Radiomics and Machine Learning with Multiparametric Breast MRI for Improved Diagnostic Accuracy in Breast Cancer Diagnosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sample
2.2. Breast MRI Technique
2.3. Imaging Processing
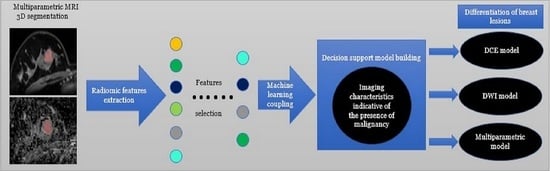
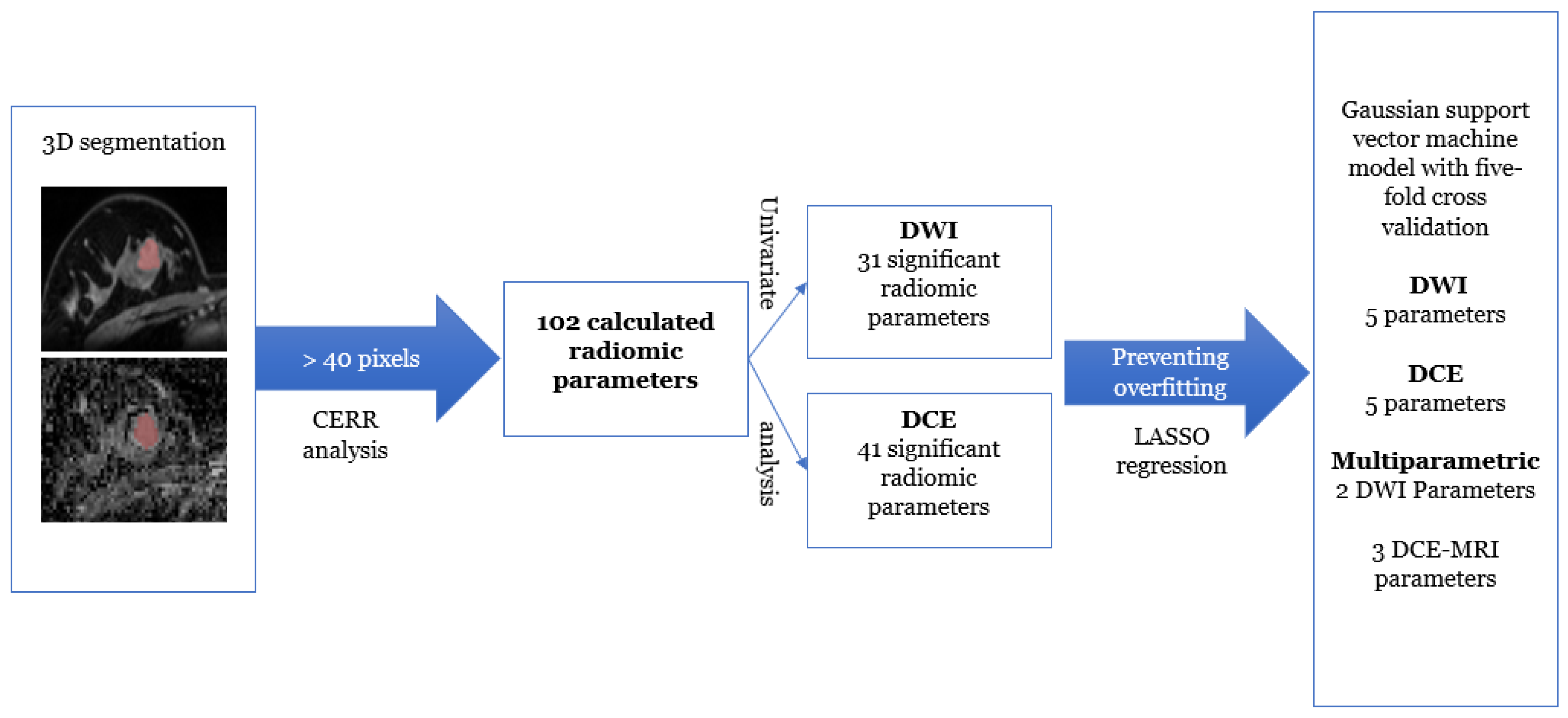
2.4. Radiomics Image Analysis
2.5. Reference Standard
2.6. Statistical Analysis and Predictive Model Building
3. Results
3.1. Patient Sample and Breast Lesion Characteristics
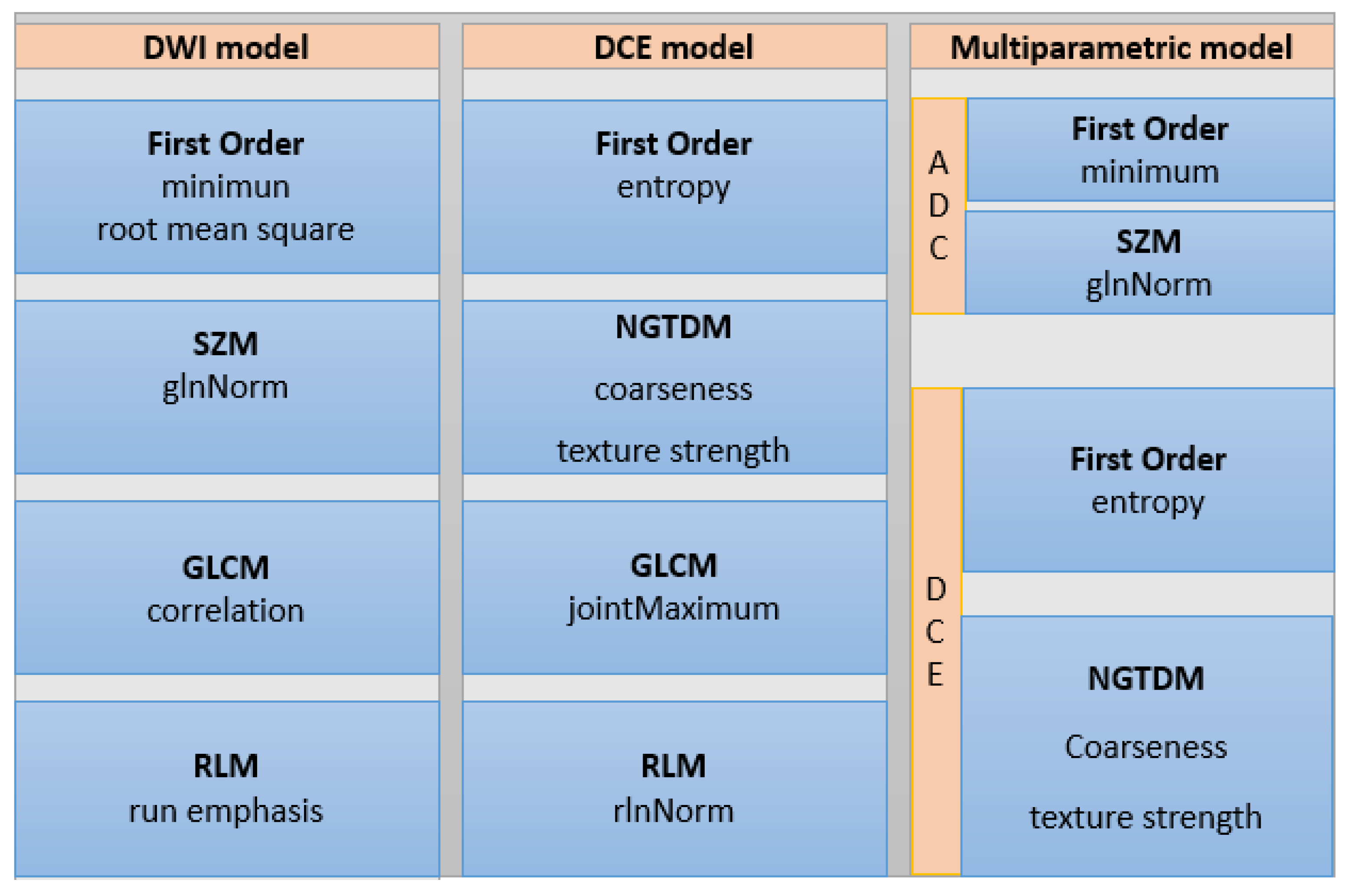
3.2. Radiomics Analysis for Breast Lesion Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mann, R.M.; Cho, N.; Moy, L. Breast MRI: State of the Art. Radiology 2019, 292, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Balleyguier, C.; Baltzer, P.A.; Bick, U.; Colin, C.; Cornford, E.; Evans, A.; Fallenberg, E.; Forrai, G.; Fuchsjäger, M.H.; et al. Breast MRI: EUSOBI recommendations for women’s information. Eur. Radiol. 2015, 25, 3669–3678. [Google Scholar] [CrossRef] [Green Version]
- Mann, R.M.; Kuhl, C.K.; Moy, L. Contrast-enhanced MRI for breast cancer screening. J. Magn. Reson. Imaging 2019, 50, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, H. Meta-analysis of diagnostic accuracy of magnetic resonance imaging and mammography for breast cancer. J. Cancer Res. Ther. 2017, 13, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Helbich, T.H.; Morris, E.A. The potential of multiparametric MRI of the breast. Br. J. Radiol. 2017, 90, 20160715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veenhuizen, S.G.A.; de Lange, S.V.; Bakker, M.F.; Pijnappel, R.M.; Mann, R.M.; Monninkhof, E.M.; Emaus, M.J.; de Koekkoek-Doll, P.K.; Bisschops, R.H.C.; Lobbes, M.B.I.; et al. Supplemental Breast MRI for Women with Extremely Dense Breasts: Results of the Second Screening Round of the DENSE Trial. Radiology 2021, 299, 278–286. [Google Scholar] [CrossRef]
