Metabolomics of Interstitial Fluid, Plasma and Urine in Patients with Arterial Hypertension: New Insights into the Underlying Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Sample Collection
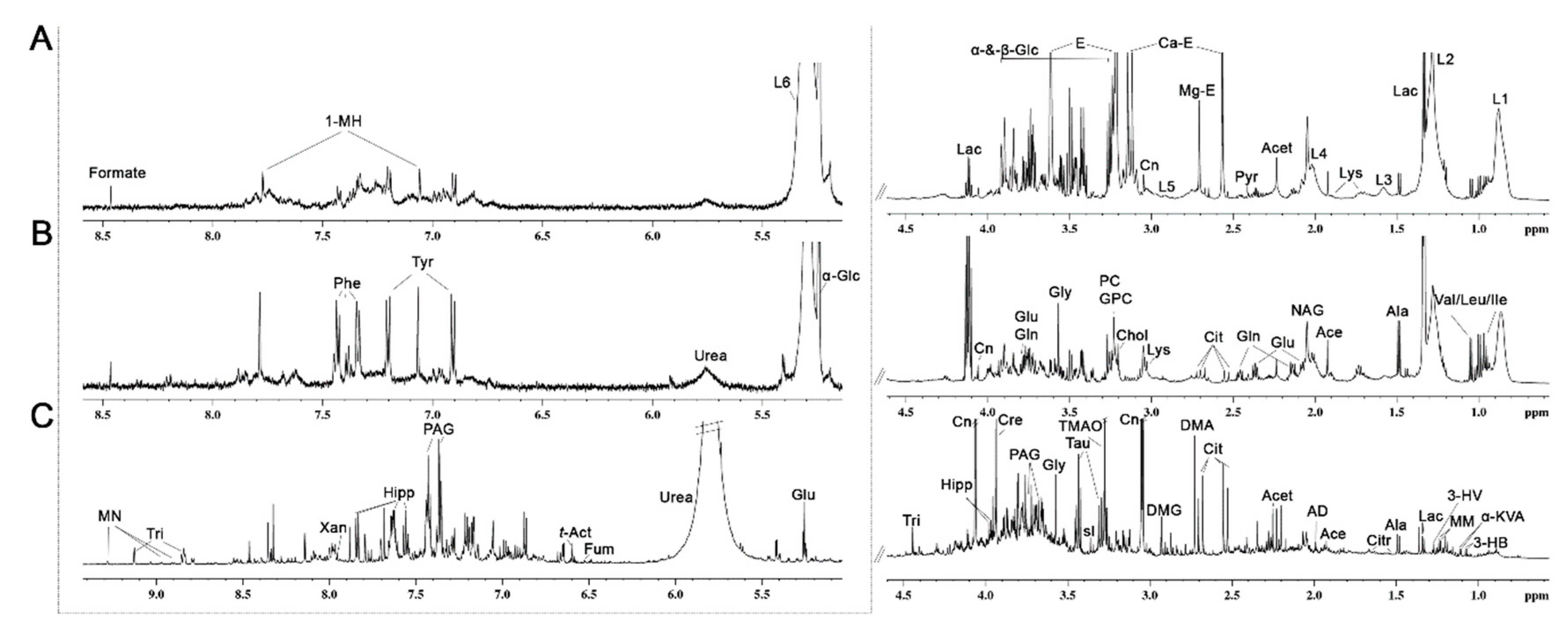
2.3. 1H NMR Analysis of Plasma, Lymph and Urine
2.4. Multivariate and Statistical Analyses
3. Results
3.1. Characteristics of the Study Subjects
3.2. Metabolic Profiling of 1H NMR Spectra of Plasma, Interstitial Fluid and Urine
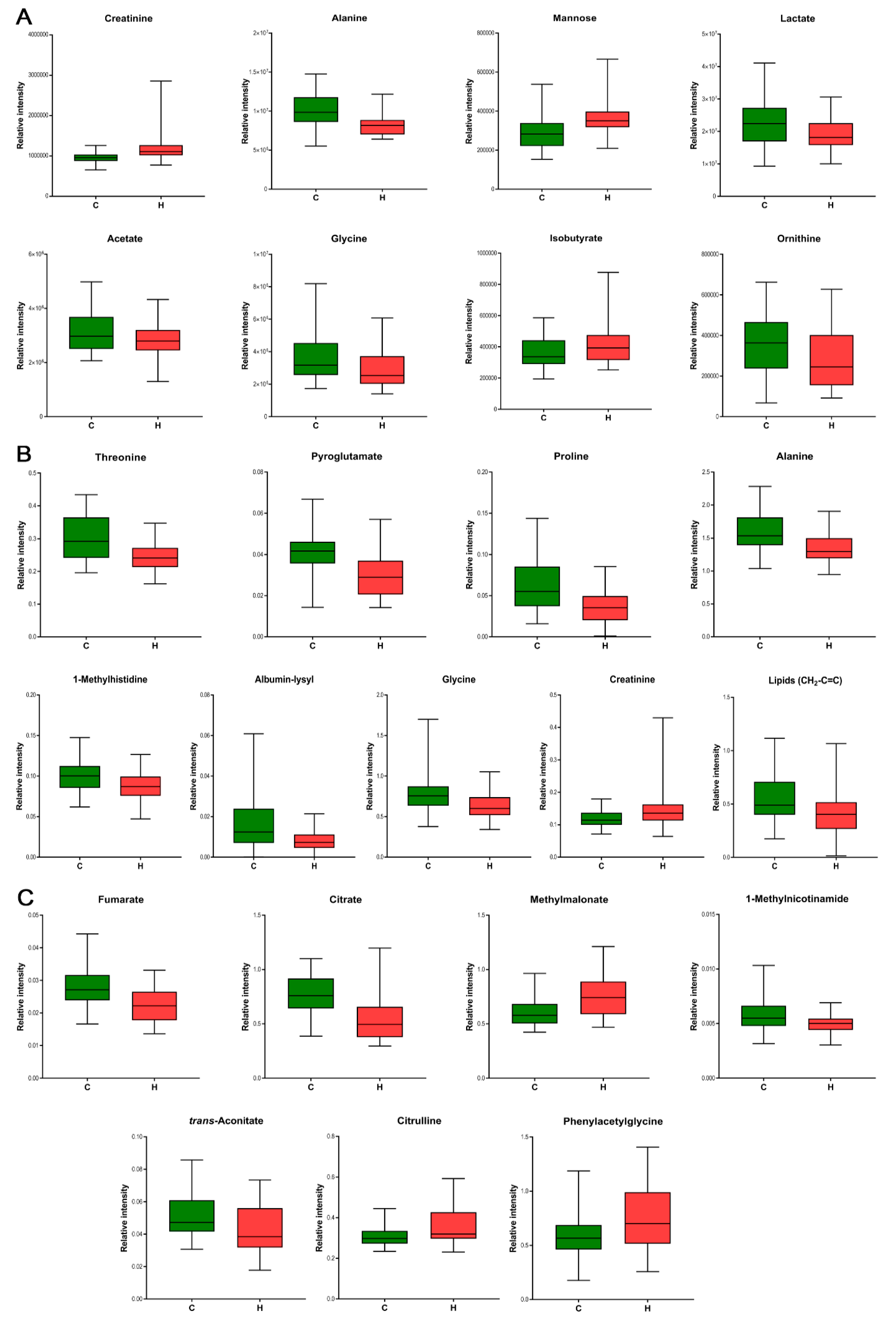
3.3. Biomarker Identification for Hypertension
4. Discussion
4.1. Differences in Metabolite Levels in the Hypertensive Group vs. Normotensive Group
4.1.1. Interstitial Fluid
4.1.2. Plasma
4.1.3. Urine
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, B.; Bentham, J.; Di Cesare, M.; Bixby, H.; Danaei, G.; Cowan, M.J.; Paciorek, C.J.; Singh, G.; Hajifathalian, K.; Bennett, J.E.; et al. Worldwide trends in blood pressure from 1975 to 2015: A pooled analysis of 1479 population-based measurement studies with 19.1 million participants. Lancet 2017, 389, 37–55. [Google Scholar] [CrossRef] [Green Version]
- Lawes, C.M.M.; Vander Hoorn, S.; Rodgers, A.; Hypertens, I.S. Global burden of blood-pressure-related disease, 2001. Lancet 2008, 371, 1513–1518. [Google Scholar] [CrossRef]
- Carretero, O.A.; Oparil, S. Essential hypertension Part I: Definition and etiology. Circulation 2000, 101, 329–335. [Google Scholar] [CrossRef]
- Dietrich, S.; Floegel, A.; Troll, M.; Kuhn, T.; Rathmann, W.; Peters, A.; Sookthai, D.; von Bergen, M.; Kaaks, R.; Adamski, J.; et al. Random Survival Forest in practice: A method for modelling complex metabolomics data in time to event analysis. Int. J. Epidemiol. 2016, 45, 1406–1420. [Google Scholar] [CrossRef] [Green Version]
- Posada-Ayala, M.; Zubiri, I.; Martin-Lorenzo, M.; Sanz-Maroto, A.; Molero, D.; Gonzalez-Calero, L.; Fernandez-Fernandez, B.; de la Cuesta, F.; Laborde, C.M.; Barderas, M.G.; et al. Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney Int. 2014, 85, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hocher, B.; Adamski, J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, F.Q.; Wang, Y.D.; Chen, J.R.; Tao, J.E.; Tian, G.; Wu, S.L.; Liu, W.B.; Cui, Q.H.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.L.; Zheng, L.Y.; Luo, R.; Zhao, X.S.; Han, Z.H.; Wang, Y.L.; Yang, Y.X. A H-1 NMR-based metabonomic investigation of time-dependent metabolic trajectories in a high salt-induced hypertension rat model. RSC Adv. 2015, 5, 281–290. [Google Scholar] [CrossRef]
- Yang, M.; Yu, Z.; Deng, S.; Chen, X.; Chen, L.; Guo, Z.; Zheng, H.; Chen, L.; Cai, D.; Wen, B.; et al. A Targeted Metabolomics MRM-MS Study on Identifying Potential Hypertension Biomarkers in Human Plasma and Evaluating Acupuncture Effects. Sci. Rep. 2016, 6, 25871. [Google Scholar] [CrossRef] [Green Version]
- Hiltunen, T.P.; Rimpela, J.M.; Mohney, R.P.; Stirdivant, S.M.; Kontula, K.K. Effects of four different antihypertensive drugs on plasma metabolomic profiles in patients with essential hypertension. PLoS ONE 2017, 12, e0187729. [Google Scholar] [CrossRef] [Green Version]
