Identification of Tumor-Specific MRI Biomarkers Using Machine Learning (ML)
Abstract
:1. Introduction
2. Imaging Biomarkers
3. MRI Biomarkers
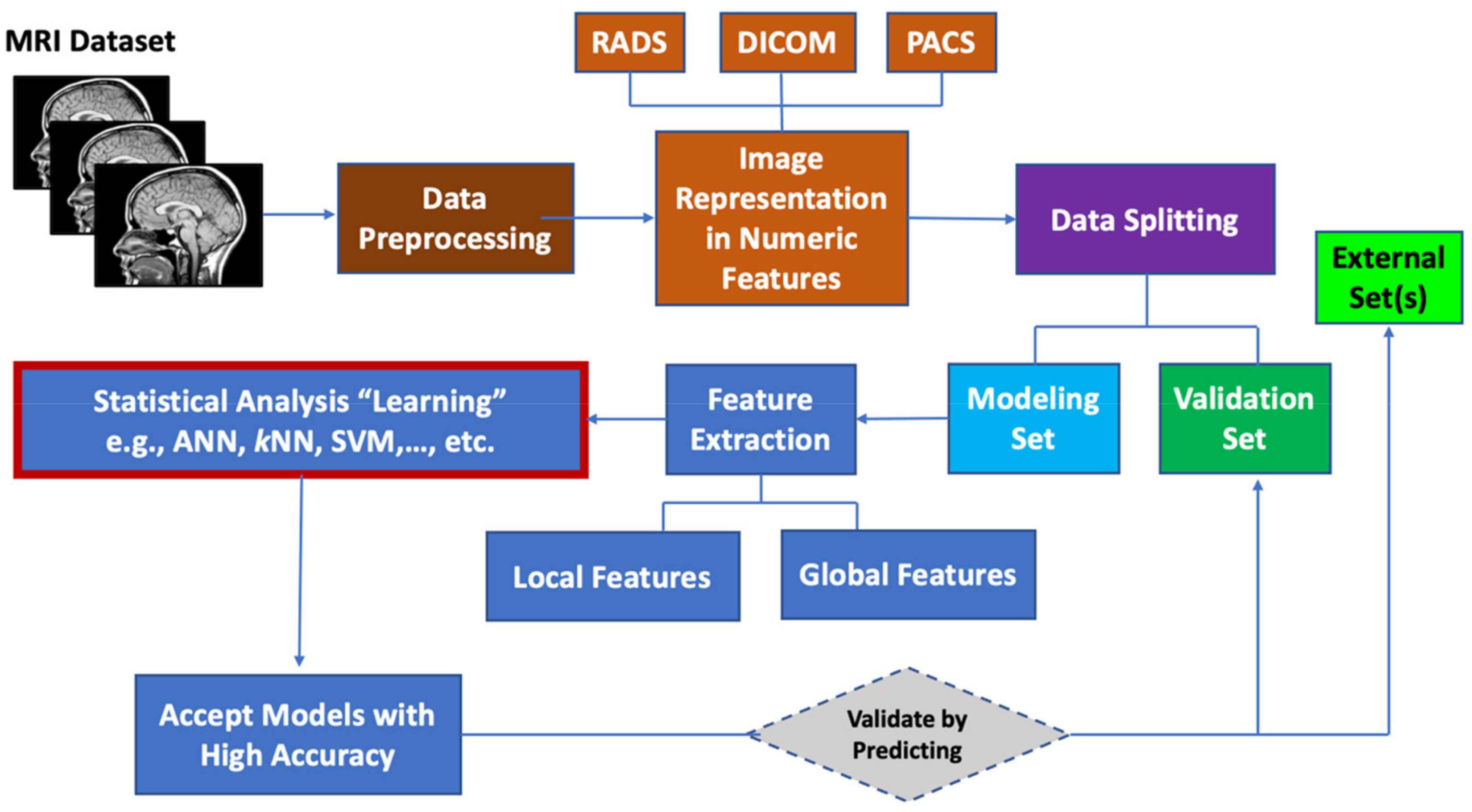
4. MRI Data Preprocessing
5. Machine Learning for MRI Data
5.1. Image Representation by Numeric Features
5.2. Feature Extraction
5.3. Data Set Division for Model Building, Model Tuning and External Validation
5.4. Machine Learning Algorithms
5.4.1. Artificial Neural Networks
5.4.2. Logistic Regression (LR)
5.4.3. Contrastive Learning
5.4.4. Deep Learning
5.4.5. k-Nearest Neighbors (kNN)
5.4.6. Support Vector Machines (SVM)
5.4.7. Random Forests
5.4.8. Self-Supervised Learning
5.4.9. Naïve Bayes
5.4.10. Decision Trees
5.4.11. Other Machine Learning Methods
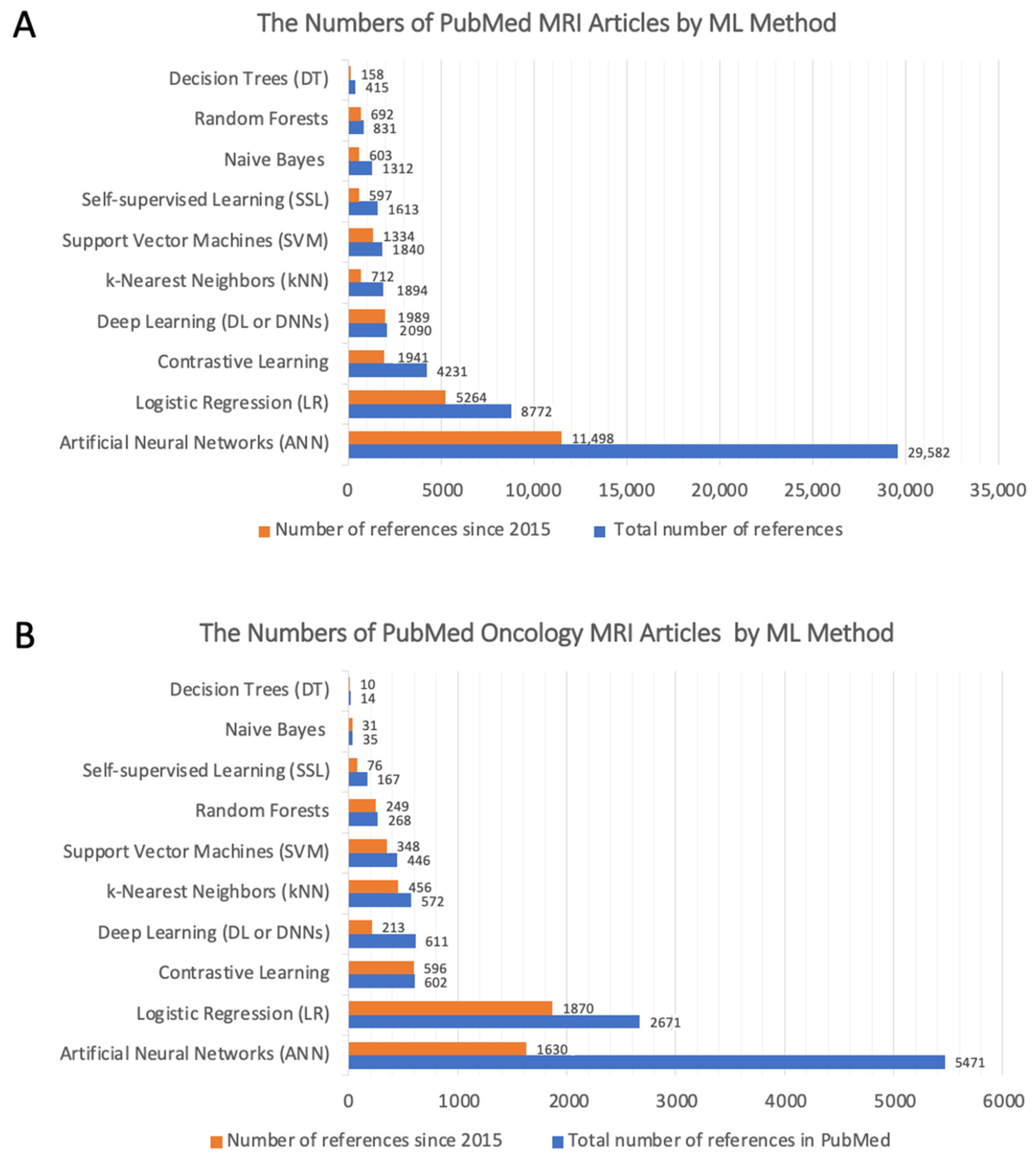
5.5. Which ML Method Is Best for Identifying Diagnostic MRI Biomarkers
5.6. Assessment of Model Performance
6. Types of MRI Biomarkers According to Clinical Use
6.1. Diagnostic Biomarkers
6.2. Prognostic Biomarkers
6.3. Response Biomarkers
7. Types of MRI Biomarkers Based on Quantitative Ability
7.1. Semi-Quantitative Recording Systems
7.2. Quantitative Recording Systems
7.3. Quantitative Imaging Biomarkers
8. Radiomic Signature Biomarkers
9. MRI Biomarker Standardization
10. Selected Examples on MRI Biomarkers in Solid Tumors
10.1. MRI Biomarkers for Prostate Cancer
10.2. MRI Biomarkers for Brain Tumors
11. Assigning and Interpreting of Proper Imaging Biomarkers to Confirm Decision-Making
12. Progress in Quantitative Imaging Biomarkers as Decision-Making Tools in Clinical Practice
13. The Challenges for Prioritizing MRI Biomarkers
14. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dregely, I.; Prezzi, D.; Kelly-Morland, C.; Roccia, E.; Neji, R.; Goh, V. Imaging Biomarkers in Oncology: Basics and Application to MRI. J. Magn. Reson. Imaging 2018, 48, 13–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercado, C.L. BI-RADS Update. Radiol. Clin. N. Am. 2014, 52, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Boellaard, R.; Delgado-Bolton, R.; Oyen, W.J.G.; Giammarile, F.; Tatsch, K.; Eschner, W.; Verzijlbergen, F.J.; Barrington, S.F.; Pike, L.C.; Weber, W.A.; et al. FDG PET/CT: EANM Procedure Guidelines for Tumour Imaging: Version 2.0. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 328–354. [Google Scholar] [CrossRef] [PubMed]
- Barentsz, J.O.; Weinreb, J.C.; Verma, S.; Thoeny, H.C.; Tempany, C.M.; Shtern, F.; Padhani, A.R.; Margolis, D.; Macura, K.J.; Haider, M.A.; et al. Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use. Eur. Urol. 2016, 69, 41–49. [Google Scholar] [CrossRef]
- DeSouza, N.M.; Achten, E.; Alberich-Bayarri, A.; Bamberg, F.; Boellaard, R.; Clément, O.; Fournier, L.; Gallagher, F.; Golay, X.; Heussel, C.P.; et al. Validated Imaging Biomarkers as Decision-Making Tools in Clinical Trials and Routine Practice: Current Status and Recommendations from the EIBALL* Subcommittee of the European Society of Radiology (ESR). Insights Imaging 2019, 10, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Leithner, D.; Helbich, T.H.; Bernard-Davila, B.; Marino, M.A.; Avendano, D.; Martinez, D.F.; Jochelson, M.S.; Kapetas, P.; Baltzer, P.A.T.; Haug, A.; et al. Multiparametric 18F-FDG PET/MRI of the Breast: Are There Differences in Imaging Biomarkers of Contralateral Healthy Tissue between Patients with and without Breast Cancer? J. Nucl. Med. 2020, 61, 20–25. [Google Scholar] [CrossRef]
- Jalali, S.; Chung, C.; Foltz, W.; Burrell, K.; Singh, S.; Hill, R.; Zadeh, G. MRI Biomarkers Identify the Differential Response of Glioblastoma Multiforme to Anti-Angiogenic Therapy. Neuro-Oncol. 2014, 16, 868–879. [Google Scholar] [CrossRef] [Green Version]
- Moffa, G.; Galati, F.; Collalunga, E.; Rizzo, V.; Kripa, E.; D’Amati, G.; Pediconi, F. Can MRI Biomarkers Predict Triple-Negative Breast Cancer? Diagnostics 2020, 10, 1090. [Google Scholar] [CrossRef]
- Grand, D.; Navrazhina, K.; Frew, J.W. A Scoping Review of Non-Invasive Imaging Modalities in Dermatological Disease: Potential Novel Biomarkers in Hidradenitis Suppurativa. Front. Med. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Just, N. Improving Tumour Heterogeneity MRI Assessment with Histograms. Br. J. Cancer 2014, 111, 2205–2213. [Google Scholar] [CrossRef] [Green Version]
- Padhani, A.R.; Liu, G.; Mu-Koh, D.; Chenevert, T.L.; Thoeny, H.C.; Takahara, T.; Dzik-Jurasz, A.; Ross, B.D.; van Cauteren, M.; Collins, D.; et al. Diffusion-Weighted Magnetic Resonance Imaging as a Cancer Biomarker: Consensus and Recommendations. In Neoplasia; Elsevier B.V.: Amsterdam, The Netherlands, 2009; Volume 11, pp. 102–125. [Google Scholar]
- Qiao, J.; Xue, S.; Pu, F.; White, N.; Jiang, J.; Liu, Z.R.; Yang, J.J. Molecular Imaging of EGFR/HER2 Cancer Biomarkers by Protein MRI Contrast Agents Topical Issue on Metal-Based MRI Contrast Agents. J. Biol. Inorg. Chem. 2014, 19, 259–270. [Google Scholar] [CrossRef] [Green Version]
- Watson, M.J.; George, A.K.; Maruf, M.; Frye, T.P.; Muthigi, A.; Kongnyuy, M.; Valayil, S.G.; Pinto, P.A. Risk Stratification of Prostate Cancer: Integrating Multiparametric MRI, Nomograms and Biomarkers. Future Oncol. 2016, 12, 2417–2430. [Google Scholar] [CrossRef] [Green Version]
- Kurhanewicz, J.; Vigneron, D.B.; Ardenkjaer-Larsen, J.H.; Bankson, J.A.; Brindle, K.; Cunningham, C.H.; Gallagher, F.A.; Keshari, K.R.; Kjaer, A.; Laustsen, C.; et al. Hyperpolarized 13C MRI: Path to Clinical Translation in Oncology. Neoplasia 2019, 21, 1–16. [Google Scholar] [CrossRef]
- O’Flynn, E.A.M.; Nandita, M.D. Functional Magnetic Resonance: Biomarkers of Response in Breast Cancer. Breast Cancer Res. 2011, 13, 204. [Google Scholar] [CrossRef] [Green Version]
- Lopci, E.; Franzese, C.; Grimaldi, M.; Zucali, P.A.; Navarria, P.; Simonelli, M.; Bello, L.; Scorsetti, M.; Chiti, A. Imaging Biomarkers in Primary Brain Tumours. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 597–612. [Google Scholar] [CrossRef]
- Weaver, O.; Leung, J.W.T. Biomarkers and Imaging of Breast Cancer. Am. J. Roentgenol. 2018, 210, 271–278. [Google Scholar] [CrossRef]
- O’Connor, J.P.B.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging Biomarker Roadmap for Cancer Studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef]
- Booth, T.C.; Williams, M.; Luis, A.; Cardoso, J.; Ashkan, K.; Shuaib, H. Machine Learning and Glioma Imaging Biomarkers. Clin. Radiol. 2020, 75, 20–32. [Google Scholar] [CrossRef] [Green Version]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and Other Tools). Updated Sept. 25 2017, 55. Available online: https://www.ncbi.nlm.nih.gov/books/NBK326791/ (accessed on 1 March 2021).
- Waldstein, S.M.; Seeböck, P.; Donner, R.; Sadeghipour, A.; Bogunović, H.; Osborne, A.; Schmidt-Erfurth, U. Unbiased Identification of Novel Subclinical Imaging Biomarkers Using Unsupervised Deep Learning. Sci. Rep. 2020, 10, 12954. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- About Biomarkers and Qualification|FDA. Available online: https://www.fda.gov/drugs/biomarker-qualification-program/about-biomarkers-and-qualification (accessed on 16 February 2021).
- European Society of Radiology. White Paper on Imaging Biomarkers. Insights Imaging 2010, 1, 42–45. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Xu, A. Application of Dual-Source CT Perfusion Imaging and MRI for the Diagnosis of Primary Liver Cancer. Oncol. Lett. 2017, 14, 5753–5758. [Google Scholar] [CrossRef] [Green Version]
- Heuvelmans, M.A.; Walter, J.E.; Vliegenthart, R.; van Ooijen, P.M.A.; de Bock, G.H.; de Koning, H.J.; Oudkerk, M. Disagreement of Diameter and Volume Measurements for Pulmonary Nodule Size Estimation in CT Lung Cancer Screening. Thorax 2018, 73, 779–781. [Google Scholar] [CrossRef]
- Van Riel, S.J.; Ciompi, F.; Jacobs, C.; Winkler Wille, M.M.; Scholten, E.T.; Naqibullah, M.; Lam, S.; Prokop, M.; Schaefer-Prokop, C.; van Ginneken, B. Malignancy Risk Estimation of Screen-Detected Nodules at Baseline CT: Comparison of the PanCan Model, Lung-RADS and NCCN Guidelines. Eur. Radiol. 2017, 27, 4019–4029. [Google Scholar] [CrossRef] [Green Version]