- Gao, Y.; Reig, B.; Heacock, L.; Bennett, D.L.; Heller, S.L.; Moy, L. Magnetic Resonance Imaging in Screening of Breast Cancer. Radiol. Clin. N. Am. 2021, 59, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, F.J.; Pinker-Domenig, K. Diagnosis and Staging of Breast Cancer: When and How to Use Mammography, Tomosynthesis, Ultrasound, Contrast-Enhanced Mammography, and Magnetic Resonance Imaging. In Diseases of the Chest, Breast, Heart and Vessels 2019–2022: Diagnostic and Interventional Imaging; Hodler, J., Kubik-Huch, R.A., von Schulthess, G.K., Eds.; IDKD Springer Series; Springer: Cham, Switzerland, 2019; pp. 155–166. [Google Scholar] [CrossRef]
- Comstock, C.E.; Gatsonis, C.; Newstead, G.M.; Snyder, B.S.; Gareen, I.F.; Bergin, J.T.; Rahbar, H.; Sung, J.S.; Jacobs, C.; Harvey, J.A.; et al. Comparison of Abbreviated Breast MRI vs Digital Breast Tomosynthesis for Breast Cancer Detection Among Women With Dense Breasts Undergoing Screening. JAMA 2020, 323, 746–756. [Google Scholar] [CrossRef]
- Lakhani, S.R.; Jacquemier, J.; Sloane, J.P.; Gusterson, B.A.; Anderson, T.J.; van de Vijver, M.J.; Farid, L.M.; Venter, D.; Antoniou, A.; Storfer-Isser, A.; et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J. Natl. Cancer Inst. 1998, 90, 1138–1145. [Google Scholar] [CrossRef]
- Marino, M.A.; Riedl, C.C.; Bernathova, M.; Bernhart, C.; Baltzer, P.A.T.; Helbich, T.H.; Pinker, K. Imaging Phenotypes in Women at High Risk for Breast Cancer on Mammography, Ultrasound, and Magnetic Resonance Imaging Using the Fifth Edition of the Breast Imaging Reporting and Data System. Eur. J. Radiol. 2018, 106, 150–159. [Google Scholar] [CrossRef]
- Meissnitzer, M.; Dershaw, D.D.; Feigin, K.; Bernard-Davila, B.; Barra, F.; Morris, E.A. MRI appearance of invasive subcentimetre breast carcinoma: Benign characteristics are common. Br. J. Radiol. 2017, 90, 20170102. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Moy, L.; Sutton, E.J.; Mann, R.M.; Weber, M.; Thakur, S.B.; Jochelson, M.S.; Bago-Horvath, Z.; Morris, E.A.; Baltzer, P.A.; et al. Diffusion-Weighted Imaging With Apparent Diffusion Coefficient Mapping for Breast Cancer Detection as a Stand-Alone Parameter: Comparison With Dynamic Contrast-Enhanced and Multiparametric Magnetic Resonance Imaging. Investig. Radiol. 2018, 53, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Iima, M.; Honda, M.; Sigmund, E.E.; Ohno Kishimoto, A.; Kataoka, M.; Togashi, K. Diffusion MRI of the breast: Current status and future directions. J. Magn. Reson. Imaging 2020, 52, 70–90. [Google Scholar] [CrossRef]