- Menni, C.; Graham, D.; Kastenmuller, G.; Alharbi, N.H.; Alsanosi, S.M.; McBride, M.; Mangino, M.; Titcombe, P.; Shin, S.Y.; Psatha, M.; et al. Metabolomic identification of a novel pathway of blood pressure regulation involving hexadecanedioate. Hypertension 2015, 66, 422–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Quan, H.; Athirakul, K.; Wetsel, W.C.; Torres, G.E.; Stevens, R.; Chen, Y.T.; Coffman, T.M.; Caron, M.G. Hypertension and impaired glycine handling in mice lacking the orphan transporter XT2. Mol. Cell. Biol. 2004, 24, 4166–4173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, A.A.; Dunn, R.L.; Marchand, M.C.; Langley-Evans, S.C. Increased systolic blood pressure in rats induced by a maternal low-protein diet is reversed by dietary supplementation with glycine. Clin. Sci. 2002, 103, 633–639. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Flores, M.; Cruz, M.; Duran-Reyes, G.; Munguia-Miranda, C.; Loza-Rodriguez, H.; Pulido-Casas, E.; Torres-Ramirez, N.; Gaja-Rodriguez, O.; Kumate, J.; Baiza-Gutman, L.A.; et al. Oral supplementation with glycine reduces oxidative stress in patients with metabolic syndrome, improving their systolic blood pressure. Can. J. Physiol. Pharmacol. 2013, 91, 855–860. [Google Scholar] [CrossRef]
- Bartus, M.; Lomnicka, M.; Kostogrys, R.B.; Kazmierczak, P.; Watala, C.; Slominska, E.M.; Smolenki, R.T.; Pisulewski, P.M.; Adamus, J.; Gebicki, J.; et al. 1-Methylnicotinamide (MNA) prevents endothelial dysfunction in hypertriglyceridemic and diabetic rats. Pharmacol. Rep. 2008, 60, 127–138. [Google Scholar]
- Loo, R.L.; Zou, X.; Appel, L.J.; Nicholson, J.K.; Holmes, E. Characterization of metabolic responses to healthy diets and association with blood pressure: Application to the Optimal Macronutrient Intake Trial for Heart Health (OmniHeart), a randomized controlled study. Am. J. Clin. Nutr. 2018, 107, 323–334. [Google Scholar] [CrossRef]
- Holmes, E.; Loo, R.L.; Stamler, J.; Bictash, M.; Yap, I.K.; Chan, Q.; Ebbels, T.; De Iorio, M.; Brown, I.J.; Veselkov, K.A.; et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008, 453, 396–400. [Google Scholar] [CrossRef]
- Brindle, J.T.; Nicholson, J.K.; Schofield, P.M.; Grainger, D.J.; Holmes, E. Application of chemometrics to H-1 NMR spectroscopic data to investigate a relationship between human serum metabolic profiles and hypertension. Analyst 2003, 128, 32–36. [Google Scholar] [CrossRef]
- Dietrich, S.; Floegel, A.; Weikert, C.; Pischon, T.; Boeing, H.; Drogan, D. Identification of Serum Metabolites Associated with Incident Hypertension in the European Prospective Investigation into Cancer and Nutrition-Potsdam Study. Hypertension 2016, 68, 471–477. [Google Scholar] [CrossRef]
- Ameta, K.; Gupta, A.; Kumar, S.; Sethi, R.; Kumar, D.; Mahdi, A.A. Essential hypertension: A filtered serum based metabolomics study. Sci. Rep. 2017, 7, 2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaleska, M.; Olszewski, W.L.; Durlik, M.; Miller, N.E. Signaling Proteins Are Represented in Tissue Fluid/Lymph from Soft Tissues of Normal Human Legs at Concentrations Different from Serum. Lymphat. Res. Biol. 2013, 11, 203–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chachaj, A.; Pula, B.; Chabowski, M.; Grzegrzolka, J.; Szahidewicz-Krupska, E.; Karczewski, M.; Janczak, D.; Dziegiel, P.; Podhorska-Okolow, M.; Mazur, G.; et al. Role of the Lymphatic System in the Pathogenesis of Hypertension in Humans. Lymphat. Res. Biol. 2018, 16, 140–146. [Google Scholar] [CrossRef]
