Comparison of Four Commercial Kits for Isolation of Urinary Cell-Free DNA and Sample Storage Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Storage
2.2. cfDNA Isolation
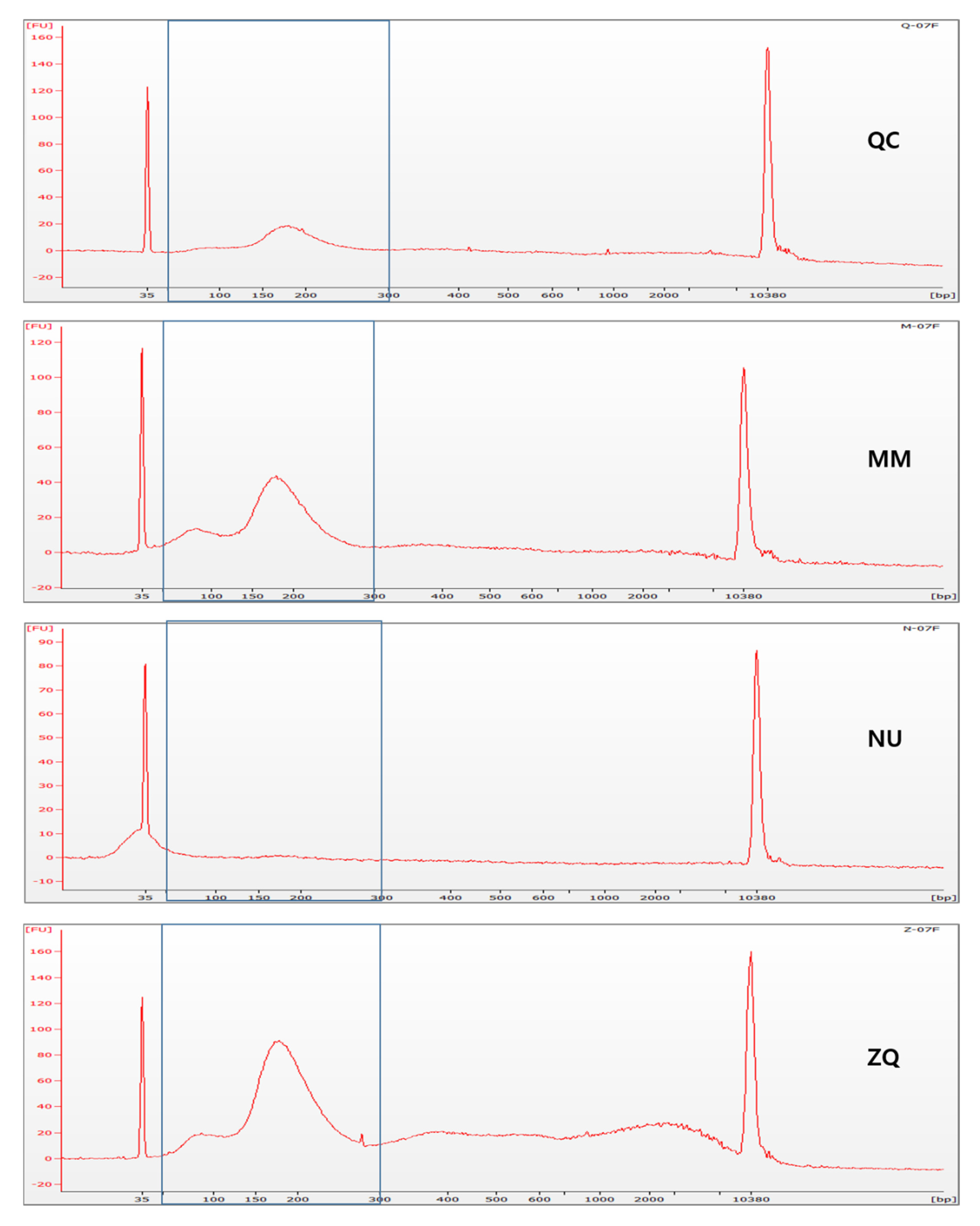
2.3. Analysis of DNA Fragments Using a Bioanalyzer
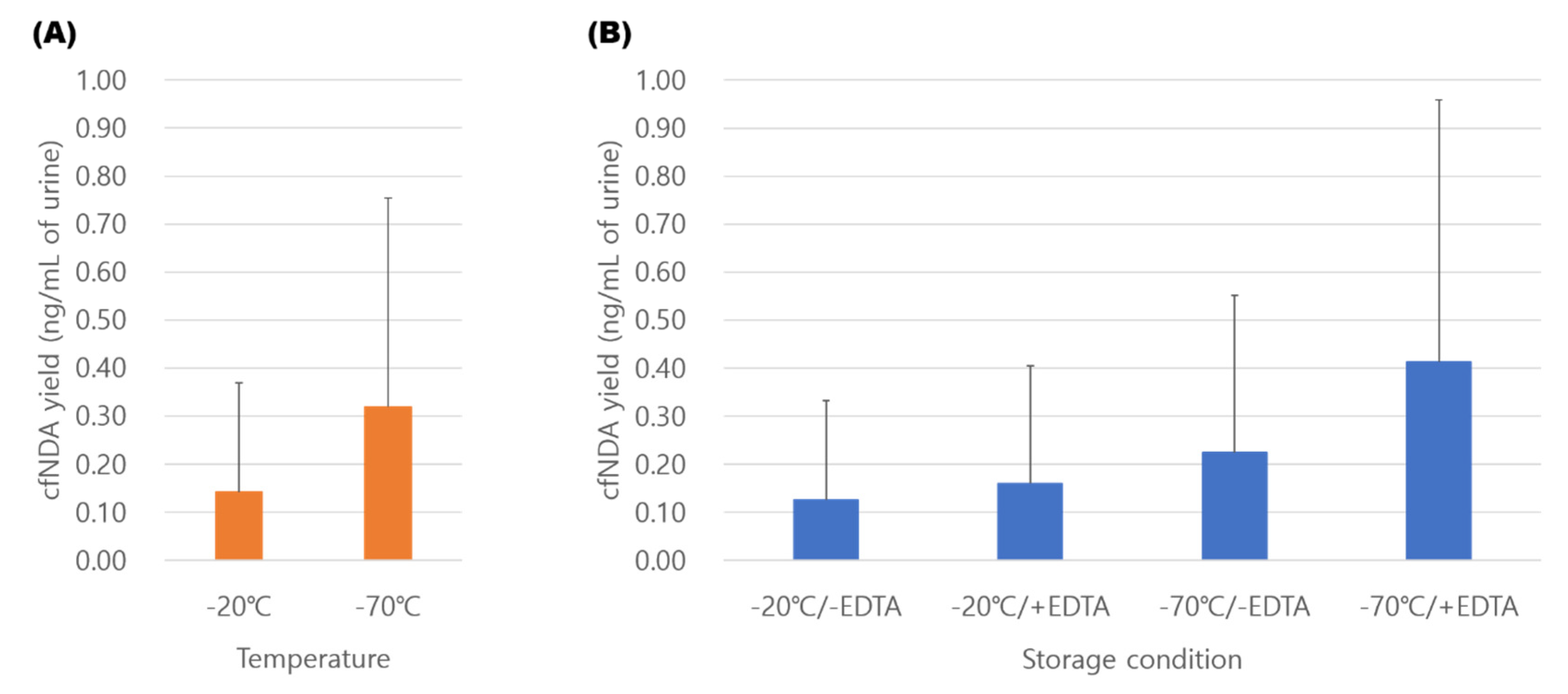
2.4. Comparison of Sample Storage Conditions
3. Results
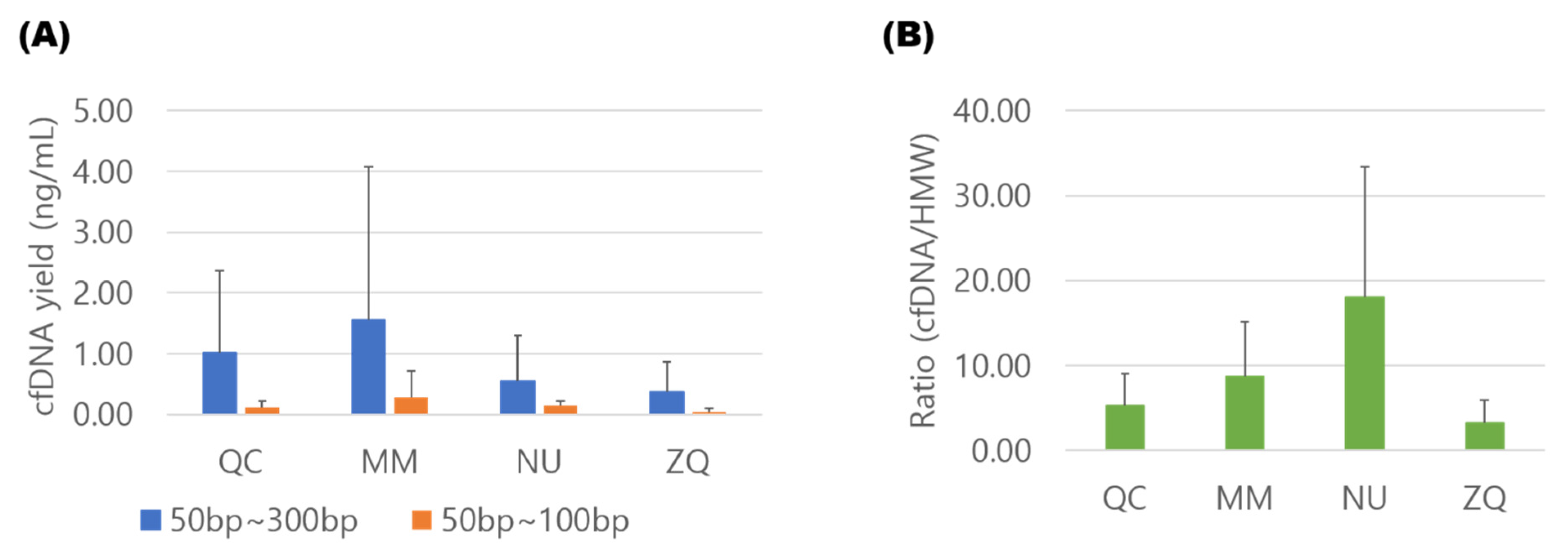
3.1. Comparison of cfDNA Isolation Efficiency
3.2. Contamination of Cellular Genomic DNA
3.3. Comparison of Storage Conditions for Urinary cfDNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef] [Green Version]
- Salvianti, F.; Pinzani, P.; Verderio, P.; Ciniselli, C.M.; Massi, D.; De Giorgi, V.; Grazzini, M.; Pazzagli, M.; Orlando, C. Multiparametric analysis of cell-free DNA in melanoma patients. PLoS ONE 2012, 7, e49843. [Google Scholar] [CrossRef] [Green Version]
- Christensen, E.; Birkenkamp-Demtröder, K.; Nordentoft, I.; Høyer, S.; van der Keur, K.; van Kessel, K.; Zwarthoff, E.; Agerbæk, M.; Ørntoft, T.F.; Jensen, J.B.; et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur. Urol. 2017, 71, 961–969. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Kang, Q.; Henry, N.L.; Paoletti, C.; Jiang, H.; Vats, P.; Chinnaiyan, A.M.; Hayes, D.F.; Merajver, S.D.; Rae, J.M.; Tewari, M. Comparative analysis of circulating tumor DNA stability in K3EDTA, Streck, and CellSave blood collection tubes. Clin. Biochem. 2016, 49, 1354–1360. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Aucamp, J.; Pretorius, P.J. Cell-free DNA: Preanalytical variables. Clin. Chim. Acta 2015, 450, 243–253. [Google Scholar] [CrossRef]
- Parpart-Li, S.; Bartlett, B.; Popoli, M.; Adleff, V.; Tucker, L.; Steinberg, R.; Georgiadis, A.; Phallen, J.; Brahmer, J.; Azad, N.; et al. The Effect of Preservative and Temperature on the Analysis of Circulating Tumor DNA. Clin. Cancer Res. 2017, 23, 2471–2477. [Google Scholar] [CrossRef] [Green Version]