- Matoba, M.; Tsuji, H.; Shimode, Y.; Nagata, H.; Tonami, H. Diagnostic Performance of Adaptive 4D Volume Perfusion CT for Detecting Metastatic Cervical Lymph Nodes in Head and Neck Squamous Cell Carcinoma. Am. J. Roentgenol. 2018, 211, 1106–1111. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, M.; Chen, S.; Lei, X.; Zhang, X.; Huan, Y. A Meta-Analysis of Use of Prostate Imaging Reporting and Data System Version 2 (PI-RADS V2) with Multiparametric MR Imaging for the Detection of Prostate Cancer. Eur. Radiol. 2017, 27, 5204–5214. [Google Scholar] [CrossRef]
- Timmers, J.M.H.; van Doorne-Nagtegaal, H.J.; Zonderland, H.M.; van Tinteren, H.; Visser, O.; Verbeek, A.L.M.; den Heeten, G.J.; Broeders, M.J.M. The Breast Imaging Reporting and Data System (Bi-Rads) in the Dutch Breast Cancer Screening Programme: Its Role as an Assessment and Stratification Tool. Eur. Radiol. 2012, 22, 1717–1723. [Google Scholar] [CrossRef] [Green Version]
- Van der Pol, C.B.; Lim, C.S.; Sirlin, C.B.; McGrath, T.A.; Salameh, J.P.; Bashir, M.R.; Tang, A.; Singal, A.G.; Costa, A.F.; Fowler, K.; et al. Accuracy of the Liver Imaging Reporting and Data System in Computed Tomography and Magnetic Resonance Image Analysis of Hepatocellular Carcinoma or Overall Malignancy—A Systematic Review. Gastroenterology 2019, 156, 976–986. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, L.H.; Seymour, L.; Litière, S.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Standardisation and Disease-Specific Adaptations: Perspectives from the RECIST Working Group. Eur. J. Cancer 2016, 62, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Wahl, R.L.; Jacene, H.; Kasamon, Y.; Lodge, M.A. From RECIST to PERCIST: Evolving Considerations for PET Response Criteria in Solid Tumors. J. Nuc. Med. 2009, 50, 122S–150S. [Google Scholar] [CrossRef] [Green Version]
- Nael, K.; Bauer, A.H.; Hormigo, A.; Lemole, M.; Germano, I.M.; Puig, J.; Stea, B. Multiparametric MRI for Differentiation of Radiation Necrosis from Recurrent Tumor in Patients with Treated Glioblastoma. Am. J. Roentgenol. 2018, 210, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Bastiaannet, E.; Groen, B.; Jager, P.L.; Cobben, D.C.P.; van der Graaf, W.T.A.; Vaalburg, W.; Hoekstra, H.J. The Value of FDG-PET in the Detection, Grading and Response to Therapy of Soft Tissue and Bone Sarcomas; a Systematic Review and Meta-Analysis. Cancer Treat. Rev. 2004, 30, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Chang, S.J.; Chang, S.C.; Yuan, M.K. The Value of Positron Emission Tomography in Early Detection of Lung Cancer in High-Risk Population: A Systematic Review. Clin. Respir. J. 2013, 7, 1–6. [Google Scholar] [CrossRef]
- Parekh, V.S.; Macura, K.J.; Harvey, S.; Kamel, I.; EI-Khouli, R.; Bluemke, D.A.; Jacobs, M.A. Multiparametric Deep Learning Tissue Signatures for a Radiological Biomarker of Breast Cancer: Preliminary Results. Med. Phys. 2018, 47, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.J.; Gnanasegaran, G.; Buscombe, J.; Navalkissoor, S. Single Photon Emission Computed Tomography/Computed Tomography in the Evaluation of Neuroendocrine Tumours: A Review of the Literature. Nucl. Med. Commun. 2013, 34, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, U.; Ferencik, M.; Udelson, J.E.; Picard, M.H.; Truong, Q.A.; Patel, M.R.; Huang, M.; Pencina, M.; Mark, D.B.; Heitner, J.F.; et al. Prognostic Value of Noninvasive Cardiovascular Testing in Patients with Stable Chest Pain: Insights from the PROMISE Trial (Prospective Multicenter Imaging Study for Evaluation of Chest Pain). Circulation 2017, 135, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, V.; Campana, D.; Tomassetti, P.; Fanti, S. 68Ga-Labelled Peptides for Diagnosis of Gastroenteropancreatic NET. Eur. J. Nuc. Med. Mol. Imaging 2012, 39, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, J.E.; Howe, J.R. Imaging in Neuroendocrine Tumors: An Update for the Clinician. Int. J. Endocr. Oncol. 2015, 2, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Zacho, H.D.; Nielsen, J.B.; Afshar-Oromieh, A.; Haberkorn, U.; deSouza, N.; de Paepe, K.; Dettmann, K.; Langkilde, N.C.; Haarmark, C.; Fisker, R.V.; et al. Prospective Comparison of 68Ga-PSMA PET/CT, 18F-Sodium Fluoride PET/CT and Diffusion Weighted-MRI at for the Detection of Bone Metastases in Biochemically Recurrent Prostate Cancer. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1884–1897. [Google Scholar] [CrossRef]
- Gabriel, M.; Decristoforo, C.; Kendler, D.; Dobrozemsky, G.; Heute, D.; Uprimny, C.; Kovacs, P.; von Guggenberg, E.; Bale, R.; Virgolini, I.J. 68Ga-DOTA-Tyr3-Octreotide PET in Neuroendocrine Tumors: Comparison with Somatostatin Receptor Scintigraphy and CT. J. Nucl. Med. 2007, 48, 508–518. [Google Scholar] [CrossRef]
- Park, S.Y.; Zacharias, C.; Harrison, C.; Fan, R.E.; Kunder, C.; Hatami, N.; Giesel, F.; Ghanouni, P.; Daniel, B.; Loening, A.M.; et al. Gallium 68 PSMA-11 PET/MR Imaging in Patients with Intermediate- or High-Risk Prostate Cancer. Radiology 2018, 288, 495–505. [Google Scholar] [CrossRef] [Green Version]
- Delgado, A.F.; Delgado, A.F. Discrimination between Glioma Grades II and III Using Dynamic Susceptibility Perfusion MRI: A Meta-Analysis. Am. J. Neuroradiol. 2017, 38, 1348–1355. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Liu, C.; Zhao, L.; Jiang, J.; Zhang, J.; Li, S.; Zhu, W.; Wang, J. Amide Proton Transfer Imaging Allows Detection of Glioma Grades and Tumor Proliferation: Comparison with Ki-67 Expression and Proton MR Spectroscopy Imaging. Am. J. Neuroradiol. 2017, 38, 1702–1709. [Google Scholar] [CrossRef] [Green Version]
- Hayano, K.; Shuto, K.; Koda, K.; Yanagawa, N.; Okazumi, S.; Matsubara, H. Quantitative Measurement of Blood Flow Using Perfusion CT for Assessing Clinicopathologic Features and Prognosis in Patients with Rectal Cancer. Dis. Colon Rectum 2009, 52, 1624–1629. [Google Scholar] [CrossRef]
- Win, T.; Miles, K.A.; Janes, S.M.; Ganeshan, B.; Shastry, M.; Endozo, R.; Meagher, M.; Shortman, R.I.; Wan, S.; Kayani, I.; et al. Tumor Heterogeneity and Permeability as Measured on the CT Component of PET/CT Predict Survival in Patients with Non-Small Cell Lung Cancer. Clin. Cancer Res. 2013, 19, 3591–3599. [Google Scholar] [CrossRef] [Green Version]
- Lund, K.V.; Simonsen, T.G.; Kristensen, G.B.; Rofstad, E.K. Pretreatment Late-Phase DCE-MRI Predicts Outcome in Locally Advanced Cervix Cancer. Acta Oncol. 2017, 56, 675–681. [Google Scholar] [CrossRef] [Green Version]
- Fasmer, K.E.; Bjørnerud, A.; Ytre-Hauge, S.; Grüner, R.; Tangen, I.L.; Werner, H.M.J.; Bjørge, L.; Salvesen, Ø.O.; Trovik, J.; Krakstad, C.; et al. Preoperative Quantitative Dynamic Contrast-Enhanced MRI and Diffusion-Weighted Imaging Predict Aggressive Disease in Endometrial Cancer. Acta Radiol. 2018, 59, 1010–1017. [Google Scholar] [CrossRef]
- Yu, J.; Xu, Q.; Huang, D.Y.; Song, J.C.; Li, Y.; Xu, L.L.; Shi, H.B. Prognostic Aspects of Dynamic Contrast-Enhanced Magnetic Resonance Imaging in Synchronous Distant Metastatic Rectal Cancer. Eur. Radiol. 2017, 27, 1840–1847. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, S.M. Radiomics in Oncological PET/CT: Clinical Applications. Nucl. Med. Mol. Imaging 2018, 52, 170–189. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The Process and the Challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.; Devaraj, A. Radiomics of Pulmonary Nodules and Lung Cancer. Transl. Lung Cancer Res. 2017, 6, 86–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Oikonomou, A.; Wong, A.; Haider, M.A.; Khalvati, F. Radiomics-Based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Q.; Liang, C.H.; He, L.; Tian, J.; Liang, C.S.; Chen, X.; Ma, Z.L.; Liu, Z.Y. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J. Clin. Oncol. 2016, 34, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Jackson, A.; Parker, G.J.M.; Roberts, C.; Jayson, G.C. Dynamic Contrast-Enhanced MRI in Clinical Trials of Antivascular Therapies. Nat. Rev. Clin. Oncol. 2012, 9, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Younes, A.; Hilden, P.; Coiffier, B.; Hagenbeek, A.; Salles, G.; Wilson, W.; Seymour, J.F.; Kelly, K.; Gribben, J.; Pfreunschuh, M.; et al. International Working Group Consensus Response Evaluation Criteria in Lymphoma (RECIL 2017). Ann. Oncol. 2017, 28, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Dalm, S.U.; Verzijlbergen, J.F.; de Jong, M. Review: Receptor Targeted Nuclear Imaging of Breast Cancer. Int. J. Mol. Sci. 2017, 18, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakht, M.K.; Oh, S.W.; Youn, H.; Cheon, G.J.; Kwak, C.; Kang, K.W. Influence of Androgen Deprivation Therapy on the Uptake of PSMA-Targeted Agents: Emerging Opportunities and Challenges. Nucl. Med. Mol. Imaging 2017, 51, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Hayano, K.; Okazumi, S.; Shuto, K.; Matsubara, H.; Shimada, H.; Nabeya, Y.; Kazama, T.; Yanagawa, N.; Ochiai, T. Perfusion CT Can Predict the Response to Chemoradiation Therapy and Survival in Esophageal Squamous Cell Carcinoma: Initial Clinical Results. Oncol. Rep. 2007, 18, 901–908. [Google Scholar] [CrossRef] [Green Version]