- Machnik, A.; Neuhofer, W.; Jantsch, J.; Dahlmann, A.; Tammela, T.; Machura, K.; Park, J.K.; Beck, F.X.; Muller, D.N.; Derer, W.; et al. Macrophages regulate salt-dependent volume and blood pressure by a vascular endothelial growth factor-C-dependent buffering mechanism. Nat. Med. 2009, 15, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Machnik, A.; Dahlmann, A.; Kopp, C.; Goss, J.; Wagner, H.; van Rooijen, N.; Eckardt, K.U.; Muller, D.N.; Park, J.K.; Luft, F.C.; et al. Mononuclear phagocyte system depletion blocks interstitial tonicity-responsive enhancer binding protein/vascular endothelial growth factor C expression and induces salt-sensitive hypertension in rats. Hypertension 2010, 55, 755–761. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.C.; D’Alessandro, A.; Clement, C.C.; Santambrogio, L. Lymph formation, composition and circulation: A proteomics perspective. Int. Immunol. 2015, 27, 219–227. [Google Scholar] [CrossRef] [Green Version]
- Milasan, A.; Ledoux, J.; Martel, C. Lymphatic network in atherosclerosis: The underestimated path. Future Sci. OA 2015, 1. [Google Scholar] [CrossRef] [Green Version]
- Olszewski, W.L. The lymphatic system in body homeostasis: Physiological conditions. Lymphat. Res. Biol. 2003, 1, 11–21. [Google Scholar] [CrossRef]
- Curigliano, G.; Lenihan, D.; Fradley, M.; Ganatra, S.; Barac, A.; Blaes, A.; Herrmann, J.; Porter, C.; Lyon, A.R.; et al. Management of cardiac disease in cancer patients throughout oncological treatment: ESMO consensus recommendations. Ann. Oncol. 2020, 31, 171–190. [Google Scholar] [CrossRef] [Green Version]
- Lenz, E.M.; Wilson, I.D. Analytical strategies in metabonomics. J. Proteome Res. 2007, 6, 443–458. [Google Scholar] [CrossRef]
- Dona, A.C.; Jimenez, B.; Schafer, H.; Humpfer, E.; Spraul, M.; Lewis, M.R.; Pearce, J.T.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal. Chem. 2014, 86, 9887–9894. [Google Scholar] [CrossRef] [PubMed]
- Loren, C.E.; Dahl, C.P.; Do, L.; Almaas, V.M.; Geiran, O.R.; Morner, S.; Hellman, U. Low Molecular Mass Myocardial Hyaluronan in Human Hypertrophic Cardiomyopathy. Cells 2019, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björkblom, B.; Wibom, C.; Jonsson, P.; Mörén, L.; Andersson, U.; Johannesen, T.B.; Langseth, H.; Antti, H.; Melin, B. Metabolomic screening of pre-diagnostic serum samples identifies association between α- and γ-tocopherols and glioblastoma risk. Oncotarget 2016, 7, 37043–37053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Tavintharan, S.; Sum, C.F.; Woon, K.; Lim, S.C.; Ong, C.N. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J. Clin. Endocrinol. Metab. 2013, 98, E1060–E1065. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Liu, R.; Yu, J.; Liu, X.; Zhao, X.; Li, Y.; Liu, L.; Sun, C. Fasting serum alphahydroxybutyrate and pyroglutamic acid as important metabolites for detecting isolated post-challenge diabetes based on organic acid profiles. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1100–1101, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hou, E.; Wang, L.; Wang, Y.; Yang, L.; Zheng, X.; Xie, G.; Sun, Q.; Liang, M.; Tian, Z. Reconstruction and analysis of correlation networks based on GC-MS metabolomics data for young hypertensive men. Anal. Chim. Acta 2015, 854, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Jung, S.; Kim, S.Y.; Lee, S.H.; Lee, J.H. Prehypertension-Associated Elevation in Circulating Lysophosphatidlycholines, Lp-PLA(2) Activity, and Oxidative Stress. PLoS ONE 2014, 9, e96735. [Google Scholar] [CrossRef]