- Meddeb, R.; Pisareva, E.; Thierry, A.R. Guidelines for the Preanalytical Conditions for Analyzing Circulating Cell-Free DNA. Clin. Chem. 2019, 65, 623–633. [Google Scholar] [CrossRef]
- Ge, G.; Peng, D.; Guan, B.; Zhou, Y.; Gong, Y.; Shi, Y.; Hao, X.; Xu, Z.; Qi, J.; Lu, H.; et al. Urothelial Carcinoma Detection Based on Copy Number Profiles of Urinary Cell-Free DNA by Shallow Whole-Genome Sequencing. Clin. Chem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Yoon, H.; Park, S.; Kim, J.S.; Ahn, Y.H.; Kwon, K.; Lee, D.; Kim, K.H. Urinary Exosomal and cell-free DNA Detects Somatic Mutation and Copy Number Alteration in Urothelial Carcinoma of Bladder. Sci. Rep. (Nat. Publ. Group) 2018, 8, 14707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.Y.; Linehan, J.A.; Wilson, T.G.; Hoon, D.S.B. Emerging Utility of Urinary Cell-free Nucleic Acid Biomarkers for Prostate, Bladder, and Renal Cancers. Eur. Urol. Focus 2017, 3, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Huang, C.C.; Dittmar, R.; Du, M.; Wang, Y.; Liu, H.; Shenoy, N.; Wang, L.; Kohli, M. Copy number variations in urine cell free DNA as biomarkers in advanced prostate cancer. Oncotarget 2016, 7, 35818–35831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, Z.; Li, K.; Yang, T.; Dai, Y.; Chandra, M.; Ning, J.; Wang, Y.; Xu, R.; Gao, T.; Xie, Y.; et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin Transl. Med. 2020, 9, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.H.; Wang, M.; Block, T.M.; Landt, O.; Botezatu, I.; Serdyuk, O.; Lichtenstein, A.; Melkonyan, H.; Tomei, L.D.; Umansky, S. Transrenal DNA as a diagnostic tool: Important technical notes. Ann. N.Y. Acad. Sci. 2004, 1022, 81–89. [Google Scholar] [CrossRef]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Roeyen, G.; Lardon, F.; Rolfo, C.; Pauwels, P. A Comparison of Cell-Free DNA Isolation Kits: Isolation and Quantification of Cell-Free DNA in Plasma. J. Mol. Diagn. 