- Bittencourt, L.K.; de Hollanda, E.S.; de Oliveira, R.V. Multiparametric MR Imaging for Detection and Locoregional Staging of Prostate Cancer. Top. Magn. Reson. Imaging 2016, 25, 109–117. [Google Scholar] [CrossRef]
- Galbán, C.J.; Hoff, B.A.; Chenevert, T.L.; Ross, B.D. Diffusion MRI in Early Cancer Therapeutic Response Assessment. NMR Biomed. 2017, 30, e3458. [Google Scholar] [CrossRef]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative Imaging Biomarkers Alliance (QIBA) Recommendations for Improved Precision of DWI and DCE-MRI Derived Biomarkers in Multicenter Oncology Trials. J. Magnet. Reson. Imaging 2019, 49, e101–e121. [Google Scholar] [CrossRef]
- Zeng, Q.; Shi, F.; Zhang, J.; Ling, C.; Dong, F.; Jiang, B. A Modified Tri-Exponential Model for Multi-b-Value Diffusion-Weighted Imaging: A Method to Detect the Strictly Diffusion-Limited Compartment in Brain. Front. Neurosci. 2018, 12, 102. [Google Scholar] [CrossRef]
- Langkilde, F.; Kobus, T.; Fedorov, A.; Dunne, R.; Tempany, C.; Mulkern, R.V.; Maier, S.E. Evaluation of Fitting Models for Prostate Tissue Characterization Using Extended-Range b-Factor Diffusion-Weighted Imaging. Magn. Reson. Med. 2018, 79, 2346–2358. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D.; Jacobson, S.; Kechris, K.; Kinney, G.L.; Foreman, M.G.; Doerschuk, C.M.; Make, B.J.; Curtis, J.L.; Rennard, S.I.; Barr, R.G.; et al. Biomarkers Predictive of Exacerbations in the SPIROMICS and COPDGene Cohorts. Am. J. Respir. Crit. Care Med. 2017, 195, 473–481. [Google Scholar] [CrossRef]
- Winfield, J.M.; Tunariu, N.; Rata, M.; Miyazaki, K.; Jerome, N.P.; Germuska, M.; Blackledge, M.D.; Collins, D.J.; de Bono, J.S.; Yap, T.A.; et al. Extracranial Soft-Tissue Tumors: Repeatability of Apparent Diffusion Coefficient Estimates from Diffusion-Weighted MR Imaging. Radiology 2017, 284, 88–99. [Google Scholar] [CrossRef]
- Taylor, A.J.; Salerno, M.; Dharmakumar, R.; Jerosch-Herold, M. T1 Mapping Basic Techniques and Clinical Applications. JACC Cardiovasc. Imaging 2016, 9, 67–81. [Google Scholar] [CrossRef] [Green Version]
- Toussaint, M.; Gilles, R.J.; Azzabou, N.; Marty, B.; Vignaud, A.; Greiser, A.; Carlier, P.G. Characterization of Benign Myocarditis Using Quantitative Delayed-Enhancement Imaging Based on MOLLI T1 Mapping. Medicine 2015, 94. [Google Scholar] [CrossRef]
- Jurcoane, A.; Wagner, M.; Schmidt, C.; Mayer, C.; Gracien, R.M.; Hirschmann, M.; Deichmann, R.; Volz, S.; Ziemann, U.; Hattingen, E. Within-Lesion Differences in Quantitative MRI Parameters Predict Contrast Enhancement in Multiple Sclerosis. J. Magn. Reson. Imaging 2013, 38, 1454–1461. [Google Scholar] [CrossRef]
- Salisbury, M.L.; Lynch, D.A.; van Beek, E.J.R.; Kazerooni, E.A.; Guo, J.; Xia, M.; Murray, S.; Anstrom, K.J.; Yow, E.; Martinez, F.J.; et al. Idiopathic Pulmonary Fibrosis: The Association between the Adaptive Multiple Features Method and Fibrosis Outcomes. Am. J. Respir. Crit. Care Med. 2017, 195, 921–929. [Google Scholar] [CrossRef] [Green Version]
- Katsube, T.; Okada, M.; Kumano, S.; Hori, M.; Imaoka, I.; Ishii, K.; Kudo, M.; Kitagaki, H.; Murakami, T. Estimation of Liver Function Using T1 Mapping on Gd-EOB-DTPA-Enhanced Magnetic Resonance Imaging. Investig. Radiol. 2011, 46, 277–283. [Google Scholar] [CrossRef]
- Mozes, F.E.; Tunnicliffe, E.M.; Moolla, A.; Marjot, T.; Levick, C.K.; Pavlides, M.; Robson, M.D. Mapping Tissue Water T1 in the Liver Using the MOLLI T1 Method in the Presence of Fat, Iron and B0 Inhomogeneity. NMR Biomed. 2019, 32. [Google Scholar] [CrossRef] [Green Version]
- Adam, S.Z.; Nikolaidis, P.; Horowitz, J.M.; Gabriel, H.; Hammond, N.A.; Patel, T.; Yaghmai, V.; Miller, F.H. Chemical Shift MR Imaging of the Adrenal Gland: Principles, Pitfalls, and Applications. Radiographics 2016, 36, 414–432. [Google Scholar] [CrossRef] [PubMed]
- Tietze, A.; Blicher, J.; Mikkelsen, I.K.; Østergaard, L.; Strother, M.K.; Smith, S.A.; Donahue, M.J. Assessment of Ischemic Penumbra in Patients with Hyperacute Stroke Using Amide Proton Transfer (APT) Chemical Exchange Saturation Transfer (CEST) MRI. NMR Biomed. 2014, 27, 163–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamoorthy, G.; Nanga, R.P.R.; Bagga, P.; Hariharan, H.; Reddy, R. High Quality Three-Dimensional GagCEST Imaging of in Vivo Human Knee Cartilage at 7 Tesla. Magn. Reson. Med. 2017, 77, 1866–1873. [Google Scholar] [CrossRef]
- Donahue, M.J.; Donahue, P.C.M.; Rane, S.; Thompson, C.R.; Strother, M.K.; Scott, A.O.; Smith, S.A. Assessment of Lymphatic Impairment and Interstitial Protein Accumulation in Patients with Breast Cancer Treatment-Related Lymphedema Using CEST MRI. Magn. Reson. Med. 2016, 75, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Lindeman, L.R.; Randtke, E.A.; High, R.A.; Jones, K.M.; Howison, C.M.; Pagel, M.D. A Comparison of Exogenous and Endogenous CEST MRI Methods for Evaluating in Vivo PH. Magn. Reson. Med. 2018, 79, 2766–2772. [Google Scholar] [CrossRef]
- Tang, A.; Bashir, M.R.; Corwin, M.T.; Cruite, I.; Dietrich, C.F.; Do, R.K.G.; Ehman, E.C.; Fowler, K.J.; Hussain, H.K.; Jha, R.C.; et al. Evidence Supporting LI-RADS Major Features for CT- and MR Imaging-Based Diagnosis of Hepatocellular Carcinoma: A Systematic Review. Radiology 2018, 286, 29–48. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, D.G.; Bruix, J.; Sherman, M.; Sirlin, C.B. LI-RADS (Liver Imaging Reporting and Data System): Summary, Discussion, and Consensus of the LI-RADS Management Working Group and Future Directions. Hepatology 2015, 61, 1056–1065. [Google Scholar] [CrossRef]
- Degani, H.; Gusis, V.; Weinstein, D.; Fields, S.; Strano, S. Mapping Pathophysiological Features of Breast Tumors by MRI at High Spatial Resolution. Nat. Med. 1997, 3, 780–782. [Google Scholar] [CrossRef]
- Uecker, M.; Zhang, S.; Voit, D.; Karaus, A.; Merboldt, K.D.; Frahm, J. Real-Time MRI at a Resolution of 20 Ms. NMR Biomed. 2010, 23, 986–994. [Google Scholar] [CrossRef] [Green Version]
- Van Wijk, D.F.; Strang, A.C.; Duivenvoorden, R.; Enklaar, D.-J.F.; Zwinderman, A.H.; van der Geest, R.J.; Kastelein, J.J.P.; de Groot, E.; Stroes, E.S.G.; Nederveen, A.J. Increasing the Spatial Resolution of 3T Carotid MRI Has No Beneficial Effect for Plaque Component Measurement Reproducibility. PLoS ONE 2015, 10, e0130878. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Trebeschi, S.; Drago, S.G.; Birkbak, N.J.; Kurilova, I.; Cǎlin, A.M.; Delli Pizzi, A.; Lalezari, F.; Lambregts, D.M.J.; Rohaan, M.W.; Parmar, C.; et al. Predicting Response to Cancer Immunotherapy Using Noninvasive Radiomic Biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [Green Version]
- Bengio, Y.; Courville, A.; Vincent, P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Li, B.; Li, W.; Zhao, D. Global and Local Features Based Medical Image Classification. J. Med. Imaging Health Inform. 2015, 5, 748–754. [Google Scholar] [CrossRef]
- Tamura, H.; Mori, S.; Yamawaki, T. Textural Features Corresponding to Visual Perception. IEEE Trans. Syst. Man Cybern. 1978, 8, 460–473. [Google Scholar] [CrossRef]
- Haralick, R.M.; Dinstein, I.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 67, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Rubner, Y.; Puzicha, J.; Tomasi, C.; Buhmann, J.M. Empirical Evaluation of Dissimilarity Measures for Color and Texture. Comput. Vis. Image Underst. 2001, 84, 25–43. [Google Scholar] [CrossRef]
- Castelli, V.; Bergman, L.D.; Kontoyiannis, I.; Li, C.S.; Robinson, J.T.; Turek, J.J. Progressive Search and Retrieval in Large Image Archives. IBM J. Res. Dev. 1998, 42, 253–267. [Google Scholar] [CrossRef] [Green Version]