- Phang, J.M.; Pandhare, J.; Liu, Y. The metabolism of proline as microenvironmental stress substrate. J. Nutr. 2008, 138, 2008S–2015S. [Google Scholar] [CrossRef]
- Kaul, S.; Sharma, S.S.; Mehta, I.K. Free radical scavenging potential of L-proline: Evidence from in vitro assays. Amino Acids 2008, 34, 315–320. [Google Scholar] [CrossRef]
- Habte-Tsion, H.M.; Ge, X.; Liu, B.; Xie, J.; Ren, M.; Zhou, Q.; Miao, L.; Pan, L.; Chen, R. A deficiency or an excess of dietary threonine level affects weight gain, enzyme activity, immune response and immune-related gene expression in juvenile blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2015, 42, 439–446. [Google Scholar] [CrossRef]
- Faure, M.; Chone, F.; Mettraux, C.; Godin, J.P.; Bechereau, F.; Vuichoud, J.; Papet, I.; Breuille, D.; Obled, C. Threonine utilization for synthesis of acute phase proteins, intestinal proteins, and mucins is increased during sepsis in rats. J. Nutr. 2007, 137, 1802–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, H.; Katsumura, Y.; Lin, M.; Muroya, Y.F.H.; Katsumura, Y.; Lin, M.; Muroya, Y.; Hata, K.; Fujii, K.; Yokoya, A.; et al. Free radical scavenging and radioprotective effects of carnosine and anserine. Radiat. Phys. Chem. 2009, 78, 1192–1197. [Google Scholar] [CrossRef]
- Zhong, Z.; Wheeler, M.D.; Li, X.; Froh, M.; Schemmer, P.; Yin, M.; Bunzendaul, H.; Bradford, B.; Lemasters, J.J. L-Glycine: A novel antiinflammatory, immunomodulatory, and cytoprotective agent. Curr. Opin. Clin. Nutr. Metab. Care 2003, 6, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Tzoulaki, I.; Iliou, A.; Mikros, E.; Elliott, P. An Overview of Metabolic Phenotyping in Blood Pressure Research. Curr. Hypertens. Rep. 2018, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, S.; Ichiyama, T.; Sonaka, I.; Ohsaki, A.; Okada, S.; Wakiguchi, H.; Kudo, K.; Kittaka, S.; Hara, M.; Furukawa, S. Cysteine, histidine and glycine exhibit anti-inflammatory effects in human coronary arterial endothelial cells. Clin. Exp. Immunol. 2012, 167, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Rodino-Janeiro, B.K.; Gonzalez-Peteiro, M.; Ucieda-Somoza, R.; Gonzalez-Juanatey, J.R.; Alvarez, E. Glycated albumin, a precursor of advanced glycation end-products, up-regulates NADPH oxidase and enhances oxidative stress in human endothelial cells: Molecular correlate of diabetic vasculopathy. Diabetes Metab. Res. Rev. 2010, 26, 550–558. [Google Scholar] [CrossRef]
- Dzieciatkowska, M.; D’Alessandro, A.; Moore, E.E.; Wohlauer, M.; Banerjee, A.; Silliman, C.C.; Hansen, K.C. Lymph Is Not a Plasma Ultrafiltrate: A Proteomic Analysis of Injured Patients. Shock 2014, 42, 485–498. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Villar, V.A.M.; Jose, P.A.; Zeng, C. Renal Dopamine Receptors and Oxidative Stress: Role in Hypertension. Antioxid. Redox Signal. 2020. [Google Scholar] [CrossRef]
- Higashi, Y.; Maruhashi, T.; Noma, K.; Kihara, Y. Oxidative stress and endothelial dysfunction: Clinical evidence and therapeutic implications. Trends Cardiovas. Med. 2014, 24, 165–169. [Google Scholar] [CrossRef]
- Matsutomo, T.; Ushijima, M.; Kodera, Y.; Nakamoto, M.; Takashima, M.; Morihara, N.; Tamura, K. Metabolomic study on the antihypertensive effect of S-1-propenylcysteine in spontaneously hypertensive rats using liquid chromatography coupled with quadrupole-Orbitrap mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1046, 147–155. [Google Scholar] [CrossRef]
