | 2 | * | ||||||||
| Micromonas RCC299 | 2 | * | ||||||||
| Ostreococcus lucimarinus CCE9901 | 2 | →flv3-XP_001416099-flv1→ | ||||||||
| Ostreococcus tauri | 2 (+2)# | →flv3-3 ORF's-flv1→ | ||||||||
| Volvox carteri f. nagariensis | 2 | * | ||||||||
| Land plants | ||||||||||
| Dysgonomonas mossii DSM 22836 | 1 | |||||||||
| Physcomitrella patens subsp. patens | 2 | * | ||||||||
| Selaginella moellendorffii | 2 | * | ||||||||
| Arabidopsis thaliana | 0 | |||||||||
| Picea sitchensis*** | 1 | |||||||||
| Symbiodinium ## | ||||||||||
| Symbiodinium Avir (clade A1) | 2 | * | ||||||||
| Symbiodinium FlAp1 (clade B1) | 2 | * | ||||||||
| Symbiodinium Mf1.5b (clade B1) | 2 | * | ||||||||
| Symbiodinium Pd44b ( clade F1) | 2 | * | ||||||||
The filamentous heterocystous cyanobacteria are marked with a gray background; * FDP-encoding gene orthologs are present but lacking gene organization; ** The FDP-encoding gene is splited into two genes, coding for a Class A FDP and flavin reductase, and situated sequentially in one polycistron; *** possess a Class A FDP; # possess two absolutely identical extra copies; ## [21]
2.2. Expression and Regulation of flv Genes
A joint analysis of global transcriptomics and proteomics data can provide useful insights into the function of specific proteins and their regulation by environmental cues. Synechocystis sp. PCC 6803 has four genes (sll1521, sll0219, sll0550 and sll0217) encoding FDPs (Flv1, Flv2, Flv3, and Flv4, respectively). Genome-wide DNA microarray data have shown that the transcription of the flv2, flv4 and flv3 genes of Synechocystis sp. PCC 6803 is strongly affected by various environmental factors (Table 2). Among all studied conditions, the most remarkable changes in flv3, flv2 and flv4 transcript levels were observed under Ci-limitation, Fe-depletion and different dark/light regimes. The transcript abundance of the flv1 gene in Synechocystis sp. PCC 6803 is significantly lower under standard growth conditions [2]. Such a low abundance of flv1 transcript and also of the Flv1 protein might result from the presence of antisense-RNA (as-RNA) for the flv1 gene (as-flv1) [22]. However, more detailed expression studies of as-flv1 under different environmental conditions would be required to make a strong conclusion. In contrast to other flvs, the flv1 transcript level does not respond to high light or Ci-limitation (Table 2). Instead, a low induction in flv1 transcript amount was observed under oxidative (methyl viologen and H2O2 treatment) and heat stress conditions (Table 2). Analysis of the flv1 expression level by RT-PCR demonstrated 3.5 and 4.2 fold increases after 1 h and 12 h nitrosative stress treatments, respectively [23]. A differential expression pattern of flv1 and flv3 in Synechocystis sp. PCC 6803 is also consistent with the fact that, in this species, the flv1 and flv3 genes are spread out in the genome (Table 1).
Differing from Synechocystis sp. PCC 6803, the flv1a and flv3a genes in Anabaena sp. PCC 7120 are clustered in the genome and this might suggest an operon organization (Table 1). The transcript level of flv1a is only slightly lower (3–4 times) than that of flv3a and the expression levels of both transcripts have been shown to respond positively to both high light and low CO2 treatment [24,25]. However, RNA-seq data [19,26] and transcript level analysis of the respective flv deletion mutants [27] suggest that flv1a and flv3a in Anabaena sp. PCC 7120 are likely transcribed independently. The extra pair of flv genes in Anabaena sp. PCC 7120, the flv1b and flv3b genes, which are arranged together in an operon and are likely co-transcribed [19], demonstrate clearly different expression profiles from the other flv genes in Synechocystis sp. PCC 6803 and Anabaena sp. PCC 7120. The transcript levels of flv1b and flv3b do not respond to changes in carbon or light regime, but substantially increase under N2-fixing conditions. The response is similar to that of the nifH gene, which encodes a subunit of the nitrogenase enzyme in heterocysts [24]. Interestingly, the flv1b transcript level remains somewhat lower than that of flv3b.
The significant up-regulation of Flv3 and Flv2 proteins under low CO2 levels, observed in Isobaric tags for relative and absolute quantitation (iTRAQ) shotgun analysis, is consistent with the above-mentioned microarray studies in Synechocystis sp. PCC 6803 (Table 1 and Table 3). Interestingly, the Flv3, and to some extent, also the Flv1 proteins, exhibited a low expression level under chemoheterotrophic condition (Table 3), implying that the function of these proteins is not essential in darkness. Moreover, iTRAQ observations demonstrated an accumulation of two extra FDPs (Flv1B, Flv3B) in the heterocyst-enriched cell fractions of N2-fixing, filamentous Anabaena sp. PCC 7120 and Nostoc punctiforme [28,29].

Table 2.
Expression of flv genes in Synechocystis sp. PCC 6803 under various environmental conditions.
| Conditions | Log2 fold change | Database | Data accession | References | |||||
|---|---|---|---|---|---|---|---|---|---|
| flv1 | flv3 | flv2 | flv4 | ||||||
| From darkness (12h) to light (4h) | 1.64 | KEGG | ex0001365/70 & ex0000868/73 | [30] Kucho et al., 2005 | |||||
| From darkness (12h) to light (12h) | 0.98 | 0.53 | KEGG | ex0001377/82 & ex0000880/5 | [30] Kucho et al., 2005 | ||||
| From darkness (12h) to light (24h) | −0.89 | KEGG | ex0001395/400 & ex0000898/903 | [30] Kucho et al., 2005 | |||||
| From darkness (12h) to light (28h) | −0.67 | KEGG | ex0001401/6 & ex0000904/09 | [30] Kucho et al., 2005 | |||||
| From darkness (12h) to light (32h) | −0.76 | KEGG | ex0001407/12 & ex0000910/5 | [30] Kucho et al., 2005 | |||||
| From darkness (12h) to light (44h) | −0.90 | KEGG | ex0001425/30 & ex0000928/33 | [30] Kucho et al., 2005 | |||||
| From light to darkness (30 min) | −0.81 | −2.64 | −1.62 | GEO | GSE45667 | [31] Lehmann et al., 2013 | |||
| From light to darkness (5.5 h) | −2.88 | −2.03 | GEO | GSE45667 | [31] Lehmann et al., 2013 | ||||
| From light to darkness (11.5h) | −0.68 | −2.78 | −1.85 | GEO | GSE45667 | [31] Lehmann et al., 2013 | |||
| From light to darkness (1h) | −079 | GEO | GSE16162 | [22] Mitschke et al., 2011 | |||||
| From darkness to light (30 min) | −0.68 | −3.54 | −2.85 | GEO | GSE45667 | [31] Lehmann et al., 2013 | |||
| From darkness to light (5.5h) | −0.88 | −3.30 | −2.56 | GEO | GSE45667 | [31] Lehmann et al., 2013 | |||
| HL_15min | 0.54 | 0.70 | ArrayExpress | E-TABM-333 | [32] Singh et al., 2008 | ||||
| HL_1h | 0.93 | 1.00 | ArrayExpress | E-TABM-333 | [32] Singh et al., 2008 | ||||
| HL_2h | 2.04 | 2.40 | ArrayExpress | E-TABM-333 | [32] Singh et al., 2008 | ||||
| HL_3h | 0.64 | 0.81 | ArrayExpress | E-TABM-333 | [32] Singh et al., 2008 | ||||
