The Feasibility of Patient-Specific Circulating Tumor DNA Monitoring throughout Multi-Modality Therapy for Locally Advanced Esophageal and Rectal Cancer: A Potential Biomarker for Early Detection of Subclinical Disease
Abstract
:1. Introduction
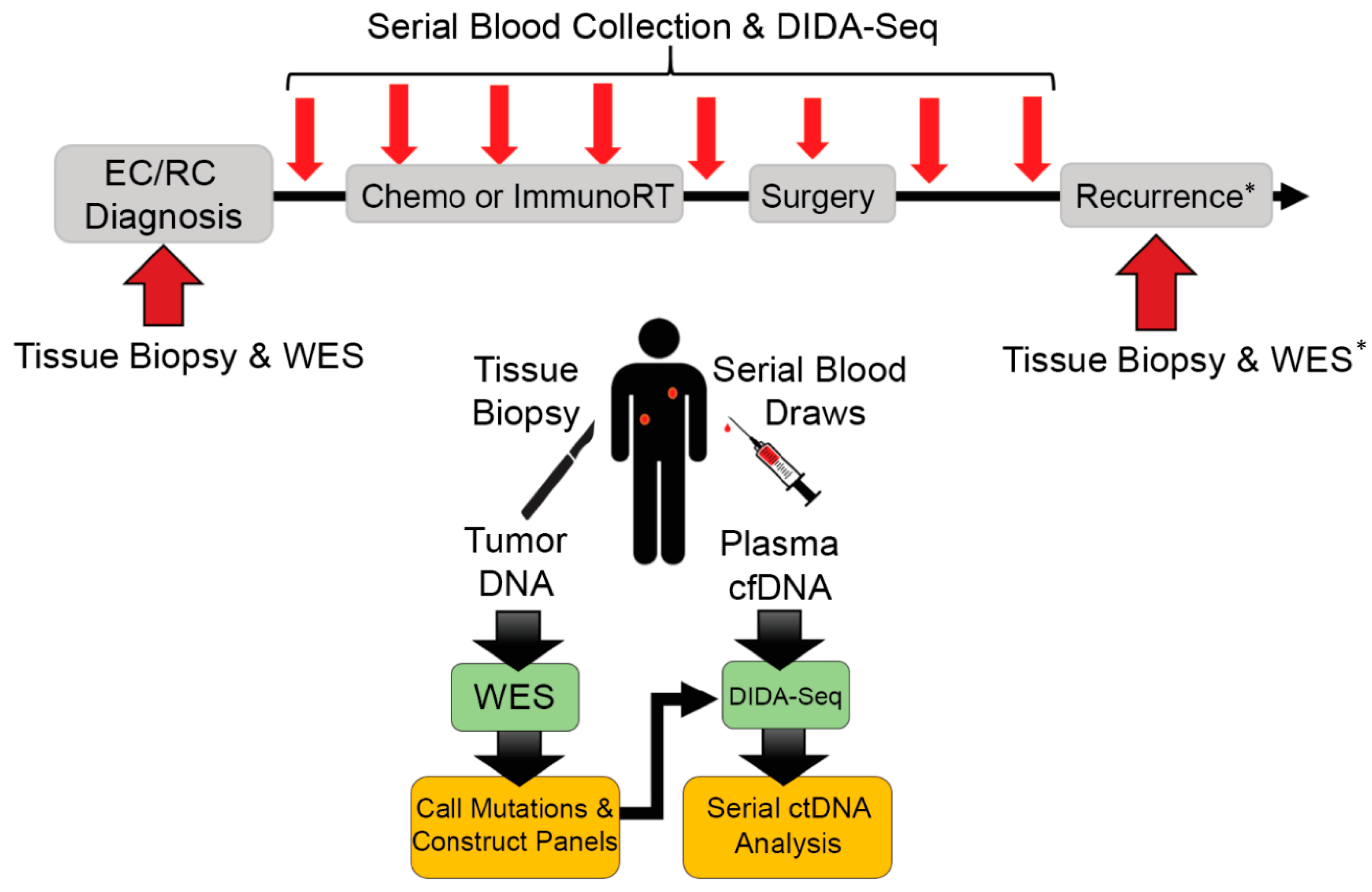
2. Materials and Methods
2.1. Patient Enrollment, Tissue Processing, and DNA Extraction
2.2. Whole-Exome Sequencing Library Preparation
2.3. Somatic Mutation Calling and Design of Tumor-Specific Capture Panels
2.4. DIDA-Seq Library Preparation and Sequencing
2.5. Evaluation of Tumor-Specific Capture Panel Performance and CtDNA Prevalence
2.6. Significance Tests for CtDNA Measurements
3. Results
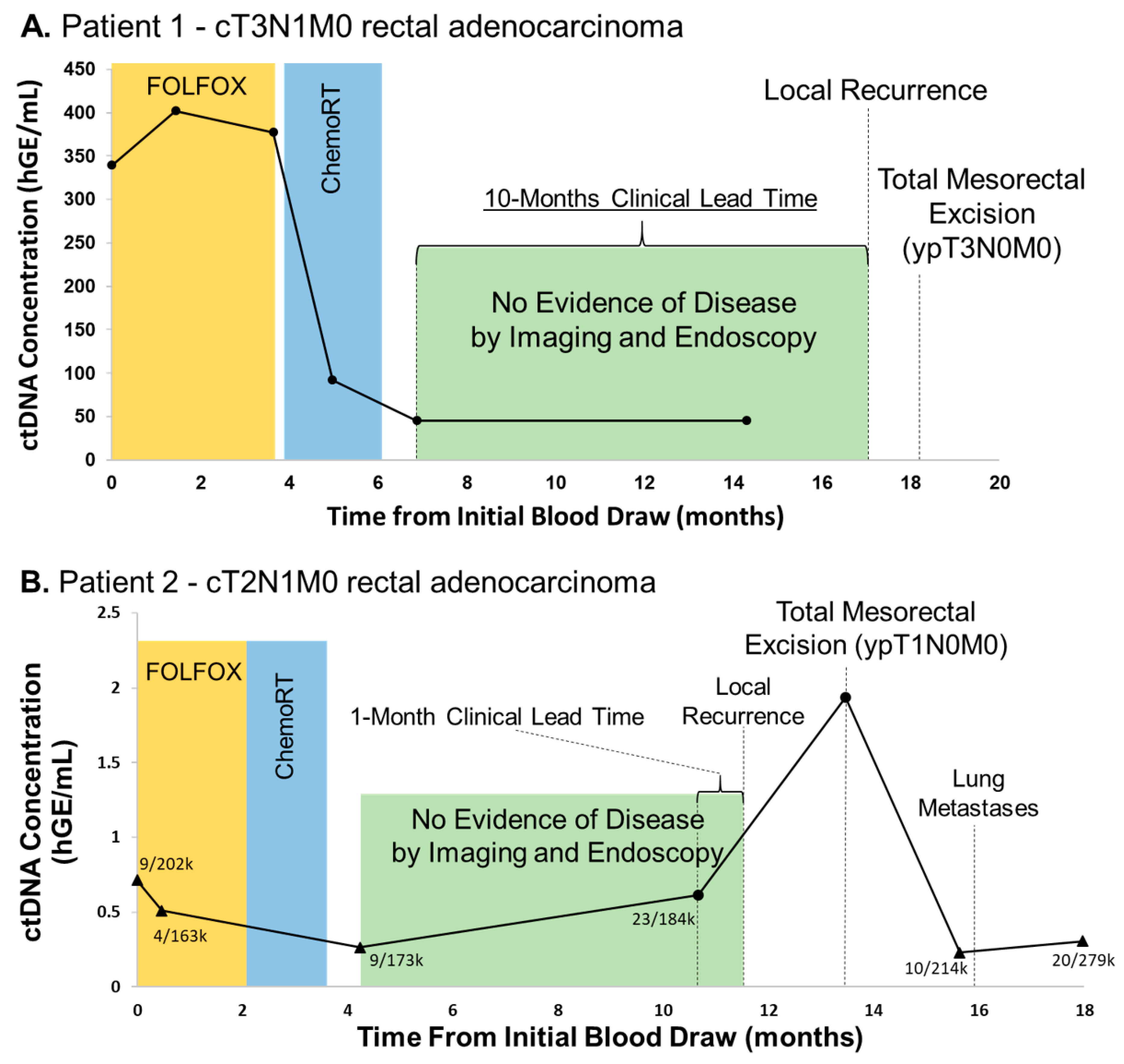
3.1. Elevated ctDNA Levels Are Associated with Recurrence in Rectal Adenocarcinoma with Clinically-Useful Lead Time
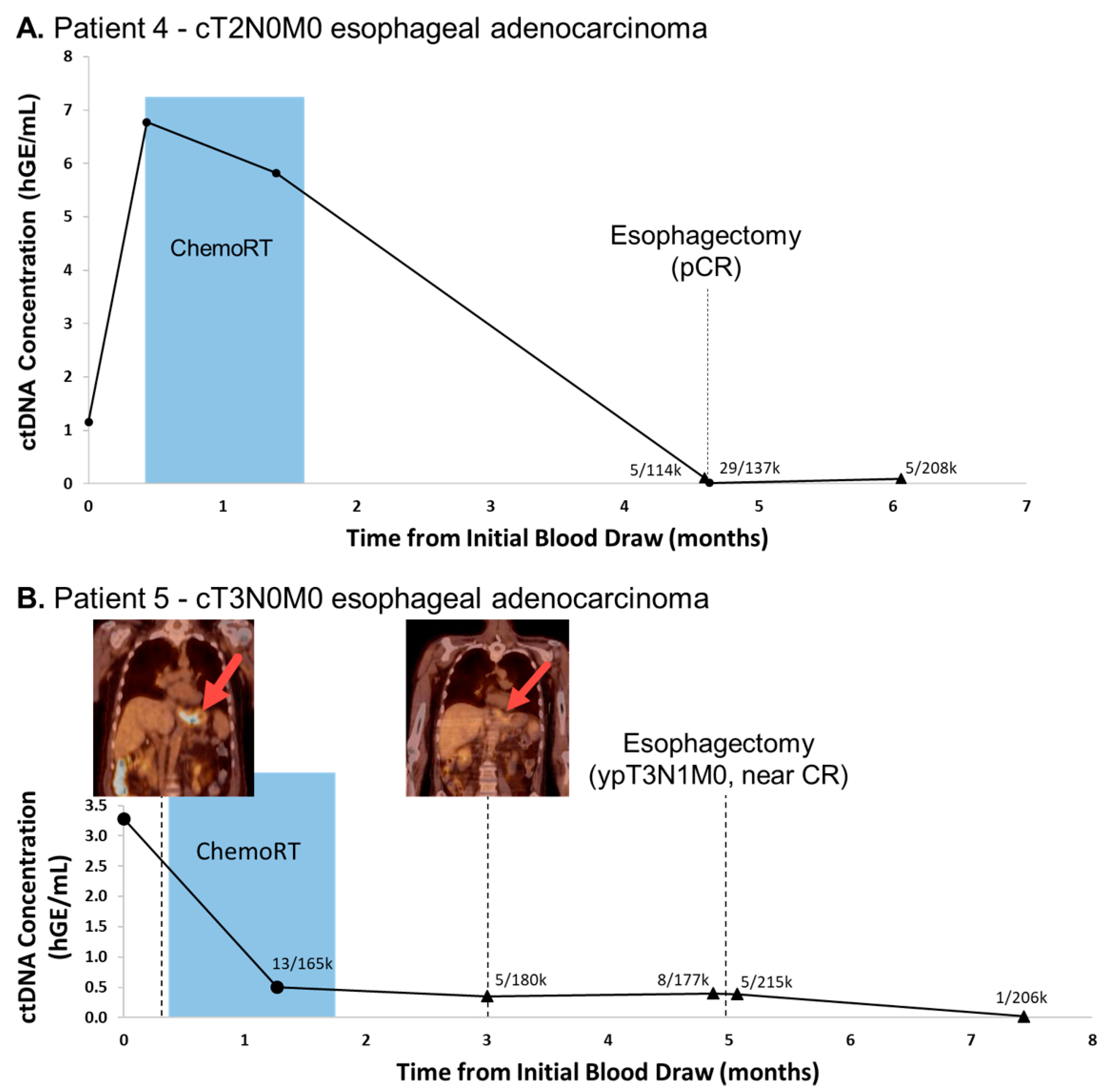
3.2. CtDNA Levels Are Associated with Tumor Burden and Progression in Oligometastatic Esophageal Cancer
3.3. Undetectable ctDNA Is Associated with Pathologic Complete Response (pCR) Following Tri-Modality Therapy for Esophageal Adenocarcinoma
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Noordman, B.J.; Wijnhoven, B.P.; Lagarde, S.M.; Boonstra, J.J.; Coene, P.P.; Dekker, J.W.T.; Doukas, M.; Van Der Gaast, A.; Heisterkamp, J.; Kouwenhoven, E.A.; et al. Neoadjuvant chemoradiotherapy plus surgery versus active surveillance for oesophageal cancer: A stepped-wedge cluster randomised trial. BMC Cancer 2018, 18, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Van Der Valk, M.J.M.; Hilling, D.E.; Bastiaannet, E.; Kranenbarg, E.M.-K.; Beets, G.L.; Figueiredo, N.L.; Habr-Gama, A.; Perez, R.O.; Renehan, A.G.; Van De Velde, C.J.H.; et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): An international multicentre registry study. Lancet 2018, 391, 2537–2545. [Google Scholar] [CrossRef] [Green Version]