- Laerke, H.N.; Mikkelsen, L.S.; Jorgensen, H.; Jensen, S.K. Effect of beta-Glucan Supplementation on Acute Postprandial Changes in Fatty Acid Profile of Lymph and Serum in Pigs. Int. J. Mol. Sci. 2014, 15, 13881–13891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenson, R.S.; Brewer, H.B., Jr.; Davidson, W.S.; Fayad, Z.A.; Fuster, V.; Goldstein, J.; Hellerstein, M.; Jiang, X.C.; Phillips, M.C.; Rader, D.J.; et al. Cholesterol efflux and atheroprotection: Advancing the concept of reverse cholesterol transport. Circulation 2012, 125, 1905–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rader, D.J.; Alexander, E.T.; Weibel, G.L.; Billheimer, J.; Rothblat, G.H. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 2009, 50, S189–S194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.H.; Liu, J.; Deng, Q.J.; Qi, Y.; Wang, M.; Wang, Y.; Zhang, X.G.; Zhao, D. Association between plasma essential amino acids and atherogenic lipid profile in a Chinese population: A cross-sectional study. Atherosclerosis 2019, 286, 7–13. [Google Scholar] [CrossRef]
- Au, A.; Cheng, K.K.; Wei, L.K. Metabolomics, Lipidomics and Pharmacometabolomics of Human Hypertension. Adv. Exp. Med. Biol. 2017, 956, 599–613. [Google Scholar] [CrossRef]
- Li, P.; Wu, G.Y. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 2018, 50, 29–38. [Google Scholar] [CrossRef]
- Stakos, D.A.; Tziakas, D.N.; Chalikias, G.K.; Mitrousi, K.; Tsigalou, C.; Boudoulas, H. Associations Between Collagen Synthesis and Degradation and Aortic Function in Arterial Hypertension. Am. J. Hypertens. 2010, 23, 488–494. [Google Scholar] [CrossRef] [Green Version]
- Melendez-Hevia, E.; De Paz-Lugo, P.; Cornish-Bowden, A.; Cardenas, M.L. A weak link in metabolism: The metabolic capacity for glycine biosynthesis does not satisfy the need for collagen synthesis. J. Biosci. 2009, 34, 853–872. [Google Scholar] [CrossRef]
- Kohn, J.C.; Lampi, M.C.; Reinhart-King, C.A. Age-related vascular stiffening: Causes and consequences. Front. Genet. 2015, 6, 112. [Google Scholar] [CrossRef] [Green Version]
- El Hafidi, M.; Perez, I.; Banos, G. Is glycine effective against elevated blood pressure? Curr. Opin. Clin. Nutr. Metab. Care 2006, 9, 26–31. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomlinson, C.; Rafii, M.; Ball, R.O.; Pencharz, P.B. Arginine can be synthesized from enteral proline in healthy adult humans. J. Nutr. 2011, 141, 1432–1436. [Google Scholar] [CrossRef] [PubMed]
- Bailie, G.R.; Uhlig, K.; Levey, A.S. Clinical practice guidelines in nephrology: Evaluation, classification, and stratification of chronic kidney disease. Pharmacotherapy 2005, 25, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Wannamethee, S.G.; Shaper, A.G.; Perry, I.J. Serum creatinine concentration and risk of cardiovascular disease: A possible marker for increased risk of stroke. Stroke 1997, 28, 557–563. [Google Scholar] [CrossRef]
- Sibilitz, K.L.; Benn, M.; Nordestgaard, B.G. Creatinine, eGFR and association with myocardial infarction, ischemic heart disease and early death in the general population. Atherosclerosis 2014, 237, 67–75. [Google Scholar] [CrossRef]
- Zhong, L.; Zhang, J.P.; Nuermaimaiti, A.G.; Yunusi, K.X. Study on plasmatic metabolomics of Uygur patients with essential hypertension based on nuclear magnetic resonance technique. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 3673–3680. [Google Scholar]