- Ngo, C.W.; Pong, T.C.; Chin, R.T. Exploiting Image Indexing Techniques in DCT Domain. Pattern Recognit. 2001, 34, 1841–1851. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.S.; Huang, T.S. Edge-Based Structural Features for Content-Based Image Retrieval. Pattern Recognit. Lett. 2001, 22, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Güld, M.O.; Keysers, D.; Deselaers, T.; Leisten, M.; Schubert, H.; Ney, H.; Lehmann, T.M. Comparison of Global Features for Categorization of Medical Images. In Proceedings of the Medical Imaging 2004, San Diego, CA, USA, 15–17 February 2004. [Google Scholar]
- Local Feature Detection and Extraction—MATLAB & Simulink. Available online: https://www.mathworks.com/help/vision/ug/local-feature-detection-and-extraction.html (accessed on 16 February 2021).
- Hajjo, R.; Grulke, C.M.; Golbraikh, A.; Setola, V.; Huang, X.-P.; Roth, B.L.; Tropsha, A. Development, Validation, and Use of Quantitative Structure-Activity Relationship Models of 5-Hydroxytryptamine (2B) Receptor Ligands to Identify Novel Receptor Binders and Putative Valvulopathic Compounds among Common Drugs. J. Med. Chem. 2010, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajjo, R.; Setola, V.; Roth, B.L.; Tropsha, A. Chemocentric Informatics Approach to Drug Discovery: Identification and Experimental Validation of Selective Estrogen Receptor Modulators as Ligands of 5-Hydroxytryptamine-6 Receptors and as Potential Cognition Enhancers. J. Med. Chem. 2012, 55, 5704–5719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forghani, R.; Savadjiev, P.; Chatterjee, A.; Muthukrishnan, N.; Reinhold, C.; Forghani, B. Radiomics and Artificial Intelligence for Biomarker and Prediction Model Development in Oncology. Comput. Struct. Biotechnol. J. 2019, 17, 995–1008. [Google Scholar] [CrossRef]
- Kononenko, I. Machine Learning for Medical Diagnosis: History, State of the Art and Perspective. Artif. Intell. Med. 2001, 23, 89–109. [Google Scholar] [CrossRef] [Green Version]
- Arora, S.; Khandeparkar, H.; Khodak, M.; Plevrakis, O.; Saunshi, N. A Theoretical Analysis of Contrastive Unsupervised Representation Learning. arXiv 2019, arXiv:1902.09229. [Google Scholar]
- Lundervold, A.S.; Lundervold, A. An Overview of Deep Learning in Medical Imaging Focusing on MRI. Z. Fur Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Mehdy, M.M.; Ng, P.Y.; Shair, E.F.; Saleh, N.I.M.; Gomes, C. Artificial Neural Networks in Image Processing for Early Detection of Breast Cancer. Comput. Math. Methods Med. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Joo, S.; Yang, Y.S.; Moon, W.K.; Kim, H.C. Computer-Aided Diagnosis of Solid Breast Nodules: Use of an Artificial Neural Network Based on Multiple Sonographic Features. IEEE Trans. Med. Imaging 2004, 23, 1292–1300. [Google Scholar] [CrossRef]
- Tolles, J.; Meurer, W.J. Logistic Regression: Relating Patient Characteristics to Outcomes. JAMA 2016, 316, 533–534. [Google Scholar] [CrossRef]
- Bühlmann, P.; van de Geer, S. Statistics for High-Dimensional Data; Springer Series in Statistics; Springer: Berlin/Heidelberg, Germany, 2011; ISBN 978-3-642-20191-2. [Google Scholar]
- Chaitanya, K.; Erdil, E.; Karani, N.; Konukoglu, E. Contrastive Learning of Global and Local Features for Medical Image Segmentation with Limited Annotations. arXiv 2020, arXiv:2006.10511. [Google Scholar]
- Chen, T.; Kornblith, S.; Swersky, K.; Norouzi, M.; Hinton, G. Big Self-Supervised Models Are Strong Semi-Supervised Learners. arXiv 2020, arXiv:2006.10029. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2323. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Xie, L.; Han, J.; Guo, X. The Application of Deep Learning in Cancer Prognosis Prediction. Cancers 2020, 12, 603. [Google Scholar] [CrossRef] [Green Version]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized Treatment Recommender System Using a Cox Proportional Hazards Deep Neural Network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef]
- Ching, T.; Zhu, X.; Garmire, L.X. Cox-Nnet: An Artificial Neural Network Method for Prognosis Prediction of High-Throughput Omics Data. PLoS Comput. Biol. 2018, 14, e1006076. [Google Scholar] [CrossRef]
- Joshi, R.; Reeves, C.R. Beyond the Cox Model: Artificial Neural Networks for Survival Analysis Part II; Systems Engineering; Coventry University: Coventry, UK, 2006; ISBN 1846000130. [Google Scholar]
- Mudassar, B.A.; Mukhopadhyay, S. FocalNet—Foveal Attention for Post-Processing DNN Outputs. In Proceedings of the International Joint Conference on Neural Networks, Budapest, Hungary, 14–19 July 2019. [Google Scholar]
- Zheng, W.; Tropsha, A. Novel Variable Selection Quantitative Structure--Property Relationship Approach Based on the k-Nearest-Neighbor Principle. J. Chem. Inf. Comput. Sci. 2000, 40, 185–194. [Google Scholar] [CrossRef]
- Vapnik, V.N.; Vapnik, V.N. Introduction: Four Periods in the Research of the Learning Problem. In The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 2000; pp. 1–15. [Google Scholar]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Dahinden, C.; Dahinden, C.; Guyon, I. An Improved Random Forests Approach with Application to the Performance Prediction Challenge Datasets. Hands Pattern Recognit. 2009, 1, 223–230. [Google Scholar]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Self-Supervised Learning for Medical Image Analysis Using Image Context Restoration. Med. Image Anal. 2019, 58, 101539. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2009; ISBN 978-0-387-84857-0. [Google Scholar]
- Piryonesi, S.M.; El-Diraby, T.E. Role of Data Analytics in Infrastructure Asset Management: Overcoming Data Size and Quality Problems. J. Transp. Eng. Part B Pavements 2020, 146, 04020022. [Google Scholar] [CrossRef]
- Rokach, L.; Maimon, O. Data Mining with Decision Trees; World Scientific: Singapore, 2014. [Google Scholar]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing Medical Imaging Data for Machine Learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Chaudhari, A.S.; Sandino, C.M.; Cole, E.K.; Larson, D.B.; Gold, G.E.; Vasanawala, S.S.; Lungren, M.P.; Hargreaves, B.A.; Langlotz, C.P. Prospective Deployment of Deep Learning in MRI: A Framework for Important Considerations, Challenges, and Recommendations for Best Practices. J. Magn. Reson. Imaging 2020. [Google Scholar] [CrossRef]
- Golbraikh, A.; Shen, M.; Tropsha, A. Enrichment: A New Estimator of Classification Accuracy of QSAR Models. In Abstracts of Papers of the American Chemical Society; American Chemical Society: Washington, DC, USA, 2002; pp. U494–U495. [Google Scholar]
- Matsuo, H.; Nishio, M.; Kanda, T.; Kojita, Y.; Kono, A.K.; Hori, M.; Teshima, M.; Otsuki, N.; Nibu, K.I.; Murakami, T. Diagnostic Accuracy of Deep-Learning with Anomaly Detection for a Small Amount of Imbalanced Data: Discriminating Malignant Parotid Tumors in MRI. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Li, M.; Shang, Z.; Yang, Z.; Zhang, Y.; Wan, H. Machine Learning Methods for MRI Biomarkers Analysis of Pediatric Posterior Fossa Tumors. Biocybern. Biomed. Eng. 2019, 39, 765–774. [Google Scholar] [CrossRef]
- Yurttakal, A.H.; Erbay, H.; İkizceli, T.; Karaçavuş, S. Detection of Breast Cancer via Deep Convolution Neural Networks Using MRI Images. Multimed. Tools Appl. 2020, 79, 15555–15573. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial Intelligence in Cancer Imaging: Clinical Challenges and Applications. CA A Cancer J. Clin. 2019, 69, 127. [Google Scholar] [CrossRef] [Green Version]
- Banaei, N.; Moshfegh, J.; Mohseni-Kabir, A.; Houghton, J.M.; Sun, Y.; Kim, B. Machine Learning Algorithms Enhance the Specificity of Cancer Biomarker Detection Using SERS-Based Immunoassays in Microfluidic Chips. RSC Adv. 2019, 9, 1859–1868. [Google Scholar] [CrossRef] [Green Version]
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Schnall, M.D.; Shtern, F.; Tempany, C.M.; et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef] [PubMed]
- El-Shater Bosaily, A.; Parker, C.; Brown, L.C.; Gabe, R.; Hindley, R.G.; Kaplan, R.; Emberton, M.; Ahmed, H.U.; Emberton, M.; Ahmed, H.; et al. PROMIS—Prostate MR Imaging Study: A Paired Validating Cohort Study Evaluating the Role of Multi-Parametric MRI in Men with Clinical Suspicion of Prostate Cancer. Contemp. Clin. Trials 2015, 42, 26–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic Accuracy of Multi-Parametric MRI and TRUS Biopsy in Prostate Cancer (PROMIS): A Paired Validating Confirmatory Study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [Green Version]