- Markar, S.R.; Karthikesalingam, A.; Thrumurthy, S.; Low, D.E. Volume-Outcome Relationship in Surgery for Esophageal malignancy: Systematic Review and Meta-analysis 2000–2011. J. Gastrointest. Surg. 2012, 16, 1055–1063. [Google Scholar] [CrossRef]
- Reece, M.; Saluja, H.; Hollington, P.; Karapetis, C.S.; Vatandoust, S.; Young, G.P.; Symonds, E.L. The Use of Circulating Tumor DNA to Monitor and Predict Response to Treatment in Colorectal Cancer. Front. Genet. 2019, 10, 1118. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12, eaaz8084. [Google Scholar] [CrossRef] [PubMed]
- Zviran, A.; Schulman, R.C.; Shah, M.; Hill, S.T.K.; Deochand, S.; Khamnei, C.C.; Maloney, D.; Patel, K.; Liao, W.; Widman, A.J.; et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020, 26, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Cervena, K.; Vodicka, P.; Vymetalkova, V. Diagnostic and prognostic impact of cell-free DNA in human cancers: Systematic review. Mutat. Res. Mutat. Res. 2019, 781, 100–129. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Gremel, G.; Marshall, A.; Myers, K.; Fisher, N.; Dunn, J.; Dhomen, N.; Corrie, P.; Middleton, M.; Lorigan, P.; et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann. Oncol. 2018, 29, 490–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Ye, X.; Zhang, M.Z.; Sun, Y.; Wang, J.Y.; Ni, J.; Zhang, H.P.; Zhang, L.; Luo, J.; Zhang, J.; et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci. Rep. 2016, 6, 20913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, T.M.; Boniface, C.; Johnson-Camacho, K.M.; Tabatabaei, S.; Melendez, D.; Kelley, T.; Gray, J.W.; Corless, C.L.; Spellman, P.T. Circulating tumor DNA dynamics using patient-customized assays are associated with outcome in neoadjuvantly treated breast cancer. Mol. Case Stud. 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.B.; Sougnez, C.; Gabriel, S.B.; Meyerson, M.L.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Sequence Read Archive (sra) Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 2009. Available online: https://www.ncbi.nlm.nih.gov/sra/ (accessed on 5 May 2020).