- Kelm, M. The L-arginine-nitric oxide pathway in hypertension. Curr. Hypertens. Rep. 2003, 5, 80–86. [Google Scholar] [CrossRef]
- Wittemans, L.B.L.; Lotta, L.A.; Oliver-Williams, C.; Stewart, I.D.; Surendran, P.; Karthikeyan, S.; Day, F.R.; Koulman, A.; Imamura, F.; Zeng, L.; et al. Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat. Commun. 2019, 10, 1060. [Google Scholar] [CrossRef]
- Ding, Y.; Svingen, G.F.; Pedersen, E.R.; Gregory, J.F.; Ueland, P.M.; Tell, G.S.; Nygard, O.K. Plasma Glycine and Risk of Acute Myocardial Infarction in Patients With Suspected Stable Angina Pectoris. J. Am. Heart Assoc. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- El Hafidi, M.; Perez, I.; Zamora, J.; Soto, V.; Carvajal-Sandoval, G.; Banos, G. Glycine intake decreases plasma free fatty acids, adipose cell size, and blood pressure in sucrose-fed rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 287, R1387–R1393. [Google Scholar] [CrossRef] [Green Version]
- Newsholme, P.; Brennan, L.; Rubi, B.; Maechler, P. New insights into amino acid metabolism, beta-cell function and diabetes. Clin. Sci. 2005, 108, 185–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuttle, K.R.; Milton, J.E.; Packard, D.P.; Shuler, L.A.; Short, R.A. Dietary amino acids and blood pressure: A cohort study of patients with cardiovascular disease. Am. J. Kidney Dis. 2012, 59, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Chen, J.; Chacko, B.K.; Traylor, J.G., Jr.; Orr, A.W.; Patel, R.P. Role of endothelial N-glycan mannose residues in monocyte recruitment during atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, e51–e59. [Google Scholar] [CrossRef] [Green Version]
- Mardinoglu, A.; Stancakova, A.; Lotta, L.A.; Kuusisto, J.; Boren, J.; Bluher, M.; Wareham, N.J.; Ferrannini, E.; Groop, P.H.; Laakso, M.; et al. Plasma Mannose Levels Are Associated with Incident Type 2 Diabetes and Cardiovascular Disease. Cell. Metab. 2017, 26, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Tzoulaki, I.; Castagne, R.; Boulange, C.L.; Karaman, I.; Chekmeneva, E.; Evangelou, E.; Ebbels, T.M.D.; Kaluarachchi, M.R.; Chadeau-Hyam, M.; Mosen, D.; et al. Serum metabolic signatures of coronary and carotid atherosclerosis and subsequent cardiovascular disease. Eur. Heart J. 2019, 5490734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Heiss, G.; Alexander, D.; Grams, M.E.; Boerwinkle, E. Associations Between the Serum Metabolome and All-Cause Mortality Among African Americans in the Atherosclerosis Risk in Communities (ARIC) Study. Am. J. Epidemiol. 2016, 183, 650–656. [Google Scholar] [CrossRef] [Green Version]
- Barba, I.; de Leon, G.; Martin, E.; Cuevas, A.; Aguade, S.; Candell-Riera, J.; Barrabes, J.A.; Garcia-Dorado, D. Nuclear magnetic resonance-based metabolomics predicts exercise-induced ischemia in patients with suspected coronary artery disease. Magn. Reson. Med. 2008, 60, 27–32. [Google Scholar] [CrossRef]
- Crawford, S.O.; Ambrose, M.S.; Hoogeveen, R.C.; Brancati, F.L.; Ballantyne, C.M.; Young, J.H. Association of lactate with blood pressure before and after rapid weight loss. Am. J. Hypertens. 2008, 21, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Menni, C.; Migaud, M.; Glastonbury, C.A.; Beaumont, M.; Nikolaou, A.; Small, K.S.; Brosnan, M.J.; Mohney, R.P.; Spector, T.D.; Valdes, A.M. Metabolomic profiling to dissect the role of visceral fat in cardiometabolic health. Obesity (Silver Spring) 2016, 24, 1380–1388. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Yu, B.; Alexander, D.; Mosley, T.H.; Heiss, G.; Nettleton, J.A.; Boerwinkle, E. Metabolomics and Incident Hypertension Among Blacks: The Atherosclerosis Risk in Communities Study. Hypertension 2013, 62, 398–403. [Google Scholar] [CrossRef] [Green Version]