| HL_4h | 1.26 | 1.47 | ArrayExpress | E-TABM-333 | [32] Singh et al., 2008 | ||||
| HL(2)_15min | 2.91 | 2.42 | KEGG | ex0000140/3 & ex0000160/1 | [33] Hihara et al., 2001 | ||||
| HL(2)_1h | 0.55 | KEGG | ex0000144/7 & ex0000152/3 | [33] Hihara et al., 2001 | |||||
| HL | 1.40 | 0.94 | GEO | GSE16162 | [22] Mitschke et al., 2011 | ||||
| 3h illumination with red and blue light | −3.42 | −2.22 | ArrayExpress | E-TABM-339 | [34] Singh et al., 2009 | ||||
| 6h illumination with red and blue light | 1.14 | 0.74 | ArrayExpress | E-TABM-339 | [35] Singh et al., 2009 | ||||
| Methyl viologen_high light | −0.93 | 1.28 | 1.36 | KEGG | ex0001349,54,55 | [36] Kobayashi et al., 2004 | |||
| Methyl viologen_moderate light | 1.12 | KEGG | ex0001441/4 | [36] Kobayashi et al., 2004 | |||||
| 15 min treatment with 3mM H2O2 | 0.72 | −2.12 | 0.50 | GEO | GSE3703 | [37] Houot et al., 2007 | |||
| 30 min treatment with 3mM H2O2 | 1.28 | 0.73 | 0.70 | GEO | GSE3703 | [37] Houot et al., 2007 | |||
| 22C_20min | −0.66 | KEGG | ex0000002/3_ex0000012/3_14/5 | [38] Suzuki et al., 2001 | |||||
| 24C_20min | −1.20 | −0.55 | KEGG | ex0001878/9 | [39] Prakash et al., 2010 | ||||
| 24C_60min | −0.86 | −0.66 | KEGG | ex0001880/1 | [39] Prakash et al., 2010 | ||||
| 24C_180min | −1.15 | −0.93 | KEGG | ex0001882/3 | [39] Prakash et al., 2010 | ||||
| 22C_20min_(2) | 1.17 | −1.28 | KEGG | ex0001839/40 | [40] Panichkin et al., 2006 | ||||
| heat_30min | 0.59 | 0.66 | 0.62 | GEO | GSE21133 | [41] Rowland et al., 2010 | |||
| heat_1h | 0.68 | 0.67 | 0.81 | GEO | GSE21133 | [41] Rowland et al., 2010 | |||
| heat_2h | 0.69 | 0.59 | 0.76 | GEO | GSE21133 | [41] Rowland et al., 2010 | |||
| heat_4h | 0.57 | 0.76 | GEO | GSE21133 | [41] Rowland et al., 2010 | ||||
| heat_8h | 0.60 | 0.60 | 0.75 | GEO | GSE21133 | [41] Rowland et al., 2010 | |||
| Ci_depletion | 1.39 | 3.07 | 1.80 | GEO | GSE16162 | [22] Mitschke et al., 2011 | |||
| CO2_limitation_1h | −0.55 | GEO | GSE1695 | [42] Wang et al., 2004 | |||||
| CO2_limitation_3h | 0.66 | −0.56 | GEO | GSE1695 | [42] Wang et al., 2004 | ||||
| CO2_limitation_3.3h | 1.29 | 7.26 | 5.70 | GEO | GSE1695 | [42] Wang et al., 2004 | |||
| CO2_limitation_6h | 2.00 | 7.16 | 5.62 | GEO | GSE1695 | [42] Wang et al., 2004 | |||
| CO2_limitation_12h | 2.15 | 6.95 | 5.61 | GEO | GSE1695 | [42] Wang et al., 2004 | |||
| CO2_limitation_24h | 2.44 | 6.88 | 6.51 | [43] Eisenhut et al., 2007 | |||||
| high_CO2_24h_vs_low_CO2_3h | −1.00 | −3.77 | −3.27 | GEO | GSE31672 | [44] Hackenberg et al., 2012 | |||
| high_CO2_vs_low_CO2_24h | −2.29 | −5.09 | −4.32 | GEO | GSE31672 | [44] Hackenberg et al., 2012 | |||
| Cd_15min | 1.07 | GEO | GSE3682 | [37] Houot et al., 2007 | |||||
| Cd_1.5h | −0.67 | GEO | GSE3682 | [37] Houot et al., 2007 | |||||
| Cd_3h | 0.81 | −1.05 | GEO | GSE3682 | [37] Houot et al., 2007 | ||||
| Cd_5h | 0.79 | 1.34 | GEO | GSE3682 | [37] Houot et al., 2007 | ||||
| Cd_6h | 0,71 | 1,64 | GEO | GSE3682 | [37] Houot et al., 2007 | ||||
| Cd_16h | GEO | GSE3682 | [37] Houot et al., 2007 | ||||||
| Zn_excess_240min | −0.53 | 2.39 | 1.56 | GEO | GSE3716 | [37] Houot et al., 2007 | |||
| shift from 2mM to 0.5 μM Fe- 96h | −0.58 | −1.77 | −0.94 | GEO | GSE3717 | [37] Houot et al., 2007 | |||
| shift from 1mM to 0.5 μM Fe- 96h | 0.79 | 1.88 | 1.64 | GEO | GSE3717 | [37] Houot et al., 2007 | |||
| Fe depletion 3h | −1.10 | −5.67 | −6.02 | GEO | GSE39804 | [45] Hernández-Prieto et al., 2012 | |||
| Fe depletion 12h | −1.18 | −6.14 | −5.80 | GEO | GSE39804 | [45] Hernández-Prieto et al., 2012 | |||
| Fe depletion 24h | −0.80 | −3.23 | −3.19 | GEO | GSE39804 | [45] Hernández-Prieto et al., 2012 | |||
| Fe depletion 48h | −1.17 | −5.83 | −6.28 | GEO | GSE39804 | [45] Hernández-Prieto et al., 2012 | |||
| Fe depletion 72h | −0.89 | −6.04 | −6.08 | GEO | GSE39804 | [45] Hernández-Prieto et al., 2012 | |||
| Fe_high_4h | 1.19 | −0.62 | GEO | GSE3715 | [37] Houot et al., 2007 | ||||
| Novobiocin | −1.13 | KEGG | ex0001825/6 | [46] Prakash et al., 2009 | |||||
| Novobiocin + heat stress | −1.50 | −1.87 | KEGG | ex0001831/34 | [46] Prakash et al., 2009 | ||||
| Novobiocin + low temperature | −1.03 | −0.72 | KEGG | ex0001827/30 | [46] Prakash et al., 2009 | ||||
| Novobiocim treatment + salt stress | 1.12 | −1.10 | KEGG | ex0001835/38 | [46] Prakash et al., 2009 | ||||
| NaCl | −0.71 | KEGG | ex0001687/90 | [47] Shoumskaya et al., 2005 | |||||
| 0.5 M NaCl | −0.77 | −3.66 | −3.37 | GEO | GSE37482 | [48] Dickson et al., 2012 | |||
| Micro-oxic | 0.58 | 1.14 | 1.82 | GEO | GSE24882 | [49] Summerfield et al., 2011 | |||
| Cells encapsulated in silico gel | −2.68 | −0.92 | GEO | GSE37482 | [48] Dickson et al., 2012 | ||||
| Acid stress | −0.49 | −0.49 | [50] Ohta et al., 2005 | ||||||

Table 3.
Expression of FDPs under various environmental conditions. (The Flv1B or Flv3B proteins are marked with *.)
| Conditions | Expression | Strain | Technique | Reference | |||
|---|---|---|---|---|---|---|---|
| Flv1 (A) or B* | Flv3 (A) or B* | Flv2 | Flv4 | ||||
| CO2 limitation 72 h | ↑ 2.41 | ↑ 1.64 | Synechocystis 6803 | iTRAQ shortgun | [51] Battchikova et al., 2010 | ||
| 684 mM NaCl 5 days | ↑ 5.6 | Synechocystis 6803 | 2D gel proteomics | [52] Fulda et al., 2006 | |||
| Chemoheterotrophic growth | ↓ ~3.4 | Synechocystis 6803 | 2D gel proteomics | [53] Kurian et al., 2006 | |||
| Diazotrophic: ammonium | *↑ 2.2 | * ↑ 1.6 | *↑ 1.8 | Anabaena 7120 | iTRAQ shortgun | [28] Ow et al., 2008 | |
| Heterocystis: vegetative cells | *↑ 1.8 | *↑ 3.8 | 0.80 | Anabaena 7120 | iTRAQ shortgun | [28] Ow et al., 2008 | |
| Diazotrophic: ammonium | *↑ 2.46 | Nostoc punctiforme | iTRAQ shortgun | [29] Ow et al., 2009 | |||
| Heterocyst: diazotrophic | *↑ 3.44 | *↑ 2.20 | Nostoc punctiforme | iTRAQ shortgun | [29] Ow et al., 2009 | ||
2.2.1. The flv4-flv2 Operon Is Regulated by NdhR and Antisense-RNA
The flv2 and flv4 genes in Synechocystis sp. PCC 6803 are organized in a three-cistronic flv4-sll0218-flv2 operon, hereafter designated as flv4-2 operon and coding for Flv4, the small protein Sll0218 and Flv2 proteins. The expression of the flv4-2 operon in the Synechocystis sp. PCC 6803 cells is tightly controlled. The operon is positively regulated by the transcription factor NdhR and negatively regulated by as-RNA, designated As1_flv4 [54]. Consequently, overexpression of As1_flv4 has been demonstrated to lead to a decrease in flv4-2 mRNA levels. The transient induction of the as1_flv4 promotor during the exposure of Synechocystis sp. PCC 6803 cells to an environmental challenge results in the momentary, but efficient, prevention of premature expression of flv4-2. This regulation avoids the potential for energy being wasted in unfavorable protein synthesis upon transient environmental changes.