- Partridge, S.C.; Nissan, N.; Rahbar, H.; Kitsch, A.E.; Sigmund, E.E. Diffusion-weighted breast MRI: Clinical applications and emerging techniques. J. Magn. Reson. Imaging 2017, 45, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Amornsiripanitch, N.; Bickelhaupt, S.; Shin, H.J.; Dang, M.; Rahbar, H.; Pinker, K.; Partridge, S.C. Diffusion-weighted MRI for Unenhanced Breast Cancer Screening. Radiology 2019, 293, 504–520. [Google Scholar] [CrossRef] [PubMed]
- Baltzer, P.; Mann, R.M.; Iima, M.; Sigmund, E.E.; Clauser, P.; Gilbert, F.J.; Martincich, L.; Partridge, S.C.; Patterson, A.; Pinker, K.; et al. Diffusion-weighted imaging of the breast-a consensus and mission statement from the EUSOBI International Breast Diffusion-Weighted Imaging working group. Eur. Radiol. 2020, 30, 1436–1450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkstra, H.; Dorrius, M.D.; Wielema, M.; Pijnappel, R.M.; Oudkerk, M.; Sijens, P.E. Quantitative DWI implemented after DCE-MRI yields increased specificity for BI-RADS 3 and 4 breast lesions. J. Magn. Reson. Imaging 2016, 44, 1642–1649. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Bogner, W.; Baltzer, P.; Gruber, S.; Bickel, H.; Brueck, B.; Trattnig, S.; Weber, M.; Dubsky, P.; Bago-Horvath, Z.; et al. Improved diagnostic accuracy with multiparametric magnetic resonance imaging of the breast using dynamic contrast-enhanced magnetic resonance imaging, diffusion-weighted imaging, and 3-dimensional proton magnetic resonance spectroscopic imaging. Investig. Radiol. 2014, 49, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Pinker, K.; Bickel, H.; Helbich, T.H.; Gruber, S.; Dubsky, P.; Pluschnig, U.; Rudas, M.; Bago-Horvath, Z.; Weber, M.; Trattnig, S.; et al. Combined contrast-enhanced magnetic resonance and diffusion-weighted imaging reading adapted to the “Breast Imaging Reporting and Data System” for multiparametric 3-T imaging of breast lesions. Eur. Radiol. 2013, 23, 1791–1802. [Google Scholar] [CrossRef]