- Adnan, S.; Nelson, J.W.; Ajami, N.J.; Venna, V.R.; Petrosino, J.F.; Bryan, R.M., Jr.; Durgan, D.J. Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genom. 2017, 49, 96–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryniarski, K.; Biedron, R.; Jakubowski, A.; Chlopicki, S.; Marcinkiewicz, J. Anti-inflammatory effect of 1-methylnicotinamide in contact hypersensitivity to oxazolone in mice; involvement of prostacyclin. Eur. J. Pharmacol. 2008, 578, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Chlopicki, S.; Swies, J.; Mogielnicki, A.; Buczko, W.; Bartus, M.; Lomnicka, M.; Adamus, J.; Gebicki, J. 1-Methylnicotinamide (MNA), a primary metabolite of nicotinamide, exerts anti-thrombotic activity mediated by a cyclooxygenase-2/prostacyclin pathway. Br. J. Pharmacol. 2007, 152, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 2002, 22, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Martin-Lorenzo, M.; Martinez, P.J.; Baldan-Martin, M.; Ruiz-Hurtado, G.; Prado, J.C.; Segura, J.; de la Cuesta, F.; Barderas, M.G.; Vivanco, F.; Ruilope, L.M.; et al. Citric Acid Metabolism in Resistant Hypertension Underlying Mechanisms and Metabolic Prediction of Treatment Response. Hypertension 2017, 70, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Park, J.C.; Shin, M.J.; Lee, H.; Oh, J.; Ryu, D.H.; Hwang, G.S.; Chung, J.H. (1)H nuclear magnetic resonance based metabolic urinary profiling of patients with ischemic heart failure. Clin. Biochem. 2011, 44, 293–299. [Google Scholar] [CrossRef]
- Akira, K.; Masu, S.; Imachi, M.; Mitome, H.; Hashimoto, T. A metabonomic study of biochemical changes characteristic of genetically hypertensive rats based on (1)H NMR spectroscopic urinalysis. Hypertens. Res. 2012, 35, 404–412. [Google Scholar] [CrossRef] [Green Version]
- Altmaier, E.; Fobo, G.; Heier, M.; Thorand, B.; Meisinger, C.; Römisch-Margl, W.; Waldenberger, M.; Gieger, C.; Illig, T.; Adamski, J.; et al. Metabolomics approach reveals effects of antihypertensives and lipid-lowering drugs on the human metabolism. Eur. J. Epidemiol. 2014, 29, 325–336. [Google Scholar] [CrossRef] [Green Version]


| Clinical Data | Hypertensive Group (n = 29) | Normotensive Group (n = 35) | p-Value |
|---|---|---|---|
| Age, y | 65.8 ± 8.4 | 53.4 ± 12.1 | <0.0001 |
| BMI, kg/m2 | 29.5 ± 4.9 | 26.3 ± 4.3 | 0.007 |
| Gender, % male | 13.8 | 11.4 | |
| Duration of hypertension, y | 11.5 ± 10.2 | ||
| SBP, mm Hg | 127.1 ± 10.9 | ||
| DBP, mm Hg | 80.4 ± 6.1 | ||
| Antihypertensive drugs, in total | 1.8 ± 0.7 | ||
| ACEi, % | 48.3 | ||
| ARB, % | 17.2 | ||
| Diuretic, % | 34.5 | ||
| Calcium channel blocker, % | 27.6 | ||
| β-Blocker agent, % | 37.9 | ||
| Clonidine, % | 3.3 | ||
| Familiar history of hypertension, % yes | 82.8 | ||
| Cancer type: | |||
| Breast cancer | 25 | 29 | |
| Cutaneous melanoma | 3 | 4 | |
| Axillary tumor | 1 | 2 |
| Metabolite | Matrix | p[1] | p-Value (t-Test) | p-Value Adjusted for Age | p-Value Adjusted for BMI | Pathway | Superpathway |
|---|---|---|---|---|---|---|---|
| Mannose | Plasma | 0.24 | 2.36 × 10−3 | 7.18 × 10−2 | 7.34 × 10−3 | Fructose, Mannose and Galactose Metabolism | Carbohydrate |