3. FDPs and Their Physiological Roles in Oxygenic Photosynthetic Organisms
All FDPs found in oxygenic photosynthetic organisms belong to Class C (except in Picea sitchensis). This observation suggests that the fusion of a flavin reductase-like domain at the C-terminal might have specific roles for the function and sustainability of oxygenic photosynthesis. Overexpression and characterization of the Synechocystis sp. PCC 6803 Flv3 protein and it is truncated form (consisting of the C-terminal flavin-reductase-like domain) in E. coli have demonstrated that Flv3 functions as NAD(P)H:oxygen oxidoreductase and is able to fully reduce O2 to water, without the production of ROS. Surprisingly, the recombinant Flv3 protein demonstrated a higher affinity to NADH than NADPH during in vitro studies [55].
Since most studies of FDPs from photosynthetic organisms are focused on the proteins from Synechocystis sp. PCC 6803 and Anabaena sp. PCC 7120, in this review, the function of FDPs is discussed mainly based on two functional pairs in Synechocystis sp. PCC 6803 (Flv2/Flv4, Flv1/Flv3) and the extra FDPs in Anabaena sp. PCC 7120 (Flv1B and Flv3B).
3.1. The Role of Flv1and Flv3
3.1.1. Mehler and “Mehler-like” Reactions
The FDPs in anaerobic species play a crucial protective role upon the transient exposure of the microorganism to O2. This is because of the unique capacity of FDPs to reduce O2 to water in a direct and safe way, without the formation of ROS [55].
Photoautotrophic organisms, like cyanobacteria, algae and plants, produce O2 as a by-product of their photosynthetic activity and concomitantly generate various negatively charged redox carriers in electron transfer processes occurring in the same compartment. Such combinations, especially where photosensitive pigments exist in oxic conditions, are likely to produce ROS, which are potentially hazardous to biological systems, particularly to the photosynthetic apparatus. Importantly, at lower concentrations ROS can also function as a signaling molecule to stimulate cellular defense and acclimation [56]. Hence, oxygenic phototrophic organisms need a sophisticated system to protect themselves against dangerous attacks by ROS generated during illumination of the light harvesting antenna as well as the Photosystem I (PSI) and Photosystem II (PSII) reaction centers of the photosynthetic apparatus.
Chloroplasts have developed various enzymatic (catalases, superoxide dismutase, peroxidases) and non-enzymatic (glutathione, carotenoids, tocopherol) defense systems for the efficient scavenging of ROS. Photoreduction of O2 to H2O2 by the photosynthetic electron transport chain was described for the first time in chloroplasts by Mehler [57,58], and is therefore known as the Mehler reaction. Later on, the primary product of photoreduction of O2 was identified as a superoxide anion (O2–), and H2O2 was specified as a disproportionation product resulting from the function of superoxide dismutase. H2O2 is, in turn, rapidly detoxified to water by the ascorbate peroxidase pathway. In this process, the electrons derived from water splitting by PSII subsequently flow through PSI to produce water again. Thus, it has been termed the water–water cycle, or “pseudocyclic electron flow” [59,60]. The physiological relevance of the water–water cycle in chloroplasts, however, has been the subject of heavy discussion and a clear consensus has not been reached so far. One of the reasons for this is related to methodological problems. Application of membrane inlet mass spectrometry (MIMS) and the 18O2 isotope allows differentiation between O2 produced by PSII and that consumed by O2 photoreduction. However, the precise measurement of the Mehler reaction is complicated due to concomitant O2 uptake by chlororespiration, the photorespiratory pathway and mitochondrial respiration. It has been proposed that the water–water cycle has a dual function [60]. When the electron transfer rate exceeds the capacity of utilization of electrons by CO2 assimilation, the flux of “extra” electrons from and/or downstream of PSI to O2 [61] may provide an organism with a protective mechanism for dissipation of excess electrons. Moreover, the Mehler reaction contributes to the generation of a proton gradient across the thylakoid membrane, thus down-regulating PSII and stimulating a rapid induction of non-photochemical energy quenching (NPQ) to dissipate excess photons but, at the same time, allowing the synthesis of ATP for cellular metabolism. Importantly, the protective function of the Mehler reaction in chloroplasts is considered possible as long as the ROS scavenging system functions properly and there is a balance between ROS production and scavenging. When this balance is disturbed, massive ROS formation leads to oxidative stress.
Compared to eukaryotic phytoplankton and plants, cyanobacteria are more sensitive to H2O2 [62,63]. The reason for this could be a poor ROS scavenging system. In line with this, cyanobacteria apply a different strategy from that of the plant-type Mehler reaction (herein referred to as “true” Mehler). They can photoreduce O2 with electrons mediated by PSI by means of soluble Flv1 and Flv3 proteins without the production of ROS [55,64,65,66,67]. As opposed to the “true” Mehler reaction, this is a four-electron transfer reaction and ROS is not released during the process. Due to this decisive difference, we refer to the O2 photoreduction performed by FDPs in cyanobacteria as a “Mehler-like” reaction [67]. The difference in the mechanism of O2 photoreduction between cyanobacteria (Mehler-like reaction) and plant chloroplasts (“true” Mehler reaction) was further supported by comparative studies of light induced O2 uptake between intact Synechocystis sp. PCC 6803 cells and the thylakoids of pea (Pisum sativum), revealing significant differences in the fractionation slopes of the three stable oxygen isotopes [65].
In Synechocystis sp. PCC 6803 the extent of Flv1 and Flv3-mediated electron flow to O2 varies depending on carbon and light regime. Under ambient CO2 and high light (300–500 µmol photons m–2 s–1) conditions about 20% of electrons originating from water-splitting PSII can photoreduce O2 via Flv1 and Flv3 [66]. Light-induced electron flux to O2 in the high CO2 maintained cells has been reported to be 15% to 30% [64], and the isotopic fingerprint results obtained with three stable oxygen isotopes demonstrated that electron flow to O2 can be as high as 40% of gross O2 evolution [65]. Such differences in reported results are likely due to different experimental set-ups and physiological states of the cells. Moreover, accurate experiments with high CO2 maintained ∆flv3 mutant cells revealed that about 6% of electrons originating from PSII water splitting is directed to O2, most likely due to activity of dark respiratory terminal oxidases in the light [65]. This hints that respiratory terminal oxidases also can contribute to O2 photoreduction under specific conditions. However, it is plausible that in the presence of powerful Flv1/Flv3 proteins and under standard growth conditions this contribution does not occur.
Despite the fact that Flv1/Flv3 can redirect a considerable amount of electrons to O2 under high-light conditions, the ∆flv1 and ∆flv3 single mutants and the ∆flv1/∆flv3 double mutant do not demonstrate strong high light sensitivity and their net photosynthetic activities are similar to those of wild-type cells [2,64,66,67]. One possible explanation for this phenomenon might be the compensatory effect of other alternative electron transport routes.
3.1.2. Flv1 and Flv3 Proteins are Crucial for the Survival of Cyanobacteria under Fluctuating Light Intensities
It is conceivable that Flv1 and Flv3 proteins play an important role during the induction of photosynthesis after a dark period, prior to the activation of Calvin–Benson cycle enzymes. This assumption is corroborated by the fact that at the onset of light exposure, after a long dark acclimation, the ∆flv1 and ∆flv3 mutants exhibit a lag-phase in oxidation of P700 due to a strong acceptor-side limitation of PSI and highly reduced plastoquinone (PQ)-pool. The reduced PQ-pool subsequently relaxes to normal wild-type level due to light-activation of the CO2 fixation machinery [64], and most likely, up-regulation of alternative electron transfer routes, which keep the PQ-pool under strict homeostatic control [68]. Consequently, the ∆flv1 and ∆flv3 mutant cells cultivated under diurnal dark/light regime do not show a growth phenotype.
Nevertheless, the natural light environment of photosynthetic organisms is more challenging than diurnal dark/light cycles, consisting of highly dynamic fluctuations of light intensity and quality. Natural aquatic systems are characterized by high frequency light fluctuations due to the focusing and defocusing of sun light by surface waves. An indispensable role for the Flv1 and Flv3 proteins acting as a strong electron sink becomes evident only under fluctuating light conditions, when low background light is regularly interrupted with high light pulses [67]. Under fluctuating light the Flv1, Flv3 and Flv1/Flv3 deficient mutants show extreme and, importantly, regularly repeated acceptor side limitation, which induces strong and regular over-reduction of electron-transport chain and a blockage of photosynthesis during the high-light phases. Indeed, the Flv1 and Flv3 proteins maintain the redox balance of the electron transfer chain in cyanobacteria and provide photoprotection for PSI under fluctuating light conditions. In line with this, the growth and photosynthesis of the Δflv1(a) and/or Δflv3(a) mutants of both Synechocystis sp. PCC 6803 and Anabaena sp. PCC 7120 become arrested, ultimately resulting in cell death, in the most severe and long-term fluctuating light conditions. Such phenomenon is mainly caused by PSI malfunction and concomitant oxidative stress induced by ROS generated during abrupt short-term increases in light intensity, as evidenced by high carbonylation levels of proteins. However, it is also likely linked to a shortage of ATP, as evidenced by the low light-induced energization of the membrane [67].