- De Rooij, M.; Hamoen, E.H.J.; Fütterer, J.J.; Barentsz, J.O.; Rovers, M.M. Accuracy of Multiparametric MRI for Prostate Cancer Detection: A Meta-Analysis. Am. J. Roentgenol. 2014, 202, 343–351. [Google Scholar] [CrossRef]
- Hamoen, E.H.J.; de Rooij, M.; Witjes, J.A.; Barentsz, J.O.; Rovers, M.M. Use of the Prostate Imaging Reporting and Data System (PI-RADS) for Prostate Cancer Detection with Multiparametric Magnetic Resonance Imaging: A Diagnostic Meta-Analysis. Eur. Urol. 2015, 67, 1112–1121. [Google Scholar] [CrossRef]
- Beets-Tan, R.G.H.; Beets, G.L.; Vliegen, R.F.A.; Kessels, A.G.H.; van Boven, H.; de Bruine, A.; von Meyenfeldt, M.F.; Baeten, C.G.M.I.; van Engelshoven, J.M.A. Accuracy of Magnetic Resonance Imaging in Prediction of Tumour-Free Resection Margin in Rectal Cancer Surgery. Lancet 2001, 357, 497–504. [Google Scholar] [CrossRef]
- Taylor, F.G.M.; Quirke, P.; Heald, R.J.; Moran, B.J.; Blomqvist, L.; Swift, I.R.; Sebag-Montefiore, D.; Tekkis, P.; Brown, G. Preoperative Magnetic Resonance Imaging Assessment of Circumferential Resection Margin Predicts Disease-Free Survival and Local Recurrence: 5-Year Follow-up Results of the MERCURY Study. J. Clin. Oncol. 2014, 32, 34–43. [Google Scholar] [CrossRef]
- Brown, G. Diagnostic Accuracy of Preoperative Magnetic Resonance Imaging in Predicting Curative Resection of Rectal Cancer: Prospective Observational Study. Br. Med. J. 2006, 333, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, S.B.; Vesoulis, Z.A.; Rao, R.; Liao, S.M.; Shimony, J.S.; McKinstry, R.C.; Mathur, A.M. A Validated Clinical MRI Injury Scoring System in Neonatal Hypoxic-Ischemic Encephalopathy. Pediatric Radiol. 2017, 47, 1491–1499. [Google Scholar] [CrossRef]
- Machino, M.; Ando, K.; Kobayashi, K.; Ito, K.; Tsushima, M.; Morozumi, M.; Tanaka, S.; Ota, K.; Ito, K.; Kato, F.; et al. Alterations in Intramedullary T2-Weighted Increased Signal Intensity Following Laminoplasty in Cervical Spondylotic Myelopathy Patients: Comparison between Pre- and Postoperative Magnetic Resonance Images. Spine 2018, 43, 1595–1601. [Google Scholar] [CrossRef]
- Chen, C.J.; Lyu, R.K.; Lee, S.T.; Wong, Y.C.; Wang, L.J. Intramedullary High Signal Intensity on T2-Weighted MR Images in Cervical Spondylotic Myelopathy: Prediction of Prognosis with Type of Intensity. Radiology 2001, 221, 789–794. [Google Scholar] [CrossRef]
- Khanna, D.; Ranganath, V.K.; FitzGerald, J.; Park, G.S.; Altman, R.D.; Elashoff, D.; Gold, R.H.; Sharp, J.T.; Fürst, D.E.; Paulus, H.E. Increased Radiographic Damage Scores at the Onset of Seropositive Rheumatoid Arthritis in Older Patients Are Associated with Osteoarthritis of the Hands, but Not with More Rapid Progression of Damage. Arthritis Rheum. 2005, 52, 2284–2292. [Google Scholar] [CrossRef]
- Jaremko, J.L.; Azmat, O.; Lambert, R.G.W.; Bird, P.; Haugen, I.K.; Jans, L.; Weber, U.; Winn, N.; Zubler, V.; Maksymowych, W.P. Validation of a Knowledge Transfer Tool According to the OMERACT Filter: Does Web-Based Real-Time Iterative Calibration Enhance the Evaluation of Bone Marrow Lesions in Hip Osteoarthritis? J. Rheumatol. 2017, 44, 1713–1717. [Google Scholar] [CrossRef]
- Molyneux, P.D.; Miller, D.H.; Filippi, M.; Yousry, T.A.; Radü, E.W.; Adèr, H.J.; Barkhof, F. Visual Analysis of Serial T2-Weighted MRI in Multiple Sclerosis: Intra- and Interobserver Reproducibility. Neuroradiology 1999, 41, 882–888. [Google Scholar] [CrossRef]
- Stollfuss, J.C.; Becker, K.; Sendler, A.; Seidl, S.; Settles, M.; Auer, F.; Beer, A.; Rummeny, E.J.; Woertler, K. Rectal Carcinoma: High Spatial-Resolution MR Imaging and T2 Quantification in Rectal Cancer Specimens. Radiology 2006, 241, 132–141. [Google Scholar] [CrossRef]
- Barrington, S.F.; Mikhaeel, N.G.; Kostakoglu, L.; Meignan, M.; Hutchings, M.; Müeller, S.P.; Schwartz, L.H.; Zucca, E.; Fisher, R.I.; Trotman, J.; et al. Role of Imaging in the Staging and Response Assessment of Lymphoma: Consensus of the International Conference on Malignant Lymphomas Imaging Working Group. J. Clin. Oncol. 2014, 32, 3048–3058. [Google Scholar] [CrossRef]
- Chernyak, V.; Fowler, K.J.; Kamaya, A.; Kielar, A.Z.; Elsayes, K.M.; Bashir, M.R.; Kono, Y.; Do, R.K.; Mitchell, D.G.; Singal, A.G.; et al. Liver Imaging Reporting and Data System (LI-RADS) Version 2018: Imaging of Hepatocellular Carcinoma in at-Risk Patients. Radiology 2018, 289, 816–830. [Google Scholar] [CrossRef]
- Elsayes, K.M.; Hooker, J.C.; Agrons, M.M.; Kielar, A.Z.; Tang, A.; Fowler, K.J.; Chernyak, V.; Bashir, M.R.; Kono, Y.; Do, R.K.; et al. 2017 Version of LI-RADS for CT and MR Imaging: An Update. Radiographics 2017, 37, 1994–2017. [Google Scholar] [CrossRef]
- Tessler, F.N.; Middleton, W.D.; Grant, E.G.; Hoang, J.K.; Berland, L.L.; Teefey, S.A.; Cronan, J.J.; Beland, M.D.; Desser, T.S.; Frates, M.C.; et al. ACR Thyroid Imaging, Reporting and Data System (TI-RADS): White Paper of the ACR TI-RADS Committee. J. Am. Coll. Radiol. 2017, 14, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Panebianco, V.; Narumi, Y.; Altun, E.; Bochner, B.H.; Efstathiou, J.A.; Hafeez, S.; Huddart, R.; Kennish, S.; Lerner, S.; Montironi, R.; et al. Multiparametric Magnetic Resonance Imaging for Bladder Cancer: Development of VI-RADS (Vesical Imaging-Reporting and Data System). Eur. Urol. 2018, 74, 294–306. [Google Scholar] [CrossRef] [Green Version]
- Kitajima, K.; Tanaka, U.; Ueno, Y.; Maeda, T.; Suenaga, Y.; Takahashi, S.; Deguchi, M.; Miyahara, Y.; Ebina, Y.; Yamada, H.; et al. Role of Diffusion Weighted Imaging and Contrast-Enhanced MRI in the Evaluation of Intrapelvic Recurrence of Gynecological Malignant Tumor. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Cornelis, F.; Tricaud, E.; Lasserre, A.S.; Petitpierre, F.; Bernhard, J.C.; le Bras, Y.; Yacoub, M.; Bouzgarrou, M.; Ravaud, A.; Grenier, N. Multiparametric Magnetic Resonance Imaging for the Differentiation of Low and High Grade Clear Cell Renal Carcinoma. Eur. Radiol. 2015, 25, 24–31. [Google Scholar] [CrossRef]
- Martin, M.D.; Kanne, J.P.; Broderick, L.S.; Kazerooni, E.A.; Meyer, C.A. Lung-RADS: Pushing the Limits. Radiographics 2017, 37, 1975–1993. [Google Scholar] [CrossRef]
- Sabra, M.M.; Sherman, E.J.; Tuttle, R.M. Tumor Volume Doubling Time of Pulmonary Metastases Predicts Overall Survival and Can Guide the Initiation of Multikinase Inhibitor Therapy in Patients with Metastatic, Follicular Cell-Derived Thyroid Carcinoma. Cancer 2017, 123, 2955–2964. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Yao, G.H.; Zhang, M.; Yin, L.X.; Zhang, C.; Xu, M.J.; Deng, Y.; Liu, Y.; Deng, Y.B.; Ren, W.D.; Li, Z.A.; et al. Doppler Echocardiographic Measurements in Normal ChineseAdults (EMINCA): A Prospective, Nationwide, and Multicentre Study. Eur. Heart J. Cardiovasc. Imaging 2016, 17, 512–522. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, C.P.; Kleyer, A.; Simon, D.; Stemmler, F.; d’Oliveira, I.; Weissenfels, A.; Museyko, O.; Friedberger, A.; Hueber, A.J.; Haschka, J.; et al. Methods for Segmentation of Rheumatoid Arthritis Bone Erosions in High-Resolution Peripheral Quantitative Computed Tomography (HR-PQCT). Semin. Arthritis Rheum. 2018, 47, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Marcus, C.D.; Ladam-Marcus, V.; Cucu, C.; Bouché, O.; Lucas, L.; Hoeffel, C. Imaging Techniques to Evaluate the Response to Treatment in Oncology: Current Standards and Perspectives. Crit. Rev. Oncol. Hematol. 2009, 72, 217–238. [Google Scholar] [CrossRef]
- Levine, Z.H.; Pintar, A.L.; Hagedorn, J.G.; Fenimore, C.P.; Heussel, C.P. Uncertainties in RECIST as a Measure of Volume for Lung Nodules and Liver Tumors. Med. Phys. 2012, 39, 2628–2637. [Google Scholar] [CrossRef]
- Hawnaur, J.M.; Johnson, R.J.; Buckley, C.H.; Tindall, V.; Isherwood, I. Staging, Volume Estimation and Assessment of Nodal Status in Carcinoma of the Cervix: Comparison of Magnetic Resonance Imaging with Surgical Findings. Clin. Radiol. 1994, 49, 443–452. [Google Scholar] [CrossRef]