2017, 19, 162–168. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Barrios, C.; Nieto-Alcolado, I.; Torrente, M.; Jiménez-Sánchez, C.; Calvo, V.; Gutierrez-Sanz, L.; Palka, M.; Donoso-Navarro, E.; Provencio, M.; Romero, A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl. Lung Cancer Res. 2016, 5, 665–672. [Google Scholar] [CrossRef] [Green Version]
- Warton, K.; Graham, L.J.; Yuwono, N.; Samimi, G. Comparison of 4 commercial kits for the extraction of circulating DNA from plasma. Cancer Genet. 2018, 228–229, 143–150. [Google Scholar] [CrossRef]
- Streleckiene, G.; Reid, H.M.; Arnold, N.; Bauerschlag, D.; Forster, M. Quantifying cell free DNA in urine: Comparison between commercial kits, impact of gender and inter-individual variation. Biotechniques 2018, 64, 225–230. [Google Scholar] [CrossRef]
- Oreskovic, A.; Brault, N.D.; Panpradist, N.; Lai, J.J.; Lutz, B.R. Analytical Comparison of Methods for Extraction of Short Cell-Free DNA from Urine. J. Mol. Diagn. 2019, 21, 1067–1078. [Google Scholar] [CrossRef]
- El Bali, L.; Diman, A.; Bernard, A.; Roosens, N.H.; De Keersmaecker, S.C. Comparative study of seven commercial kits for human DNA extraction from urine samples suitable for DNA biomarker-based public health studies. J. Biomol. Tech. 2014, 25, 96–110. [Google Scholar]
- Silva, M.A.; Medeiros, Z.; Soares, C.R.; Silva, E.D.; Miranda-Filho, D.B.; Melo, F.L. A comparison of four DNA extraction protocols for the analysis of urine from patients with visceral leishmaniasis. Rev. Soc. Bras. Med. Trop. 2014, 47, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Bordelon, H.; Russ, P.K.; Wright, D.W.; Haselton, F.R. A magnetic bead-based method for concentrating DNA from human urine for downstream detection. PLoS ONE 2013, 