- Clauser, P.; Krug, B.; Bickel, H.; Dietzel, M.; Pinker, K.; Neuhaus, V.F.; Marino, M.A.; Moschetta, M.; Troiano, N.; Helbich, T.H.; et al. Diffusion-weighted Imaging Allows for Downgrading MR BI-RADS 4 Lesions in Contrast-enhanced MRI of the Breast to Avoid Unnecessary Biopsy. Clin. Cancer Res. 2021, 27, 1941–1948. [Google Scholar] [CrossRef]
- Zhang, M.; Horvat, J.V.; Bernard-Davila, B.; Marino, M.A.; Leithner, D.; Ochoa-Albiztegui, R.E.; Helbich, T.H.; Morris, E.A.; Thakur, S.; Pinker, K. Multiparametric MRI model with dynamic contrast-enhanced and diffusion-weighted imaging enables breast cancer diagnosis with high accuracy. J. Magn. Reson. Imaging 2019, 49, 864–874. [Google Scholar] [CrossRef]
- Bickelhaupt, S.; Jaeger, P.F.; Laun, F.B.; Lederer, W.; Daniel, H.; Kuder, T.A.; Wuesthof, L.; Paech, D.; Bonekamp, D.; Radbruch, A.; et al. Radiomics Based on Adapted Diffusion Kurtosis Imaging Helps to Clarify Most Mammographic Findings Suspicious for Cancer. Radiology 2018, 287, 761–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bickelhaupt, S.; Paech, D.; Kickingereder, P.; Steudle, F.; Lederer, W.; Daniel, H.; Götz, M.; Gählert, N.; Tichy, D.; Wiesenfarth, M.; et al. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J. Magn. Reson. Imaging 2017, 46, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Shin, H.J.; Moon, W.K. Diffusion-Weighted Magnetic Resonance Imaging of the Breast: Standardization of Image Acquisition and Interpretation. Korean J. Radiol. 2021, 22, 9–22. [Google Scholar] [CrossRef]
- Lo Gullo, R.; Daimiel, I.; Morris, E.A.; Pinker, K. Combining molecular and imaging metrics in cancer: Radiogenomics. Insights Imaging 2020, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Reig, B.; Heacock, L.; Geras, K.J.; Moy, L. Machine learning in breast MRI. J. Magn. Reson. Imaging 2020, 52, 998–1018. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.; Primakov, S.; Beuque, M.; Woodruff, H.C.; Halilaj, I.; Wu, G.; Refaee, T.; Granzier, R.; Widaatalla, Y.; Hustinx, R.; et al. Radiomics for precision medicine: Current challenges, future prospects, and the proposal of a new framework. Methods 2021, 188, 20–29. [Google Scholar] [CrossRef]