- Soutter, W.P.; Hanoch, J.; D’Arcy, T.; Dina, R.; McIndoe, G.A.; DeSouza, N.M. Pretreatment Tumour Volume Measurement on High-Resolution Magnetic Resonance Imaging as a Predictor of Survival in Cervical Cancer. BJOG Int. J. Obstet. Gynaecol. 2004, 111, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; You, K.; Qiu, X.; Bi, Z.; Mo, H.; Li, L.; Liu, Y. Tumor Volume Predicts Local Recurrence in Early Rectal Cancer Treated with Radical Resection: A Retrospective Observational Study of 270 Patients. Int. J. Surg. 2018, 49, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Tayyab, M.; Razack, A.; Sharma, A.; Gunn, J.; Hartley, J.E. Correlation of Rectal Tumor Volumes with Oncological Outcomes for Low Rectal Cancers: Does Tumor Size Matter? Surg. Today 2015, 45, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, H.C.; Trimbos, J.B.M.Z.; Postema, S.; Anastasopoulou, A.; van der Geest, R.J.; Reiber, J.H.C.; Kenter, G.G.; Peters, A.A.W.; Pattynama, P.M.T. Tumor Diameter and Volume Assessed by Magnetic Resonance Imaging in the Prediction of Outcome for Invasive Cervical Cancer. Gynecol. Oncol. 2001, 82, 474–482. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, S.M.; Yun, M.; Cho, A. Prognostic Value of Volumetric Parameters on Staging and Posttreatment FDG PET/CT in Patients with Stage IV Non-Small Cell Lung Cancer. Clin. Nucl. Med. 2016, 41, 347–353. [Google Scholar] [CrossRef]
- Kurtipek, E.; Çayci, M.; Düzgün, N.; Esme, H.; Terzi, Y.; Bakdik, S.; Aygün, M.S.; Unlü, Y.; Burnik, C.; Bekci, T.T. 18F-FDG PET/CT Mean SUV and Metabolic Tumor Volume for Mean Survival Time in Non-Small Cell Lung Cancer. Clin. Nucl. Med. 2015, 40, 459–463. [Google Scholar] [CrossRef]
- Meignan, M.; Itti, E.; Gallamini, A.; Younes, A. FDG PET/CT Imaging as a Biomarker in Lymphoma. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 623–633. [Google Scholar] [CrossRef]
- Kanoun, S.; Tal, I.; Berriolo-Riedinger, A.; Rossi, C.; Riedinger, J.M.; Vrigneaud, J.M.; Legrand, L.; Humbert, O.; Casasnovas, O.; Brunotte, F.; et al. Influence of Software Tool and Methodological Aspects of Total Metabolic Tumor Volume Calculation on Baseline [18F] FDG PET to Predict Survival in Hodgkin Lymphoma. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Kostakoglu, L.; Chauvie, S. Metabolic Tumor Volume Metrics in Lymphoma. Semin. Nucl. Med. 2018, 48, 50–66. [Google Scholar] [CrossRef]
- Mori, S.; Oishi, K.; Faria, A.V.; Miller, M.I. Atlas-Based Neuroinformatics via MRI: Harnessing Information from Past Clinical Cases and Quantitative Image Analysis for Patient Care. Annu. Rev. Biomed. Eng. 2013, 15, 71–92. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.H.; Poudel, R.P.K.; Tsagkrasoulis, D.; Caan, M.W.A.; Steves, C.; Spector, T.D.; Montana, G. Predicting Brain Age with Deep Learning from Raw Imaging Data Results in a Reliable and Heritable Biomarker. NeuroImage 2017, 163, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Hosny, A.; Zeleznik, R.; Parmar, C.; Coroller, T.; Franco, I.; Mak, R.H.; Aerts, H.J.W.L. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin. Cancer Res. 2019, 25, 3266–3275. [Google Scholar] [CrossRef] [Green Version]
- Davnall, F.; Yip, C.S.P.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.J.; Goh, V. Assessment of Tumor Heterogeneity: An Emerging Imaging Tool for Clinical Practice? Insights Imaging 2012, 3, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and Limitations of Radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, J.P.B.; Rose, C.J.; Waterton, J.C.; Carano, R.A.D.; Parker, G.J.M.; Jackson, A. Imaging Intratumor Heterogeneity: Role in Therapy Response, Resistance, and Clinical Outcome. Clin. Cancer Res. 2015, 21, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Drukker, K.; Giger, M.L.; Joe, B.N.; Kerlikowske, K.; Greenwood, H.; Drukteinis, J.S.; Niell, B.; Fan, B.; Malkov, S.; Avila, J.; et al. Combined Benefit of Quantitative Three-Compartment Breast Image Analysis and Mammography Radiomics in the Classification of Breast Masses in a Clinical Data Set. Radiology 2019, 290, 621–628. [Google Scholar] [CrossRef]
- Zhao, B.; Tan, Y.; Tsai, W.Y.; Qi, J.; Xie, C.; Lu, L.; Schwartz, L.H. Reproducibility of Radiomics for Deciphering Tumor Phenotype with Imaging. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Peerlings, J.; Woodruff, H.C.; Winfield, J.M.; Ibrahim, A.; van Beers, B.E.; Heerschap, A.; Jackson, A.; Wildberger, J.E.; Mottaghy, F.M.; DeSouza, N.M.; et al. Stability of Radiomics Features in Apparent Diffusion Coefficient Maps from a Multi-Centre Test-Retest Trial. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Sanduleanu, S.; Woodruff, H.C.; de Jong, E.E.C.; van Timmeren, J.E.; Jochems, A.; Dubois, L.; Lambin, P. Tracking Tumor Biology with Radiomics: A Systematic Review Utilizing a Radiomics Quality Score. Radiother. Oncol. 2018, 127, 349–360. [Google Scholar] [CrossRef]
- Strickland, N.H. PACS (Picture Archiving and Communication Systems): Filmless Radiology. Arch. Dis. Child. 2000, 83, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Bidgood, W.D.; Horii, S.C.; Prior, F.W.; van Syckle, D.E. Understanding and Using DICOM, the Data Interchange Standard for Biomedical Imaging. J. Am. Med. Inform. Assoc. 1997, 4, 199–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; Bellmunt, J.; Bolla, M.; Briers, E.; Cumberbatch, M.G.; de Santis, M.; Fossati, N.; Gross, T.; Henry, A.M.; Joniau, S.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017, 71, 618–629. [Google Scholar] [CrossRef]
- Manenti, G.; Nezzo, M.; Chegai, F.; Vasili, E.; Bonanno, E.; Simonetti, G. DWI of Prostate Cancer: Optimal b -Value in Clinical Practice. Prostate Cancer 2014, 2014, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Kim, C.K.; Park, B.K.; Kwon, G.Y. Comparison of Apparent Diffusion Coefficient Calculation between Two-Point and Multipoint b Value Analyses in Prostate Cancer and Benign Prostate Tissue at 3 T: Preliminary Experience. Am. J. Roentgenol. 2014, 202, W287–W294. [Google Scholar] [CrossRef]
- Penzkofer, T.; Tempany-Afdhal, C.M. Prostate Cancer Detection and Diagnosis: The Role of MR and Its Comparison with Other Diagnostic Modalities—a Radiologist’s Perspective. NMR Biomed. 2014, 27, 3–15. [Google Scholar] [CrossRef]
- Schimmöller, L.; Quentin, M.; Arsov, C.; Hiester, A.; Buchbender, C.; Rabenalt, R.; Albers, P.; Antoch, G.; Blondin, D. MR-Sequences for Prostate Cancer Diagnostics: Validation Based on the PI-RADS Scoring System and Targeted MR-Guided in-Bore Biopsy. Eur. Radiol. 2014, 24, 2582–2589. [Google Scholar] [CrossRef]
- Panebianco, V.; Barchetti, F.; Sciarra, A.; Ciardi, A.; Indino, E.L.; Papalia, R.; Gallucci, M.; Tombolini, V.; Gentile, V.; Catalano, C. Multiparametric Magnetic Resonance Imaging vs. Standard Care in Men Being Evaluated for Prostate Cancer: A Randomized Study. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 17.e1–17.e7. [Google Scholar] [CrossRef]
- Petrillo, A.; Fusco, R.; Setola, S.V.; Ronza, F.M.; Granata, V.; Petrillo, M.; Carone, G.; Sansone, M.; Franco, R.; Fulciniti, F.; et al. Multiparametric MRI for Prostate Cancer Detection: Performance in Patients with Prostate-Specific Antigen Values between 2.5 and 10 Ng/ML. J. Magn. Reson. Imaging 2014, 39, 1206–1212. [Google Scholar] [CrossRef]
- Van der Leest, M.; Cornel, E.; Israël, B.; Hendriks, R.; Padhani, A.R.; Hoogenboom, M.; Zamecnik, P.; Bakker, D.; Setiasti, A.Y.; Veltman, J.; et al. Head-to-Head Comparison of Transrectal Ultrasound-Guided Prostate Biopsy Versus Multiparametric Prostate Resonance Imaging with Subsequent Magnetic Resonance-Guided Biopsy in Biopsy-Naïve Men with Elevated Prostate-Specific Antigen: A Large Prospective Multicenter Clinical Study (Figure Presented.). Eur. Urol. 2019, 75, 570–578. [Google Scholar] [CrossRef] [Green Version]
- Glazer, D.I.; Mayo-Smith, W.W.; Sainani, N.I.; Sadow, C.A.; Vangel, M.G.; Tempany, C.M.; Dunne, R.M. Interreader Agreement of Prostate Imaging Reporting and Data System Version 2 Using an In-Bore Mri-Guided Prostate Biopsy Cohort: A Single Institution’s Initial Experience. Am. J. Roentgenol. 2017, 209, W145–W151. [Google Scholar] [CrossRef]
- Rosenkrantz, A.B.; Ayoola, A.; Hoffman, D.; Khasgiwala, A.; Prabhu, V.; Smereka, P.; Somberg, M.; Taneja, S.S. The Learning Curve in Prostate MRI Interpretation: Self-Directed Learning versus Continual Reader Feedback. Am. J. Roentgenol. 2017, 208, W92–W100. [Google Scholar] [CrossRef]