8, e68369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsui, N.B.; Jiang, P.; Chow, K.C.; Su, X.; Leung, T.Y.; Sun, H.; Chan, K.C.; Chiu, R.W.; Lo, Y.M. High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS ONE 2012, 7, e48319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burnham, P.; Dadhania, D.; Heyang, M.; Chen, F.; Westblade, L.F.; Suthanthiran, M.; Lee, J.R.; De Vlaminck, I. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 2018, 9, 2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Wang, X.; Meng, Q.; Wang, C.; Li, F.; Zhu, Z.; Liu, S.; Shi, Y.; Huang, J.; Chen, S.; Li, C. Investigation of transrenal KRAS mutation in late stage NSCLC patients correlates to disease progression. Biomarkers 2017, 22, 654–660. [Google Scholar] [CrossRef]
- Li, F.; Huang, J.; Ji, D.; Meng, Q.; Wang, C.; Chen, S.; Wang, X.; Zhu, Z.; Jiang, C.; Shi, Y.; et al. Utility of urinary circulating tumor DNA for EGFR mutation detection in different stages of non-small cell lung cancer patients. Clin. Transl. Oncol. 2017, 19, 1283–1291. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, J.; Cui, L.; Liu, Y. Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clin. Transl. Oncol. 2017, 19, 332–340. [Google Scholar] [CrossRef]
- Archer, M.J.; Lin, B.; Wang, Z.; Stenger, D.A. Magnetic bead-based solid phase for selective extraction of genomic DNA. Anal. Biochem. 2006, 355, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Song, J.; Wang, Z.; Wang, X.H.; Wang, M.; Brenner, D.E.; Block, T.M. Removal of high-molecular-weight DNA by carboxylated magnetic beads enhances the detection of mutated K-ras DNA in urine. Ann. N. Y. Acad. Sci. 2008, 1137, 82–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, I.J.; Ju, Y.; Gordon, N.S.; Zeegers, M.P.; Cheng, K.K.; James, N.D.; Bryan, R.T.; Ward, D.G. Toward Personalised Liquid Biopsies for Urothelial Carcinoma: Characterisation of ddPCR and Urinary cfDNA for the Detection of the TERT 228G>A/T Mutation. Bladder Cancer 2018, 4, 41–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadano, D.; Yasuda, T.; Kishi, K. Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method. Clin. Chem. 1993, 39, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Cannas, A.; Kalunga, G.; Green, C.; Calvo, L.; Katemangwe, P.; Reither, K.; Perkins, M.D.; Maboko, L.; Hoelscher, M.; Talbot, E.A.; et al. Implications of storing urinary DNA from different populations for molecular analyses. PLoS ONE 2009, 4, e6985. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.H.; Zhao, S.M.; Zhao, Z.M.; Li, C.T. Genotyping of urinary samples stored with EDTA for forensic applications. Genetics and molecular research: GMR 2012, 11, 3007–3012. [Google Scholar] [CrossRef]
- Ponti, G.; Maccaferri, M.; Manfredini, M.; Kaleci, S.; Mandrioli, M.; Pellacani, G.; Ozben, T.; Depenni, R.; Bianchi, G.; Pirola, G.M.; et al. The value of fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of cell-free DNA (cfDNA) in malignant melanoma and prostate cancer patients. Clin. Chim. Acta 2018, 479, 14–19. [Google Scholar] [CrossRef]
- Hussing, C.; Kampmann, M.L.; Mogensen, H.S.; Børsting, C.; Morling, N. Quantification of massively parallel sequencing libraries—A comparative study of eight methods. Sci. Rep. (Nat. Publ. Group) 2018, 8, 1110. [Google Scholar] [CrossRef] [Green Version]
- van Ginkel, J.H.; van den Broek, D.A.; van Kuik, J.; Linders, D.; de Weger, R.; Willems, S.M.; Huibers, M.M.H. Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med. 2017, 6, 2297–2307. [Google Scholar] [CrossRef]
- Ramachandran, K.; Speer, C.G.; Fiddy, S.; Reis, I.M.; Singal, R. Free circulating DNA as a biomarker of prostate cancer: Comparison of quantitation methods. Int. J. Cancer Res. Treat. 2013, 33, 4521–4529. [Google Scholar]