- Ramos, A.H.; Lichtenstein, L.; Gupta, M.; Lawrence, M.S.; Pugh, T.J.; Saksena, G.; Meyerson, M.; Getz, G. Oncotator: Cancer Variant Annotation Tool. Hum. Mutat. 2015, 36, E2423–E2429. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, S.R.; Schmitt, M.W.; Fox, E.J.; Kohrn, B.F.; Salk, J.J.; Ahn, E.H.; Prindle, M.J.; Kuong, K.J.; Shen, J.-C.; Risques, R.-A.; et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 2014, 9, 2586–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montoya, J.A.; P., G.F.; González-Sánchez, D. Statistical inference for the Weitzman overlapping coefficient in a family of distributions. Appl. Math. Model. 2019, 71, 558–568. [Google Scholar] [CrossRef]
- Garcia-Aguilar, J.; Patil, S.; Kim, J.K.; Yuval, J.B.; Thompson, H.; Verheij, F.; Lee, M.; Saltz, L.B. Preliminary results of the organ preservation of rectal adenocarcinoma (OPRA) trial. J. Clin. Oncol. 2020, 38, 4008. [Google Scholar] [CrossRef]
- Rohatgi, P.R.; Swisher, S.G.; Correa, A.M.; Wu, T.T.; Liao, Z.; Komaki, R.; Walsh, G.; Vaporciyan, A.; Lynch, P.M.; Rice, D.C.; et al. Failure patterns correlate with the proportion of residual carcinoma after preoperative chemoradiotherapy for carcinoma of the esophagus. Cancer 2005, 104, 1349–1355. [Google Scholar] [CrossRef]
- Urba, S.G.; Orringer, M.B.; Turrisi, A.; Iannettoni, M.; Forastiere, A.; Strawderman, M. Randomized Trial of Preoperative Chemoradiation Versus Surgery Alone in Patients with Locoregional Esophageal Carcinoma. J. Clin. Oncol. 2001, 19, 305–313. [Google Scholar] [CrossRef]
- Ancona, E.; Ruol, A.; Santi, S.; Merigliano, S.; Sileni, V.C.; Koussis, H.; Zaninotto, G.; Bonavina, L.; Peracchia, A. Only pathologic complete response to neoadjuvant chemotherapy improves significantly the long term survival of patients with resectable esophageal squamous cell carcinoma: Final report of a randomized, controlled trial of preoperative chemotherapy versus surgery alone. Cancer 2001, 91, 2165–2174. [Google Scholar]