- Gatti, M.; Faletti, R.; Calleris, G.; Giglio, J.; Berzovini, C.; Gentile, F.; Marra, G.; Misischi, F.; Molinaro, L.; Bergamasco, L.; et al. Prostate Cancer Detection with Biparametric Magnetic Resonance Imaging (BpMRI) by Readers with Different Experience: Performance and Comparison with Multiparametric (MpMRI). Abdom. Radiol. 2019, 44, 1883–1893. [Google Scholar] [CrossRef]
- McGarry, S.D.; Hurrell, S.L.; Iczkowski, K.A.; Hall, W.; Kaczmarowski, A.L.; Banerjee, A.; Keuter, T.; Jacobsohn, K.; Bukowy, J.D.; Nevalainen, M.T.; et al. Radio-Pathomic Maps of Epithelium and Lumen Density Predict the Location of High-Grade Prostate Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 1179–1187. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, C.J.; Bao, M.L.; Zhang, J.; Wang, X.N.; Zhang, Y.D. Machine Learning-Based Analysis of MR Radiomics Can Help to Improve the Diagnostic Performance of PI-RADS v2 in Clinically Relevant Prostate Cancer. Eur. Radiol. 2017, 27, 4082–4090. [Google Scholar] [CrossRef]
- Wu, M.; Krishna, S.; Thornhill, R.E.; Flood, T.A.; McInnes, M.D.F.; Schieda, N. Transition Zone Prostate Cancer: Logistic Regression and Machine-Learning Models of Quantitative ADC, Shape and Texture Features Are Highly Accurate for Diagnosis. J. Magn. Reson. Imaging 2019, 50, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Wildeboer, R.R.; Mannaerts, C.K.; van Sloun, R.J.G.; Budäus, L.; Tilki, D.; Wijkstra, H.; Salomon, G.; Mischi, M. Automated Multiparametric Localization of Prostate Cancer Based on B-Mode, Shear-Wave Elastography, and Contrast-Enhanced Ultrasound Radiomics. Eur. Radiol. 2020, 30, 806–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arif, M.; Schoots, I.G.; Castillo Tovar, J.; Bangma, C.H.; Krestin, G.P.; Roobol, M.J.; Niessen, W.; Veenland, J.F. Clinically Significant Prostate Cancer Detection and Segmentation in Low-Risk Patients Using a Convolutional Neural Network on Multi-Parametric MRI. Eur. Radiol. 2020, 30, 6582–6592. [Google Scholar] [CrossRef]
- Winkel, D.J.; Breit, H.-C.; Shi, B.; Boll, D.T.; Seifert, H.-H.; Wetterauer, C. Predicting Clinically Significant Prostate Cancer from Quantitative Image Features Including Compressed Sensing Radial MRI of Prostate Perfusion Using Machine Learning: Comparison with PI-RADS v2 Assessment Scores. Quant. Imaging Med. Surg. 2020, 10, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Classification of Brain Tumors. Available online: https://www.aans.org/en/Media/Classifications-of-Brain-Tumors (accessed on 20 February 2021).
- Brain Cancer: Causes, Symptoms & Treatments|CTCA. Available online: https://www.cancercenter.com/cancer-types/brain-cancer (accessed on 20 February 2021).
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Course Tumor Boundaries. In Proceedings of the ICASSP 2019—IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1368–1372. [Google Scholar]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced Performance of Brain Tumor Classification via Tumor Region Augmentation and Partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sejdić, E. Radiological Images and Machine Learning: Trends, Perspectives, and Prospects. Comput. Biol. Med. 2019, 108, 354–370. [Google Scholar] [CrossRef] [Green Version]
- Badža, M.M.; Barjaktarović, M.C. Classification of Brain Tumors from Mri Images Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef] [Green Version]
- Ugga, L.; Perillo, T.; Cuocolo, R.; Stanzione, A.; Romeo, V.; Green, R.; Cantoni, V.; Brunetti, A. Meningioma MRI Radiomics and Machine Learning: Systematic Review, Quality Score Assessment, and Meta-Analysis. Neuroradiology 2021, 1–12. [Google Scholar] [CrossRef]
- Banzato, T.; Bernardini, M.; Cherubini, G.B.; Zotti, A. A Methodological Approach for Deep Learning to Distinguish between Meningiomas and Gliomas on Canine MR-Images. BMC Vet. Res. 2018, 14, 317. [Google Scholar] [CrossRef]
- Kanis, J.A.; Harvey, N.; Cooper, C.; Johansson, H.; Odén, A.; McCloskey, E.; The Advisory Board of the National Osteoporosis Guideline Group; Poole, K.E.; Gittoes, N.; Hope, S. A Systematic Review of Intervention Thresholds Based on FRAX: A Report Prepared for the National Osteoporosis Guideline Group and the International Osteoporosis Foundation. Arch. Osteoporos. 2016, 11, 1–48. [Google Scholar] [CrossRef] [Green Version]
- El Alaoui-Lasmaili, K.; Faivre, B. Antiangiogenic Therapy: Markers of Response, “Normalization” and Resistance. Crit. Rev. Oncol. Hematol. 2018, 128, 118–129. [Google Scholar] [CrossRef]
- Sheikhbahaei, S.; Mena, E.; Yanamadala, A.; Reddy, S.; Solnes, L.B.; Wachsmann, J.; Subramaniam, R.M. The Value of FDG PET/CT in Treatment Response Assessment, Follow-up, and Surveillance of Lung Cancer. Am. J. Roentgenol. 2017, 208, 420–433. [Google Scholar] [CrossRef]
- van Dijk, L.V.; Brouwer, C.L.; van der Laan, H.P.; Burgerhof, J.G.M.; Langendijk, J.A.; Steenbakkers, R.J.H.M.; Sijtsema, N.M. Geometric Image Biomarker Changes of the Parotid Gland Are Associated with Late Xerostomia. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 1101–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goense, L.; van Rossum, P.S.N.; Reitsma, J.B.; Lam, M.G.E.H.; Meijer, G.J.; van Vulpen, M.; Ruurda, J.P.; van Hillegersberg, R. Diagnostic Performance of 18F-FDG PET and PET/CT for the Detection of Recurrent Esophageal Cancer after Treatment with Curative Intent: A Systematic Review and Meta-Analysis. J. Nucl. Med. 2015, 56, 995–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, D.C.; Obuchowski, N.A.; Kessler, L.G.; Raunig, D.L.; Gatsonis, C.; Huang, E.P.; Kondratovich, M.; McShane, L.M.; Reeves, A.P.; Barboriak, D.P.; et al. Metrology Standards for Quantitative Imaging Biomarkers1. Radiology 2015, 277, 813–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterton, J.C.; Pylkkanen, L. Qualification of Imaging Biomarkers for Oncology Drug Development. Eur. J. Cancer 2012, 48, 409–415. [Google Scholar] [CrossRef]
- White, T.; Blok, E.; Calhoun, V.D. Data Sharing and Privacy Issues in Neuroimaging Research: Opportunities, Obstacles, Challenges, and Monsters under the Bed. Hum. Brain Mapp. 2020. [Google Scholar] [CrossRef]
- Zhuang, M.; García, D.V.; Kramer, G.M.; Frings, V.; Smit, E.F.; Dierckx, R.; Hoekstra, O.S.; Boellaard, R. Variability and Repeatability of Quantitative Uptake Metrics in 18F-FDG PET/CT of Non–Small Cell Lung Cancer: Impact of Segmentation Method, Uptake Interval, and Reconstruction Protocol. J. Nucl. Med. 2019, 60, 600–607. [Google Scholar] [CrossRef] [Green Version]
- Barrington, S.F.; Kirkwood, A.A.; Franceschetto, A.; Fulham, M.J.; Roberts, T.H.; Almquist, H.; Brun, E.; Hjorthaug, K.; Viney, Z.N.; Pike, L.C.; et al. PET-CT for Staging and Early Response: Results from the Response-Adapted Therapy in Advanced Hodgkin Lymphoma Study. Blood 2016, 127, 1531–1538. [Google Scholar] [CrossRef] [Green Version]
| Disease | Biomarker | Quantitative (Q)/Semi-Quantitative (SQ)/Non-Quantitative (NQ) | Biomarkers Uses |
|---|---|---|---|
| Malignant disease | Lung RADS, pancreatic cancer action network (PanCan), national comprehensive cancer network (NCCN) criteria [25,26] | SQ | AUC for malignancy 0.81–0.87 [27] |
| CT blood flow, perfusion, permeability measurements | Q | Sensitivity 0.73, specificity 0.70 [28] AUC 0.75, sensitivity 0.79, specificity 0.75 [29] | |
| Breast imaging (BI)-RADS [30] Prostate imaging (PI)-RADS [29] Liver imaging (LI)-RADS [31] | SQ | positive predictive value (PPV) BI-RADS 0 14.1%, BI-RADS 4 39.1%, BI-RADS 5 92.9% PI-RADS 2 pooled sensitivity 0.85, pooled specificity 0.71 pooled sensitivity for malignancy 0.93 | |
| Apparent diffusion coefficient (ADC) | Q | Liver AUC 0.82–0.95 Prostate AUC 0.84 | |
| RECIST/morphological volume | Q | Ongoing guidelines for treatment evaluation [32] | |
| Positron emission response criteria in solid tumors (PERCIST) /metabolic Volume [33] | Q | Ongoing guidelines for treatment evaluation [32] | |
| Liver cancer Recurrent glioblastoma | Dynamic contrast enhanced (DCE) metrics (perfusion parameters Ktrans, Kep, blood flow, Ve) | Q | Hepatocellular cancer AUC 0.85, sensitivity 0.85, specificity 0.81 [29] Brain- Ktrans accuracy 86% [34] |