3.1.3. Cooperation of the FDP Mediated Mehler-Like Reaction and the Photorespiratory Pathway in Cyanobacteria
Besides the Mehler reaction and dark respiration, photorespiration also consumes O2 in oxygenic photosynthetic organisms. Photorespiration is based on the oxygenation activity of RuBisCO, the key enzyme of photosynthetic CO2 assimilation, which binds either CO2 or O2 in the active site, depending on the partial pressure of these gases. During evolution, many cyanobacteria have developed unique carbon concentration mechanisms (CCM) to facilitate the carboxylation of RuBisCO and to be able to grow under ambient Ci-limiting conditions [15,16,69,70]. Therefore, for many decades it was believed that photorespiration, the oxygenation of RuBisCO, does not occur in cyanobacteria [71]. More recently, however, an active photorespiratory metabolism was also discovered in the cyanobacterium Synechocystis sp. PCC 6803 [66,72,73,74].
The photoreduction of O2 by Flv1 and Flv3 proteins was long seen as the major obstacle to the direct monitoring of photorespiratory gas-exchange in cyanobacteria. Indeed, the capacity of the Mehler-like reaction in Synechocystis sp. PCC 6803 seems to be very high. In Synechocystis sp. PCC 6803 cells under Ci-limitation, which favors oxygenation and suppresses the carboxylation of RuBisCO, about 60% of the electrons originating from PSII can be transferred to O2 during the dark-light transition [66]. Iodoacetamide (IAC), a widely used inhibitor of CO2 fixation and photorespiration [64,75] was found to have a stimulatory effect on the photoreduction of O2 under Ci-limitation, perhaps by suppressing the CO2 assimilation and thereby stimulating the electron flux to Mehler-like reaction to a larger extent. Nevertheless, it is important to note that the occurrence of the “true”-Mehler reaction in wild-type Synechocystis sp. PCC 6803 cells under these conditions cannot be fully excluded.
Application of 18O2 labeling and severe Ci-limitation of cells lacking the Mehler-like reaction, Δflv1/Δflv3, has revealed a strong photoreduction of O2, whereby 40%–60% of electrons are transferred from the photosynthetic electron transfer chain to O2. The majority of this O2 photoreduction was carried out by photorespiration, since it could be inhibited by the application of IAC [66]. This observation confirms that both the FDP-mediated Mehler-like reaction and the photorespiratory metabolism are effective sinks for electrons under conditions of Ci-limitation. In line with these results, cooperation between the photorespiration and the Mehler-like reaction was revealed in a double mutant defective in both Flv3 and the glycine decarboxylase complex subunit GsvT, which is involved in one of the photorespiratory pathways. The double mutant Δflv3/ΔgcvT could not be segregated completely and demonstrated a high-light-sensitive phenotype [76]. About 25% of O2 photoreduction in the Δflv3 and Δflv1 cells has been found to be insensitive to IAC, which might belong to uninhibited fraction of RuBisCO, or possibly to the true Mehler reaction.
Based on an increasing number of reports, it is becoming clear that the Mehler-like reaction has the potential to function as an efficient sink of electrons and thereby to dissipate excess electrons from the photosynthetic electron-transfer chain of Synechocystis sp. PCC 6803, in cooperation with photorespiration. Nevertheless, it is likely that cyanobacteria with efficient CO2 concentrating mechanisms and Flv1/Flv3 proteins do not frequently use photorespiratory pathways at full capacity.
3.2. Flavodiiron Proteins in Filamentous Heterocystous Cyanobacteria
The Basic Local Alignment Search Tool (BLAST) and genome sequence analyses have demonstrated that filamentous heterocystous cyanobacteria contain four to six genes encoding FDPs (Table 1). Nostoc punctiforme is the only exception, showing five flv genes. In Anabaena sp. PCC 7120, a N2-fixing filamentous heterocystous model cyanobacterium, two genes, all4444 and all4446, share high sequence similarity with flv2 and flv4 of Synechocystis sp. PCC 6803, respectively, and are designated in the same way (flv2 and flv4 of Anabaena sp. PCC 7120). The flv2 and flv4 genes form an operon, with all4445 in between ([2], Table 1) and demonstrate a drastic increase in transcript abundance upon CO2 limitation [24]. These features strongly suggest that the proteins encoded by the flv2 and flv4 genes in Anabaena sp. PCC 7120 might have a role in the photoprotection of PSII, similar to the corresponding proteins in Synechocystis sp. PCC 6803 (see Section 3.3).
Four other Anabaena sp. PCC 7120 genes: all3891, all0177, all3895 and all0178 (hereafter designated as flv1a, flv1b, flv3a and flv3b, respectively) are homologous to sll1521 and sll0550 (flv1 and flv3) of Synechocystis sp. PCC 6803. The genes encoding Flv1A and Flv3A proteins are organized in the Anabaena sp. PCC 7120 genome as flv3a-3ORFs-flv1a and only expressed in vegetative cells [24]. Transcript levels of flv1a and flv3a are regulated by CO2 concentration and light intensity, similar to flv3 in Synechocystis sp. PCC 6803 [24]. The Anabaena sp. PCC 7120 mutants deficient in Flv1A and Flv3A proteins demonstrate a strong bleaching phenotype under fluctuating light conditions, which is similar to the Synechocystis sp. PCC 6803 Δflv1 and Δflv3 mutants [67]. Taken together, it is conceivable that Anabaena sp. PCC 7120 Flv1A and Flv3A proteins also function in a cyanobacterial Mehler-like reaction, reducing O2 to water in vegetative cells.
As mentioned above, the two “extra” genes, flv1b and flv3b, form an operon in Anabaena sp. PCC 7120 [19] (Table 1). These genes are transcribed under N2-fixing conditions, and the respective proteins are localized exclusively in heterocysts [18,24,26,53] (Table 3). These records, together with a finding of Milligan and co-workers [77], strongly support the idea that light-induced O2 uptake in heterocysts of N2-fixing cyanobacteria could play an important role in the protection of nitrogenase. Indeed, a recent paper demonstrated that the Flv3B protein is responsible for the light-induced reduction of O2 in heterocysts, and participates in the maintenance of a micro-oxic environment inside heterocysts for the proper function of the N2-fixing machinery under the light [18].
Importantly, the Flv1B mutant has been shown not to contribute to the light-induced O2 uptake in heterocysts and growth of the ∆flv1b mutant did not differ from that of wild-type Anabaena sp. PCC 7120. Thus, the function of the Flv1B protein in heterocysts remains to be elucidated. Although terminal oxidases do not contribute to light-induced O2 uptake in heterocysts, as demonstrated in Δflv3b, they are likely responsible for the constitutive level of O2 consumption independently on light or dark conditions [18]. In line with this, the mutant strain lacking Flv3B demonstrated significantly increased transcript amounts of coxA3, which is part of the operon encoding the heterocyst-specific terminal oxidase. Genes encoding lactate oxidase, expected to reduce O2, as well as rubrerythrin and Mn-catalase, which both reduce H2O2 were also upregulated [18].
3.3. The Role of Flv2–Flv4
Compared to Synechocystis sp. PCC 6803 Flv1 and Flv3 proteins studied both in vitro and in vivo since 2002 [55,64,65,66,67,76] the Flv2 and Flv4 proteins have received little attention, and it has only been recently that they have been recognized as important players in PSII photoprotection [2,54,78,79,80]. The ∆flv4 mutant cells lacking all proteins encoded by the whole operon (Flv2, Sll0218 and Flv4 proteins) demonstrate slow growth and a reduced level of PSII centers. This mutant was also found to be susceptible to high light intensities under ambient CO2 (e.g., air level CO2), conditions that highly enhance the expression of flv2 and flv4 [2,78] (Table 2,Table 3). In sharp contrast to these observations, the overexpression of the flv4-2 operon in Synechocystis sp. PCC 6803 resulted in improved photochemistry of PSII and the resistance of cells to high light intensity, as compared to control strains and knock-out mutants [79]. These findings clearly suggest a role for the Flv2/Flv4 heterodimer in the photoprotection of PSII [2,79]. Heterodimer organization of the Flv2 and Flv4 proteins is discussed in the Section 3.4.