- Berger, A.C.; Farma, J.; Scott, W.J.; Freedman, G.; Weiner, L.; Cheng, J.D.; Wang, H.; Goldberg, M. Complete Response to Neoadjuvant Chemoradiotherapy in Esophageal Carcinoma Is Associated with Significantly Improved Survival. J. Clin. Oncol. 2005, 23, 4330–4337. [Google Scholar] [CrossRef] [PubMed]
- Chirieac, L.R.; Swisher, S.G.; Ajani, J.A.; Komaki, R.R.; Correa, A.M.; Morris, J.S.; Roth, J.A.; Rashid, A.; Hamilton, S.R.; Wu, T.-T. Posttherapy pathologic stage predicts survival in patients with esophageal carcinoma receiving preoperative chemoradiation. Cancer 2005, 103, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Kleinberg, L.; Knisely, J.P.; Heitmiller, R.; Zahurak, M.; Salem, R.; Burtness, B.; Heath, E.I.; Forastiere, A.A. Mature survival results with preoperative cisplatin, protracted infusion 5-fluorouracil, and 44-Gy radiotherapy for esophageal cancer. Int. J. Radiat. Oncol. 2003, 56, 328–334. [Google Scholar] [CrossRef]
- Scheer, R.V.; Fakiris, A.J.; Johnstone, P.A. Quantifying the Benefit of a Pathologic Complete Response After Neoadjuvant Chemoradiotherapy in the Treatment of Esophageal Cancer. Int. J. Radiat. Oncol. 2011, 80, 996–1001. [Google Scholar] [CrossRef]
- Bailey, S.H.; Bull, D.A.; Harpole, D.H.; Rentz, J.J.; Neumayer, L.A.; Pappas, T.N.; Daley, J.; Henderson, W.G.; Krasnicka, B.; Khuri, S.F. Outcomes after esophagectomy: A ten-year prospective cohort. Ann. Thorac. Surg. 2003, 75, 217–222. [Google Scholar] [CrossRef]
- Biere, S.S.; Maas, K.W.; Cuesta, M.; Van Der Peet, D.L. Cervical or Thoracic Anastomosis after Esophagectomy for Cancer: A Systematic Review and Meta-Analysis. Dig. Surg. 2011, 28, 29–35. [Google Scholar] [CrossRef]
- Kuo, E.Y.; Chang, Y.; Wright, C.D. Impact of hospital volume on clinical and economic outcomes for esophagectomy. Ann. Thorac. Surg. 2001, 72, 1118–1124. [Google Scholar] [CrossRef]