| Lactate | Plasma | −0.20 | 1.33 × 10−2 | 6.07 × 10−3 | 3.92 × 10−3 | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | Carbohydrate |
| Isobutyrate | Plasma | 0.18 | 3.56 × 10−2 | 2.54 × 10−2 | 2.85 × 10−2 | Gut microbiota | |
| Acetate | Plasma | −0.13 | 2.45 × 10−2 | 2.15 × 10−2 | 1.26 × 10−1 | Gut microbiota | |
| Ornithine | Plasma | −0.19 | 4.21 × 10−2 | 9.76 × 10−2 | 1.01 × 10−2 | Urea cycle; Arginine and Proline Metabolism | Amino acid |
| Creatinine | Plasma Lymph | 0.27 0.19 | 4.36 × 10−3 3.86× 10−2 | 3.97 × 10−3 3.31 × 10−2 | 9.34 × 10−3 1.37 × 10−1 | Creatine metabolism | Amino acid |
| Alanine | Plasma Lymph | −0.29 −0.30 | 2.98 × 10−4 2.15× 10−3 | 1.36 × 10−3 3.10 × 10−3 | 7.43 × 10−3 6.79 × 10−3 | Alanine and aspartate metabolism | Amino acid |
| Glycine | Plasma Lymph | −0.21 −0.25 | 2.82 × 10−2 8.96× 10−3 | 5.39 × 10−1 2.96 × 10−1 | 3.51 × 10−1 7.04 × 10−2 | Glycine, serine and threonine metabolism | Amino acid |
| Threonine | Lymph | −0.27 | 2.21× 10−4 | 5.71 × 10−3 | 3.64 × 10−4 | Glycine, Serine and Threonine Metabolism | Amino acid |
| Pyroglutamate | Lymph | −0.27 | 3.72× 10−4 | 6.54 × 10−3 | 3.07 × 10−3 | Glutathione metabolism | Amino acid |
| Proline | Lymph | −0.26 | 1.22 × 10−3 | 2.73 × 10−2 | 3.99 × 10−3 | Urea cycle; Arginine and Proline Metabolism | Amino acid |
| 1-Methylhistidine | Lymph | −0.18 | 2.54× 10−3 | 2.43 × 10−2 | 2.35 × 10−3 | Histidine metabolism | Amino acid |
| Albumin-lysyl | Lymph | −0.20 | 3.42× 10−3 | 8.77 × 10−2 | 1.72 × 10−2 | Protein | Lipid |
| Lipids (CH2-C=C) | Lymph | −0.16 | 4.79× 10−2 | 3.73 × 10−1 | 4.87 × 10−2 | Fatty Acid Metabolism | Lipid |
| Methylmalonate | Urine | 0.23 | 1.61 × 10−3 | 6.49 × 10−4 | 2.67 × 10−4 | Fatty Acid Metabolism (also BCAA Metabolism) | Lipid |
| Phenylacetylglycine | Urine | 0.16 | 3.56 × 10−2 | 5.09 × 10−2 | 2.32 × 10−2 | Acetylated Peptides | Peptide |
| Fumarate | Urine | −0.22 | 1.39 × 10−4 | 2.11 × 10−3 | 1.27 × 10−3 | Krebs cycle | Energy |
| Citrate | Urine | −0.19 | 1.40 × 10−4 | 1.89 × 10−3 | 9.58 × 10−4 | Krebs cycle | Energy |
| trans-Aconitate | Urine | −0.17 | 1.68 × 10−2 | 1.20 × 10−1 | 5.44 × 10−2 | Krebs cycle | Energy |
| Citrulline | Urine | 0.18 | 1.81 × 10−2 | 2.38 × 10−2 | 8.97 × 10−3 | Urea cycle; Arginine and Proline Metabolism | Amino acid |
| 1-Methylnicotinamide | Urine | −0.11 | 6.57 × 10−3 | 2.84 × 10−2 | 3.09 × 10−2 | Nicotinate and Nicotinamide Metabolism | Cofactors and Vitamins |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chachaj, A.; Matkowski, R.; Gröbner, G.; Szuba, A.; Dudka, I. Metabolomics of Interstitial Fluid, Plasma and Urine in Patients with Arterial Hypertension: New Insights into the Underlying Mechanisms. Diagnostics 2020, 10, 936. https://doi.org/10.3390/diagnostics10110936
Chachaj A, Matkowski R, Gröbner G, Szuba A, Dudka I. Metabolomics of Interstitial Fluid, Plasma and Urine in Patients with Arterial Hypertension: New Insights into the Underlying Mechanisms. Diagnostics. 2020; 10(11):936. https://doi.org/10.3390/diagnostics10110936
Chicago/Turabian StyleChachaj, Angelika, Rafał Matkowski, Gerhard Gröbner, Andrzej Szuba, and Ilona Dudka. 2020. "Metabolomics of Interstitial Fluid, Plasma and Urine in Patients with Arterial Hypertension: New Insights into the Underlying Mechanisms" Diagnostics 10, no. 11: 936. https://doi.org/10.3390/diagnostics10110936