| Cancer Sarcoma [35] Lung cancer [36] | 18FDG- standardized uptake value (SUV) | Q | Sarcoma-sensitivity 0.91, specificity 0.85, accuracy 0.88 Lung-sensitivity 0.68–0.95 |
| Cancer | Targeted radionuclides [37] In-octreotide [38,39] 68Gallium (Ga)-DOTA-TOC [39] and 68Ga DOTA-TATE [39,40,41] 68Ga prostate-specific membrane antigen (PSMA) [42] | NQ | Sensitivity 97%, specificity 92% for octreotide [43] Sensitivity 100%, specificity 100% for PSMA [44] |
| Brain cancer | Dynamic susceptibility contrast (DSC)-MRI | SQ | AUC = 0.77 for classifying glioma grades II and III [45] |
| Glioma | Adjuvant paclitaxel and trastuzumab (APT) trial | Q | APT accords with cancer grade and Ki67 index [46] |
| Rectal cancer Lung cancer | DCE-CT parameters Blood flow, permeability | Q | Blood flow 75% accuracy for detecting rectal cancers with lymph node metastases [47] CT permeability anticipated survival regardless of treatment in lung cancer [48] |
| Cervix cancer Endometrial cancer Rectal cancer Breast cancer | DCE-MRI parameters | Q | Cancer volume with increasing metrics is considered a significant independent factor for disease-free survival (DFS) and overall survival (OS) in cervical cancer [49] Low cancer blood flow and low rate constant for contrast agent intravasation (kep) correlated with high risk of endometrial cancer [50] Ktrans, Kep and Ve are higher in rectal cancers accompanied with metastasis [51] Ktrans, iAUC qualitative and ADC anticiptate low-risk breast cancers (AUC of combined parameters 0.78) |
| Diverse cancer types [52,53] | Radiomic signature [54] DCE-MR parameters | Q | Data endpoints, feature detection protocols, and classifiers are important factors in lung cancer prediction [55] Radiomic signature is significantly associated with lymph node (LN) status in colorectal cancer [56] Evaluating therapeutic effect subsequent to antiangiogenic agents [57] |
| Lymphoma | Deauville or response evaluation criteria in lymphoma (RECIL) score on 18F-FDG-PET | SQ | Assessment of lymphoma treatment in clinical trials employs the summation of longest diameters of three target lesions [58] |
| Breast cancer [59] Prostate cancer [60] | Receptor tyrosine-protein kinase erbB-2, CD340, and HER2 prostate-specific membrane antigen (PSMA) | SQ | Selective cancer receptor; investigation of cancer treatment on receptor expression. Assessing therapy response to antiangiogenic agents [57] |
| Oesophageal cancer | CT perfusion/blood flow | Q | Multivariate analysis detects blood flow as a predictor of response [61] |
| Gastrointestinal stromal cancers | CT density HU | Q | Decrease in cancer density of > 15% on CT associated with a sensitivity of 97% and a specificity of 100% in identifying PET responders compared to 52% and 100% by RECIST [61] |
| ML Method | Diagnostic Characteristics |
|---|---|
| Artificial Neural Network (ANN) | The mathematics behind the classification algorithm is simple. The non-linearities and weights allow the neural network (NN) to solve complex problems. Long training time is required for numerous iterations over the training data. Tendency for overfitting. Numerous additional tuning hyperparameters including # of hidden layers/hidden nodes are required for determining optimal performance. |
| Contrastive Learning | Self-supervised, task-independent deep learning technique that allows a model to learn about data, even without labels. Learns the general features of a dataset by teaching the model which data points are similar or different. Can potentially surpass supervised methods. May yield suboptimal performance on downstream tasks if the wrong transformation invariances are presumed. |
| Decision Trees (DTs) |
Easy to visualize Easy to understand. Feature selection plays a dominant role in the accuracy of the algorithm. One set of features can provide drastically different performance than a different set of features, therefore, large Random Forests can be used to alleviate this problem. Prone to overfitting. |
| Deep Learning (DL) | Can perform both image analysis (deep feature extraction) and construction of a prediction algorithm, eliminating the need for separate steps of extracting radiomic features and using that that to train a prediction model. Can learn from complex datasets and achieve high performance without requiring prior feature extraction. Permits massive parallel computations using GPUs. Requires additional hyper-parameters tune the model for better performance including the number of convolution filters, the size of the filters, and parameters involved in the pooling. Requires large training sets and it is not an optimal approach for pilot studies or internal data with small datasets. Computationally-expensive. |
| k Nearest Neighbor (kNN) | Easy to implement as it only requires the calculation of the distance between different points on the basis of data of different features. Computationally-expensive for large datasets. Does not work well with high dimensionality as this will complicate the distance calculating process to calculate distance for each dimension. Sensitive to noisy and missing data. Requires feature scaling. Prone to overfitting. |
| Logistic Regression |
Constructs linear boundaries, i.e., it assumes linearity between dependent and independent variables. However, linearly separable data is rarely found in real-world scenarios. |
| Naïve Bayes |
Models are faster to train and are simple, datasets and inferior performance on larger datasets. The Naïve Bayes classifier has generally shown to have superior performance in comparison to the Logistic Regression classifier on smaller datasets. Less potential for overfitting. Shows difficulties with complex datasets due to being linear classifiers. |
| Random Forests (RFs) | Less prone to overfitting, and it reduces overfitting in decision trees and helps to improve the accuracy. Outputs the importance of features which is a very useful for model interpretation. Works well with both categorical and continuous values, for both classification and regression problems. Tolerates missing values in the data by automating missing value interpretation. Output changes significantly with small changes in data. |
| Self-supervised Learning (SSL) |
Suitable for large unlabeled datasets, but its utility on small datasets is unknown. Reduces the relative error rate of few-shot meta-learners, even when the datasets are small and only utilizing images within the datasets. |
| Support Vector Machines (SVM) | Simple mathematics are behind the decision boundary Can be applied in higher dimensions. Time-consuming for large datasets, especially for datasets with larger margin decision boundary. Prone to overfitting. Sensitive to noisy and large datasets. |
| Category | RECIST | |
|---|---|---|
| Target Lesions | Nontarget Lesions | |
| Progressive disease (PD) | >20% ↑ in the sum of target lesions (TL) diameters. Absolute ↑ (5 mm). Appearance of new lesions. | Clear progress of surviving nontarget lesion. Appearance of new lesions. |
| Stable disease (SD) | Neither PD nor PR | Continuity of ≥ 1 nontarget lesion |
| Partial response (PR) | >30% ↓ in the sum of TL | Non-PD/CR |
| Complete response (CR) | Disappearance of TL. All nodes < 10 mm Non-pathological nodes | Disappearance of nontarget lesions. All nodes < 10 mm Non-pathological nodes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hajjo, R.; Sabbah, D.A.; Bardaweel, S.K.; Tropsha, A. Identification of Tumor-Specific MRI Biomarkers Using Machine Learning (ML). Diagnostics 2021, 11, 742. https://doi.org/10.3390/diagnostics11050742
Hajjo R, Sabbah DA, Bardaweel SK, Tropsha A. Identification of Tumor-Specific MRI Biomarkers Using Machine Learning (ML). Diagnostics. 2021; 11(5):742. https://doi.org/10.3390/diagnostics11050742
Chicago/Turabian StyleHajjo, Rima, Dima A. Sabbah, Sanaa K. Bardaweel, and Alexander Tropsha. 2021. "Identification of Tumor-Specific MRI Biomarkers Using Machine Learning (ML)" Diagnostics 11, no. 5: 742. https://doi.org/10.3390/diagnostics11050742
APA StyleHajjo, R., Sabbah, D. A., Bardaweel, S. K., & Tropsha, A. (2021). Identification of Tumor-Specific MRI Biomarkers Using Machine Learning (ML). Diagnostics, 11(5), 742. https://doi.org/10.3390/diagnostics11050742