3.3.1. An Alternative Electron Transfer Route from PSII to the Flv2/Flv4 Heterodimer
The Flv2 and Flv4 proteins are not involved in the photoreduction of O2 in Synechocystis sp. PCC 6803, at least under conditions studied so far [64,66]. Further investigations of the Flv2/Flv4 related photoprotection mechanism have suggested that these proteins are neither involved in state-transitions nor in OCP-related NPQ [2,79]. Detailed comparisons of PSII properties of wild type and mutant cells, either lacking the Flv2 and Flv4 proteins or overexpressing the flv4–flv2 operon (flv4-2/OE), have revealed a newly identified electron transfer route functioning in close proximity to the QB site. This route could alleviate excitation pressure by channeling excess electrons to a yet unknown electron acceptor under ambient CO2 conditions [2,78,79]. Indeed, the Flv2/Flv4 heterodimer stabilizes forward electron transfer and increases the charge separation rate in PSII [80]. Importantly, an increased amount of singlet oxygen (1O2) and carotenoids in the ∆flv4 mutant compared to the wild type, sharply contrasting the significantly decreased levels in the flv4-2/OE overexpression strain, strongly support the idea that the Flv2/Flv4 heterodimer protects the PSII complex by decreasing 1O2 production [79].
3.3.2. Phycobilisomes and flv4-2 Mediated Photoprotection
The Flv2/Flv4 heterodimer regulates energy transfer from phycobilisomes, more specifically from terminal emitters to the PSII reaction center [78,79]. This was evidenced by 77K fluorescence emission spectra, demonstrating intensification of the F685 nm peak in the ∆flv4 mutant during phycobilisome excitation [78]. In sharp contrast to this observation, the flv4-2/OE mutant demonstrated a lower peak compared to the wild type or ∆flv4 mutant, implying an improved energy transfer to PSII [79]. Importantly, the expression of the flv4-2 operon is dependent on the presence of phycobilisomes. The mutants lacking phycobilisomes (PAL) or containing truncated phycobilisomes (CK and ApcDF) respectively have demonstrated nearly absent, or reduced amounts of the proteins encoded by the flv4-2 operon [79]. Moreover, the deletion of the flv4-2 operon induces disconnection of about 20% of phycobilisomes and reduces the PSII dimer to monomer ratio, showing a direct correlation between PSII dimer destabilization and PBS detachment [80]. In contrast to this, the ∆OCP mutant, deficient in blue-light induced NPQ, demonstrated a significant up-regulation of Flv2 and Flv4 protein content, indicating a particularly important function of Flv2 and Flv4 proteins when the OCP mechanism is absent [79].
3.4. Do Cyanobacterial FDPs Function as a Homodimer or Heterodimer?
In anaerobic prokaryotes and eukaryotes, FDPs function as a homodimer or a homotetramer. In cyanobacteria, however, the organization of FDPs is more complex. The frequent co-occurrence of FDPs in cyanobacteria (as pairs or in operons) suggests their possible function as a heterodimer. Biochemical Blue Native (BN)-PAGE experiments have demonstrated that Flv2 and Flv4 proteins do indeed form a heterodimer [78]. Although Flv2 is able to form a homodimer in the absence of Flv4, complementation experiments have provided evidence that neither Flv2 nor Flv4 is physiologically functional as a homodimer. Further, constructed homology structural models (Figure 3) have demonstrated that the Flv2/Flv4 heterodimer has a more conserved active center for rapid electron transfer than that of the homodimers [78], supporting the first direct evidence of FDP heterodimer formation in Synechocystis sp. PCC 6803.
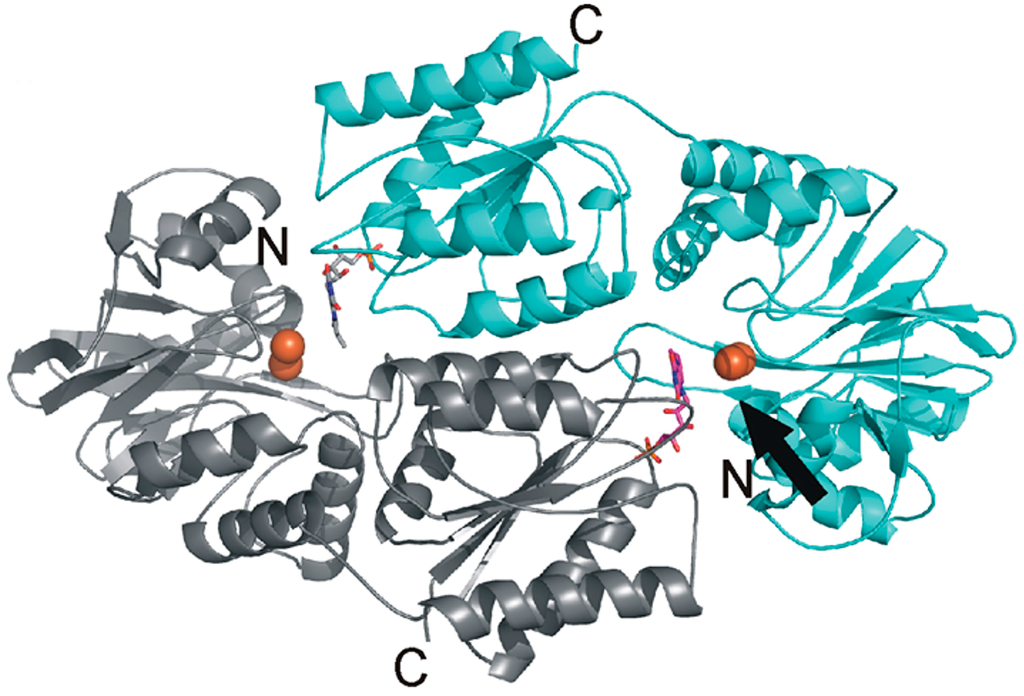
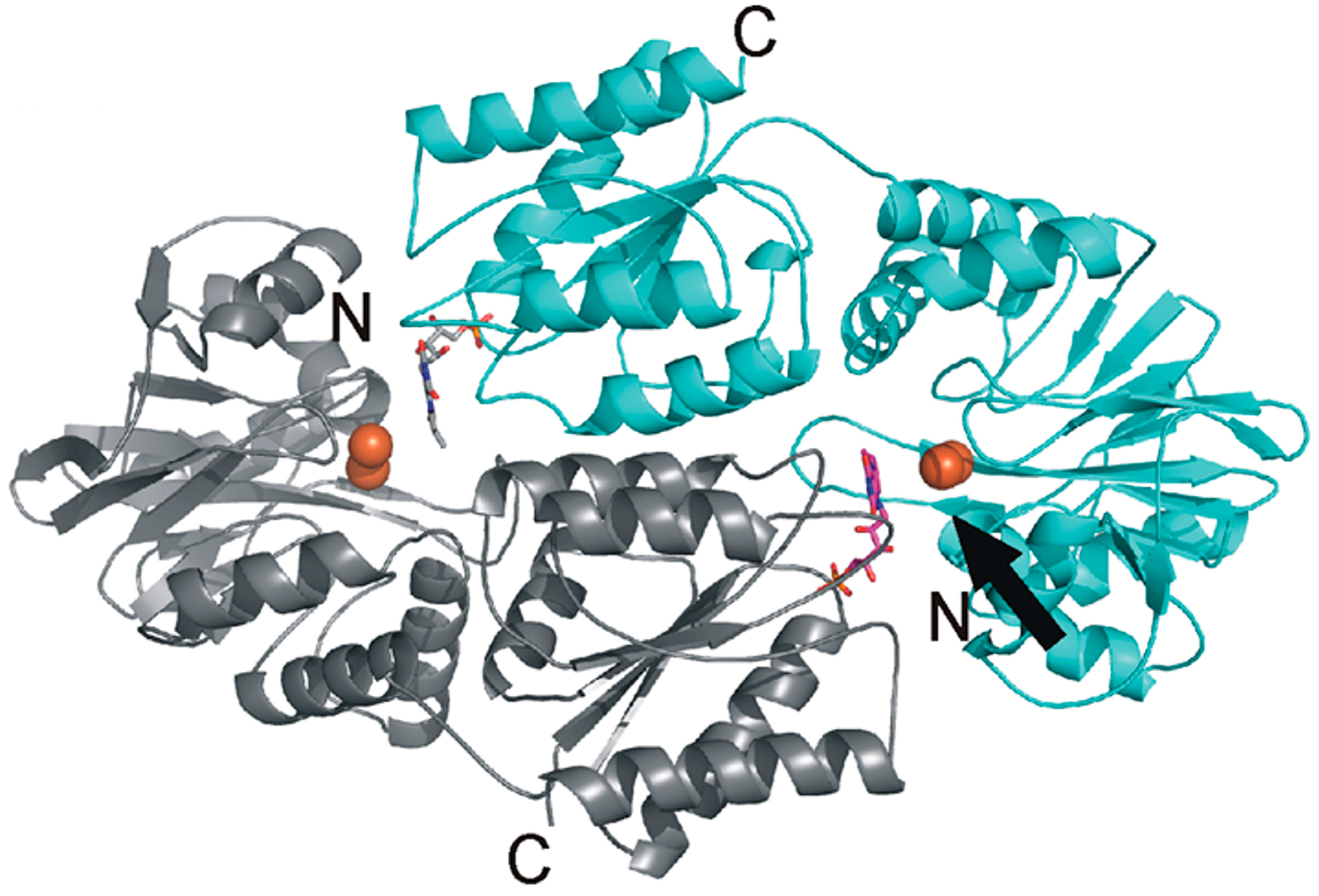
Figure 3.