- Cheng, T.H.T.; Jiang, P.; Tam, J.C.W.; Sun, X.; Lee, W.S.; Yu, S.C.Y.; Teoh, J.Y.C.; Chiu, P.K.F.; Ng, C.F.; Chow, K.M.; et al. Genomewide bisulfite sequencing reveals the origin and time-dependent fragmentation of urinary cfDNA. Clin. Biochem. 2017, 50, 496–501. [Google Scholar] [CrossRef] [PubMed]

| Qiagen | Applied Bio Systems | Norgen | Zymo Research | |
|---|---|---|---|---|
| Product name (Abbreviation) | QIAamp Circulating Nucleic Acid Kit (QC) | MagMAX cell-free DNA isolation kit (MM) | Urine cell-free circulating DNA purification kit-midi (NU) | Quick-DNA ™ Urine Kit (ZQ) |
| Method | Column | Bead | Column | Bead + column |
| Time for run | 1.5 h | 2 h | 2 h | 2 h |
| Starting volume | 4 mL | 4 mL | 10 mL | 24 mL * |
| Cost for sample | $31 | $31 | $23 | $12 |
| Cost for 1 ng urinary cfDNA isolation † | $7.6 | $5.0 | $4.1 | $2.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.Y.; Lee, E.-J.; Yoon, H.; Lee, D.H.; Kim, K.H. Comparison of Four Commercial Kits for Isolation of Urinary Cell-Free DNA and Sample Storage Conditions. Diagnostics 2020, 10, 234. https://doi.org/10.3390/diagnostics10040234
Lee EY, Lee E-J, Yoon H, Lee DH, Kim KH. Comparison of Four Commercial Kits for Isolation of Urinary Cell-Free DNA and Sample Storage Conditions. Diagnostics. 2020; 10(4):234. https://doi.org/10.3390/diagnostics10040234
Chicago/Turabian StyleLee, Eun Young, Eun-Ju Lee, Hana Yoon, Dong Hyeon Lee, and Kwang Hyun Kim. 2020. "Comparison of Four Commercial Kits for Isolation of Urinary Cell-Free DNA and Sample Storage Conditions" Diagnostics 10, no. 4: 234. https://doi.org/10.3390/diagnostics10040234
APA StyleLee, E. Y., Lee, E.-J., Yoon, H., Lee, D. H., & Kim, K. H. (2020). Comparison of Four Commercial Kits for Isolation of Urinary Cell-Free DNA and Sample Storage Conditions. Diagnostics, 10(4), 234. https://doi.org/10.3390/diagnostics10040234