- Habr-Gama, A.; Gama-Rodrigues, J.; Julião, G.P.S.; Proscurshim, I.; Sabbagh, C.; Lynn, P.B.; Perez, R.O. Local Recurrence After Complete Clinical Response and Watch and Wait in Rectal Cancer After Neoadjuvant Chemoradiation: Impact of Salvage Therapy on Local Disease Control. Int. J. Radiat. Oncol. 2014, 88, 822–828. [Google Scholar] [CrossRef]
- Maas, M.; Beets-Tan, R.G.; Lambregts, D.M.; Lammering, G.; Nelemans, P.J.; Engelen, S.M.; Van Dam, R.M.; Jansen, R.L.; Sosef, M.; Leijtens, J.W.; et al. Wait-and-See Policy for Clinical Complete Responders After Chemoradiation for Rectal Cancer. J. Clin. Oncol. 2011, 29, 4633–4640. [Google Scholar] [CrossRef]
- Smith, J.D.; Ruby, J.A.; Goodman, K.A.; Saltz, L.B.; Guillem, J.G.; Weiser, M.R.; Temple, L.K.; Nash, G.M.; Paty, P.B. Nonoperative Management of Rectal Cancer with Complete Clinical Response After Neoadjuvant Therapy. Ann. Surg. 2012, 256, 965–972. [Google Scholar] [CrossRef] [Green Version]
- Gollub, M.J.; Gultekin, D.H.; Akin, O.; Do, R.K.; Fuqua, J.L.; Gonen, M.; Kuk, D.; Weiser, M.; Saltz, L.; Schrag, D.; et al. Dynamic contrast enhanced-MRI for the detection of pathological complete response to neoadjuvant chemotherapy for locally advanced rectal cancer. Eur. Radiol. 2012, 22, 821–831. [Google Scholar] [CrossRef]
- Cheedella, N.K.S.; Suzuki, A.; Xiao, L.; Hofstetter, W.L.; Maru, D.M.; Taketa, T.; Sudo, K.; Blum, M.A.; Lin, S.H.; Welch, J.; et al. Association between clinical complete response and pathological complete response after preoperative chemoradiation in patients with gastroesophageal cancer: Analysis in a large cohort. Ann. Oncol. 2013, 24, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Tranchart, H.; Lefevre, J.H.; Svrcek, M.; Fléjou, J.-F.; Tiret, E.; Parc, Y. What is the Incidence of Metastatic Lymph Node Involvement After Significant Pathologic Response of Primary Tumor Following Neoadjuvant Treatment for Locally Advanced Rectal Cancer? Ann. Surg. Oncol. 2012, 20, 1551–1559. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Azad, T.D.; Chaudhuri, A.A.; Fang, P.; Qiao, Y.; Esfahani, M.S.; Chabon, J.J.; Hamilton, E.G.; Yang, Y.D.; Lovejoy, A.; Newman, A.M.; et al. Circulating Tumor DNA Analysis for Detection of Minimal Residual Disease After Chemoradiotherapy for Localized Esophageal Cancer. Gastroenterology 2020, 158, 494–505. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boniface, C.; Deig, C.; Halsey, C.; Kelley, T.; Heskett, M.B.; Thomas, C.R., Jr.; Spellman, P.T.; Nabavizadeh, N. The Feasibility of Patient-Specific Circulating Tumor DNA Monitoring throughout Multi-Modality Therapy for Locally Advanced Esophageal and Rectal Cancer: A Potential Biomarker for Early Detection of Subclinical Disease. Diagnostics 2021, 11, 73. https://doi.org/10.3390/diagnostics11010073
Boniface C, Deig C, Halsey C, Kelley T, Heskett MB, Thomas CR Jr., Spellman PT, Nabavizadeh N. The Feasibility of Patient-Specific Circulating Tumor DNA Monitoring throughout Multi-Modality Therapy for Locally Advanced Esophageal and Rectal Cancer: A Potential Biomarker for Early Detection of Subclinical Disease. Diagnostics. 2021; 11(1):73. https://doi.org/10.3390/diagnostics11010073
Chicago/Turabian StyleBoniface, Christopher, Christopher Deig, Carol Halsey, Taylor Kelley, Michael B. Heskett, Charles R. Thomas, Jr., Paul T. Spellman, and Nima Nabavizadeh. 2021. "The Feasibility of Patient-Specific Circulating Tumor DNA Monitoring throughout Multi-Modality Therapy for Locally Advanced Esophageal and Rectal Cancer: A Potential Biomarker for Early Detection of Subclinical Disease" Diagnostics 11, no. 1: 73. https://doi.org/10.3390/diagnostics11010073
APA StyleBoniface, C., Deig, C., Halsey, C., Kelley, T., Heskett, M. B., Thomas, C. R., Jr., Spellman, P. T., & Nabavizadeh, N. (2021). The Feasibility of Patient-Specific Circulating Tumor DNA Monitoring throughout Multi-Modality Therapy for Locally Advanced Esophageal and Rectal Cancer: A Potential Biomarker for Early Detection of Subclinical Disease. Diagnostics, 11(1), 73. https://doi.org/10.3390/diagnostics11010073