Heterodimeric organization of the Flv2 and Flv4 proteins. The functional reactive site (shown with arrow) is organized with flavin mononucleotide (FMN) (magenta) from the Flv2 monomer (gray) and diiron (orange spheres) site from the Flv4 monomer (cyan). More details in [78].
Figure 3.
Heterodimeric organization of the Flv2 and Flv4 proteins. The functional reactive site (shown with arrow) is organized with flavin mononucleotide (FMN) (magenta) from the Flv2 monomer (gray) and diiron (orange spheres) site from the Flv4 monomer (cyan). More details in [78].
Unlike Flv2 and Flv4, the Flv1 and Flv3 proteins in Synechocystis sp. PCC 6803 have been detected in the soluble protein fraction of cells [2]. The presence of a 120 kDa protein complex in the soluble protein fraction from both the wild-type and its ∆flv1 mutant, observed in the BN-PAGE gels probed with a Flv3-specific antibody, suggests that the Flv3 protein can organize a homodimer [66]. Moreover, the in vitro activity of the recombinant Flv3 protein [55] and the fact that the flv1 and flv3 genes are spread out in the genome, collectively support the homodimer organization of the Flv3 protein. However, the biochemical data is not corroborated by the in vivo functional analyses. The lack of O2 photoreduction in both the ∆flv1 and the ∆ flv3 mutants provide strong evidence for the inability of Flv1 and Flv3 to function as homodimers in the “Mehler-like” reaction in Synechocystis sp. PCC 6803 [64,66]. In line with the functional significance of the heterodimer, the accumulation of the Flv3 protein has been shown to be dependent on the presence of the Flv1 protein, with the amount of Flv3 significantly down-regulated in the ∆flv1 mutant cells [66]. Nevertheless, taking into account a low transcript abundance of flv1 and a low accumulation of the Flv1 protein in wild-type Synechocystis sp. PCC 6803, the possibility of Flv1 functioning as an auxiliary protein for the organization of functional Flv3 homodimer in vivo cannot be excluded. Indeed, further investigations are needed to clarify the details of functional dimer organization of the Flv1 and Flv3 proteins.
Likewise, the biochemical data demonstrating the organization of FDP dimers in heterocysts is still missing. However, functional data obtained thus far from the heterocysts of Anabaena sp. PCC 7120, strongly suggests that the Flv3B protein can function independently of Flv1B, probably as a homodimer [18].
Finally, it is worth mentioning that FDPs of oxygenic photosynthetic organisms have highly variable putative metal ligands [4,23]. The Cluster B FDPs, Flv3 and Flv4, contain canonical iron ligand residues, whereas Flv1 and Flv2 proteins from Cluster A do not contain canonical ligands at the diiron catalytic site. This raises a question about the absence of a functional metal site in the Flv1 and Flv2 proteins, in turn questioning the O2 reducing activity of these particular proteins [4,23].
4. Significance of FDPs During Evolution
FDP orthologs have been found in all genomes of obligatory anaerobic prokaryotes and some facultative microbes. Since oxygen is detrimental for strict anaerobes, mechanisms of O2 detoxification are crucial for their survival. It has been reported that rubredoxin:oxygen oxidoreductase, by scavenging O2 and preventing ROS formation in vivo, significantly enhances the survival of the strict anaerobe Desulfovibrio vulgaris under microaerophilic conditions [81]. The identification of FDPs in some microaerophilic protozoa led to the proposal of their ability to efficiently scavenge O2, allowing the parasites to survive in microoxic environments [6,82]. The presence of specific FDPs in oxygenic photosynthetic organisms, including almost all sequenced cyanobacteria, and some species of green algae, mosses and lycophytes, suggests that FDPs are involved in photosynthesis-related processes. Among higher plants, only Picea sitchensis possesses one single gene similar to flv3 (Table 1). Analysis of the primary structure of this putative FDP showed that, unlike in other oxygenic photosynthetic organisms, the FDP of Picea sitchensis belongs to the Class A (Figure 1), which lacks the C-terminal flavin reductase domain typical to all other oxygenic photosynthetic organisms, thus questioning the functionality of the putative enzyme.
Photosynthetic eukaryotes and α-cyanobacteria have two (one pair of) FDPs, which are closest to Synechocystis sp. PCC 6803 Flv1 and Flv3, based on sequence similarity. They are probably also involved in O2 photoreduction (the Mehler-like reaction) to dissipate excess electrons in a harmless way. The physiological role of Flv1 and Flv3 is similar to the primary O2 scavenging function of FDPs in anaerobic prokaryotes. However, no FDP homologs are typical for higher plants (Picea sitchensis being the only exception with a single putative Class A FDP), suggesting that the cyanobacterial type Mehler-like reaction was gradually eliminated during the evolution of the green lineage, and was completely substituted by the plant-type “true” Mehler reaction, along with the development of the sophisticated ROS scavenging enzyme system. Orthologs of Flv2 and Flv4 are only found in β-cyanobacteria, indicating their limited appearance in certain subgroups and environmental niches. Originally, the Flv2 and Flv4 proteins were detected only in low Ci-acclimated wild-type Synechocystis sp. PCC 6803 cells [2]. However, a recent study showed a strong accumulation of flv2 and flv4 transcripts in a carboxysome-less mutant grown under high CO2 conditions, similar to the strong accumulation observed in low CO2 grown wild-type cells [44]. These data suggest that Flv2/Flv4 is required when electron flux to carbon fixation is largely limited.
5. Concluding Remarks
The crucial role of FDPs for the evolution of oxygenic photosynthesis in cyanobacteria is an intriguing discovery. The conserved flv4-2 operon specific to β-cyanobacteria offers protection to PSII from singlet oxygen formation under strong excitation pressure by allowing electron flow from the acceptor side of PSII. The operon is expressed only under conditions where the excitation pressure on PSII becomes high, but the actual electron acceptors and the detailed electron transfer mechanism remain elusive.
Another photosystem, PSI, is also susceptible to photodamage under severe fluctuating light conditions. The Flv1 and Flv3 proteins, which are largely present in all cyanobacteria, alleviate photodamage to PSI by dissipating excess electrons down-stream of the photosynthetic electron transport chain, thus also lowering oxidative stress in the cells. The crosstalk of Flv1 and Flv3 proteins with other metabolic pathways needs more through investigation.
Acknowledgment
This work was financially supported by the Academy of Finland projects 271832 and 273870 to Eva-Mari Aro and the Kone foundation to Yagut Allahverdiyeva. MSc. Maria Ermakova is acknowledged for a critical reading of the manuscript, Fiona Lynch for language corrections.
Authors Contribution
Yagut Allahverdiyeva, Pengpeng Zhang, Eva-Mari Aro wrote the manuscript, Janne Isojärvi contributed to data mining.
Conflicts of interest
The authors declare no conflict of interest.
References
- Wasserfallen, A.; Ragettli, S.; Jouanneau, Y.; Leisinger, T. A family of flavoproteins in the domains Archaea and Bacteria. Eur. J. Biochem. 1998, 254, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Allahverdiyeva, Y.; Eisenhut, M.; Aro, E.M. Flavodiiron proteins in oxygenic photosynthetic organisms: Photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS One 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Peltier, G.; Tolleter, D.; Billon, E.; Cournac, L. Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth. Res. 2010, 106, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, V.L.; Vicente, J.B.; Saraiva, L.M.; Teixeira, M. Flavodiiron proteins and their role in cyanobacteria. In Bioenergetic Process of Cyanobacteria; Peschek, G.A., Obinger, C., Renger, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 631–653. [Google Scholar]
- Vicente, J.B.; Carrondo, M.A.; Teixeira, M.; Frazao, C. Structural studies on flavodiiron proteins. Methods Enzymol. 2008, 437, 3–19. [Google Scholar] [PubMed]
- Di Matteo, A.; Scandurra, F.M.; Testa, F.; Forte, E.; Sarti, P.; Brunori, M.; Giuffre, A. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 2008, 283, 4061–4068. [Google Scholar] [CrossRef] [PubMed]
- Frazao, C.; Silva, G.; Gomes, C.M.; Matias, P.; Coelho, R.; Sieker, L.; Macedo, S.; Liu, M.Y.; Oliveira, S.; Teixeira, M.; et al. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 2000, 7, 1041–1045. [Google Scholar] [CrossRef]
- Seedorf, H.; Hagemeier, C.H.; Shima, S.; Thauer, R.K.; Warkentin, E.; Ermler, U. Structure of coenzyme F420H2 oxidase (FprA), a di-iron flavoprotein from methanogenic Archaea catalyzing the reduction of O2 to H2O. FEBS J. 2007, 274, 1588–1599. [Google Scholar] [CrossRef] [PubMed]
- Silaghi-Dumitrescu, R.; Kurtz, D.M., Jr.; Ljungdahl, L.G.; Lanzilotta, W.N. X-ray crystal structures of Moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry 2005, 44, 6492–6501. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Oliveira, S.; LeGall, J.; Xavier, A.; Rodrigues-Pousada, C. Analysis of the Desulfovibrio gigas transcriptional unit containing rubredoxin (rd) and rubredoxin-oxygen oxidoreductase (roo) genes and upstream ORFs. Biochem. Biophys. Res. Commun. 2001, 280, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.M.; Helmick, R.A.; Gardner, P.R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002, 277, 8172–8177. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.M.; Giuffre, A.; Forte, E.; Vicente, J.B.; Saraiva, L.M.; Brunori, M.; Teixeira, M. A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J. Biol. Chem. 2002, 277, 25273–25276. [Google Scholar] [CrossRef] [PubMed]
- Seedorf, H.; Dreisbach, A.; Hedderich, R.; Shima, S.; Thauer, R.K. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F420-dependent enzyme involved in O2 detoxification. Arch. Microbiol. 2004, 182, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Silaghi-Dumitrescu, R.; Ng, K.Y.; Viswanathan, R.; Kurtz, D.M., Jr. A flavo-diiron protein from Desulfovibrio vulgaris with oxidase and nitric oxide reductase activities. Evidence for an in vivo nitric oxide scavenging function. Biochemistry 2005, 44, 3572–3579. [Google Scholar] [CrossRef] [PubMed]
- Zarzycki, J.; Axen, S.D.; Kinney, J.N.; Kerfeld, C.A. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 2013, 64, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Hanson, G.D.; Price, G.D. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 2002, 29, 161–173. [Google Scholar] [CrossRef]
- Whitehead, L.; Long, B.M.; Price, G.D.; Badge, M.R. Comparing the in vivo function of α-carboxysomes and β-carboxysomes in two model cyanobacteria. Plant Physiol. 2014, 165, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Battchikova, N.; Richaud, P.; Leino, H.; Kosourov, S.; Isojarvi, J.; Peltier, G.; Flores, E.; Cournac, L.; Allahverdiyeva, Y.; et al. Heterocyst-specific flavodiiron protein Flv3B enables oxic diazotrophic growth of the filamentous cyanobacterium Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA. 2014, 111, 11205–11210. [Google Scholar] [CrossRef]
- Mitschke, J.; Vioque, A.; Haas, F.; Hess, W.R.; Muro-Pastor, A.M. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA. 2011, 108, 20130–20135. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Archibald, J.M. Evolving a photosynthetic organelle. BMC Biol. 2012, 10. [Google Scholar] [CrossRef]
- Roberty, S.; Bailleul, B.; Berne, N.; Franck, F.; Cardol, P. PSI Mehler reaction is the main alternative photosynthetic electron pathway in Symbiodinium sp., symbiotic dinoflagellates of cnidarians. New Phytol. 2014, 204, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, J.; Georg, J.; Scholz, I.; Sharma, C.M.; Dienst, D.; Bantscheff, J.; Voss, B.; Steglich, C.; Wilde, A.; Vogel, J.; Hess, W.R. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA. 2011, 108, 2124–2129. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, V.L.; Saraiva, L.M.; Teixeira, M. Gene expression study of the flavodiiron proteins from the cyanobacterium Synechocystis sp. PCC 6803. Biochem. Soc. Trans. 2011, 39, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Battchikova, N.; Allahverdiyeva, Y.; Aro, E.M. Novel heterocyst-specific flavodiiron proteins in Anabaena sp. PCC 7120. FEBS Lett. 2013, 587, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Picossi, S.; Flores, E.; Herrero, A. ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genomics 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, B.L.; van Nieuwerburgh, F.; Head, S.R.; Golden, J.W. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Battchikova, N.; Richaud, P.; Hagemann, M.; Allahverdiyeva, Y.; Aro, E.M. Characterization of Flavodiiron protein Flv3(a)-mediated photoprotection in cyanobacteria. Manuscript under preparation.
- Ow, S.Y.; Cardona, T.; Taton, A.; Magnuson, A.; Lindblad, P.; Stensjo, K.; Wright, P.C. Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC 7120 using 8-plex isobaric peptide tags. J. Proteome Res. 2008, 7, 1615–1628. [Google Scholar] [CrossRef] [PubMed]
- Ow, S.Y.; Noirel, J.; Cardona, T.; Taton, A.; Lindblad, P.; Stensjo, K.; Wright, P.C. Quantitative overview of N2 fixation in Nostoc punctiforme ATCC 29133 through cellular enrichments and iTRAQ shotgun proteomics. J. Proteome Res. 2009, 8, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Kucho, K.; Okamoto, K.; Tsuchiya, Y.; Nomura, S.; Nango, M.; Kanehisa, M.; Ishiura, M. Global analysis of circadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2005, 187, 2190–2199. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R.; Machne, R.; Georg, J.; Benary, M.; Axmann, I.; Steuer, R. How cyanobacteria pose new problems to old methods: challenges in microarray time series analysis. BMC Bioinf. 2013, 14. [Google Scholar] [CrossRef]
- Singh, A.K.; Elvitigala, T.; Bhattacharyya-Pakrasi, M.; Aurora, R.; Ghosh, B.; Pakrasi, H.B. Integration of carbon and nitrogen metabolism with energy production is crucial to light acclimation in the cyanobacterium Synechocystis. Plant Physiol. 2008, 148, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Hihara, Y.; Kamei, A.; Kanehisa, M.; Kaplan, A.; Ikeuchi, M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 2001, 13, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Ehira, S.; Ohmori, M.; Sato, N. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2003, 10, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bhattacharyya-Pakrasi, M.; Elvitigala, T.; Ghosh, B.; Aurora, R.; Pakrasi, H.B. A systems-level analysis of the effects of light quality on the metabolism of a cyanobacterium. Plant Physiol. 2009, 151, 1596–1608. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Ishizuka, T.; Katayama, M.; Kanehisa, M.; Bhattacharyya-Pakrasi, M.; Pakrasi, H.B.; Ikeuchi, M. Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2004, 45, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Houot, L.; Floutier, M.; Marteyn, B.; Michaut, M.; Picciocchi, A.; Legrain, P.; Aude, J.C.; Cassier-Chauvat, C.; Chauvat, F. Cadmium triggers an integrated reprogramming of the metabolism of Synechocystis PCC 6803, under the control of the Slr1738 regulator. BMC Genomics 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, I.; Kanesaki, Y.; Mikami, K.; Kanehisa, M.; Murata, N. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 2001, 40, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Prakash, J.S.; Krishna, P.S.; Sirisha, K.; Kanesaki, Y.; Suzuki, I.; Shivaji, S.; Murata, N. An RNA helicase, CrhR, regulates the low-temperature-inducible expression of heat-shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiology 2010, 156, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Panichkin, V.B.; Arakawa-Kobayashi, S.; Kanaseki, T.; Suzuki, I.; Los, D.A.; Shestakov, S.V.; Murata, N. Serine/threonine protein kinase SpkA in Synechocystis sp. strain PCC 6803 is a regulator of expression of three putative pilA operons, formation of thick pili, and cell motility. J. Bacteriol. 2006, 188, 7696–7699. [Google Scholar] [CrossRef] [PubMed]
- Rowland, J.G.; Pang, X.; Suzuki, I.; Murata, N.; Simon, W.J.; Slabas, A.R. Identification of components associated with thermal acclimation of photosystem II in Synechocystis sp. PCC 6803. PLoS One 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Postier, B.L.; Burnap, R.L. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004, 279, 5739–5751. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; von Wobeser, E.A.; Jonas, L.; Schubert, H.; Ibelings, B.W.; Bauwe, H.; Matthijs, H.C.; Hagemann, M. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2007, 144, 1946–1959. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, C.; Huege, J.; Engelhardt, A.; Wittink, F.; Laue, M.; Matthijs, H.C.; Kopka, J.; Bauwe, H.; Hagemann, M. Low-carbon acclimation in carboxysome-less and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 2012, 158, 398–413. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Prieto, M.A.; Schon, V.; Georg, J.; Barreira, L.; Varela, J.; Hess, W.R.; Futschik, M.E. Iron deprivation in Synechocystis: Inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3 (Bethesda) 2012, 2, 1475–1495. [Google Scholar] [CrossRef]
- Prakash, J.S.; Sinetova, M.; Zorina, A.; Kupriyanova, E.; Suzuki, I.; Murata, N.; Los, D.A. DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium Synechocystis. Mol. Biosyst 2009, 5, 1904–1912. [Google Scholar] [CrossRef] [PubMed]
- Shoumskaya, M.A.; Paithoonrangsarid, K.; Kanesaki, Y.; Los, D.A.; Zinchenko, V.V.; Tanticharoen, M.; Suzuki, I.; Murata, N. Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J. Biol. Chem. 2005, 280, 21531–21538. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.J.; Luterra, M.D.; Ely, R.L. Transcriptomic responses of Synechocystis sp. PCC 6803 encapsulated in silica gel. Appl. Microbiol. Biotechnol. 2012, 96, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Summerfield, T.C.; Nagarajan, S.; Sherman, L.A. Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 2011, 157, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Shibata, Y.; Haseyama, Y.; Yoshino, Y.; Suzuki, T.; Kagasawa, T.; Kamei, A.; Ikeuchi, M.; Enami, I. Identification of genes expressed in response to acid stress in Synechocystis sp. PCC 6803 using DNA microarrays. Photos. Res 2005, 84, 225–230. [Google Scholar] [CrossRef]
- Battchikova, N.; Vainonen, J.P.; Vorontsova, N.; Keranen, M.; Carmel, D.; Aro, E.M. Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J. Proteome Res. 2010, 9, 5896–5912. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Mikkat, S.; Huang, F.; Huckaut, J.; Marin, K.; Norling, B.; Hagemann, M. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 2006, 6, 2733–2745. [Google Scholar] [CrossRef] [PubMed]
- Kurian, D.; Jansen, T.; Maenpaa, P. Proteomic analysis of heterotrophy in Synechocystis sp. PCC 6803. Proteomics 2006, 6, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Georg, J.; Klahn, S.; Sakurai, I.; Mustila, H.; Zhang, P.; Hess, W.R.; Aro, E.M. The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J. Biol. Chem. 2012, 287, 33153–33162. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.B.; Gomes, C.M.; Wasserfallen, A.; Teixeira, M. Module fusion in an A-type flavoprotein from the cyanobacterium Synechocystis condenses a multiple-component pathway in a single polypeptide chain. Biochem. Biophys. Res. Commun. 2002, 294, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox Regulation in Photosynthetic Organisms: Signaling, Acclimation, and Practical Implications. Antioxid. Redox Signal. 2008, 11, 861–905. [Google Scholar] [CrossRef]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts: I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 1951, 33, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Mehler, A.H.; Brown, A.H. Studies on reactions of illuminated chloroplasts: III. Simultaneous photoproduction and consumption of oxygen studied with oxygen isotopes. Arch. Biochem. Biophys. 1952, 38, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dsissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. Lond. B 2000, 355, 1419–1431. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Oxygen processing in photosynthesis: Regulation and signalling. New Phytol. 2000, 146, 359–388. [Google Scholar] [CrossRef]
- Drábková, M.; Matthijs, H.C.P.; Admiraal, W.; Maršálek, B. Selective effects of H2O2 on cyanobacterial photosynthesis. Photosynthetica 2007, 45, 363–369. [Google Scholar] [CrossRef]
- Matthijs, H.C.; Visser, P.M.; Reeze, B.; Meeuse, J.; Slot, P.C.; Wijn, G.; Talens, R.; Huisman, J. Selective suppression of harmful cyanobacteria in an entire lake with hydrogen peroxide. Water Res. 2012, 46, 1460–1472. [Google Scholar] [CrossRef] [PubMed]
- Helman, Y.; Tchernov, D.; Reinhold, L.; Shibata, M.; Ogawa, T.; Schwarz, R.; Ohad, I.; Kaplan, A. Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 2003, 13, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Helman, Y.; Barkan, E.; Eisenstadt, D.; Luz, B.; Kaplan, A. Fractionation of the three stable oxygen isotopes by oxygen-producing and oxygen-consuming reactions in photosynthetic organisms. Plant Physiol. 2005, 138, 2292–2298. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyeva, Y.; Ermakova, M.; Eisenhut, M.; Zhang, P.; Richaud, P.; Hagemann, M.; Cournac, L.; Aro, E.M. Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J. Biol. Chem. 2011, 286, 24007–24014. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyeva, Y.; Mustila, H.; Ermakova, M.; Bersanini, L.; Richaud, P.; Ajlani, G.; Battchikova, N.; Cournac, L.; Aro, E.M. Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc. Natl. Acad. Sci. USA 2013, 110, 4111–4116. [Google Scholar] [CrossRef] [PubMed]
- Schuurmans, R.M.; Schuurmans, J.M.; Bekker, M.; Kromkamp, J.C.; Matthijs, H.C.; Hellingwerf, K.J. The redox potential of the plastoquinone pool of the cyanobacterium Synechocystis species strain PCC 6803 is under strict homeostatic control. Plant Physiol. 2014, 165, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Price, G.D. CO2 concentrating mechanisms in cyanobacteria: Molecular components, their diversity and evolution. J. Exp. Bot. 2003, 54, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Reinhold, L. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 539–570. [Google Scholar] [CrossRef] [PubMed]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Kahlon, S.; Hasse, D.; Ewald, R.; Lieman-Hurwitz, J.; Ogawa, T.; Ruth, W.; Bauwe, H.; Kaplan, A.; Hagemann, M. The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 2006, 142, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Huege, J.; Schwarz, D.; Bauwe, H.; Kopka, J.; Hagemann, M. Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2008, 148, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Huege, J.; Goetze, J.; Schwarz, D.; Bauwe, H.; Hagemann, M.; Kopka, J. Modulation of the major paths of carbon in photorespiratory mutants of Synechocystis. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Wildner, G.F.; Henkel, J. Specific inhibition of the oxygenase activity of ribulose—1,5-bisphosphate carboxylase. Biochem. Biophys. Res. Commun. 1976, 69, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, C.; Engelhardt, A.; Matthijs, H.C.; Wittink, F.; Bauwe, H.; Kaplan, A.; Hagemann, M. Photorespiratory 2-phosphoglycolate metabolism and photoreduction of O2 cooperate in high-light acclimation of Synechocystis sp. strain PCC 6803. Planta 2009, 230, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Milligan, A.J.; Berman-Frank, I.; Gerchman, Y.; Dismukes, G.C.; Falkowski, P.G. Light-dependent oxygen consumption in nitrogen-fixing cyanobacteria plays a key role in nitrogenase protection1. J. Phycol. 2007, 43, 845–852. [Google Scholar] [CrossRef]
- Zhang, P.; Eisenhut, M.; Brandt, A.M.; Carmel, D.; Silen, H.M.; Vass, I.; Allahverdiyeva, Y.; Salminen, T.A.; Aro, E.M. Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 2012, 24, 1952–1971. [Google Scholar] [CrossRef] [PubMed]
- Bersanini, L.; Battchikova, N.; Jokel, M.; Rehman, A.; Vass, I.; Allahverdiyeva, Y.; Aro, E.M. Flavodiiron protein Flv2/Flv4-related photoprotective mechanism dissipates excitation pressure of PSII in cooperation with phycobilisomes in cyanobacteria. Plant Physiol. 2014, 164, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Chukhutsina, V.; Bersanini, L.; Aro, E.; van Amerongen, H. Cyanobacterial flv4-2 operon-encoded proteins optimize light harvesting and charge separation in photosystem II. Mol. Plant 2014, in press. [Google Scholar]
- Wildschut, J.D.; Lang, R.M.; Voordouw, J.K.; Voordouw, G. Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris hildenborough under microaerophilic conditions. J. Bacteriol. 2006, 188, 6253–6260. [Google Scholar] [CrossRef] [PubMed]
- Smutna, T.; Goncalves, V.L.; Saraiva, L.M.; Tachezy, J.; Teixeira, M.; Hrdy, I. Flavodiiron protein from Trichomonas vaginalis hydrogenosomes: The terminal oxygen reductase. Eukaryot. Cell. 2009, 8, 47–55. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