- Rogers, W.; Thulasi Seetha, S.; Refaee, T.A.G.; Lieverse, R.I.Y.; Granzier, R.W.Y.; Ibrahim, A.; Keek, S.A.; Sanduleanu, S.; Primakov, S.P.; Beuque, M.P.L.; et al. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020, 93, 20190948. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, H.; Ko, E.S. Radiomics in Breast Imaging from Techniques to Clinical Applications: A Review. Korean J. Radiol. 2020, 21, 779–792. [Google Scholar] [CrossRef]
- Leithner, D.; Bernard-Davila, B.; Martinez, D.F.; Horvat, J.V.; Jochelson, M.S.; Marino, M.A.; Avendano, D.; Ochoa-Albiztegui, R.E.; Sutton, E.J.; Morris, E.A.; et al. Radiomic Signatures Derived from Diffusion-Weighted Imaging for the Assessment of Breast Cancer Receptor Status and Molecular Subtypes. Mol. Imaging Biol. 2020, 22, 453–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Feng, Q.; Yang, W.; Lu, Z.; Deng, C.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur. Radiol. 2018, 28, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parekh, V.S.; Jacobs, M.A. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 2017, 3, 43. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Peng, Y.; Ou, C.; Chen, M.; Li, L. Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: A machine learning approach. PLoS ONE 2014, 9, e87387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apte, A.P.; Iyer, A.; Crispin-Ortuzar, M.; Pandya, R.; van Dijk, L.V.; Spezi, E.; Thor, M.; Um, H.; Veeraraghavan, H.; Oh, J.H.; et al. Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Med. Phys. 2018. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Hanley, J.A.; McNeil, B.J. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983, 148, 839–843. [Google Scholar] [CrossRef] [Green Version]
- Nie, K.; Chen, J.H.; Yu, H.J.; Chu, Y.; Nalcioglu, O.; Su, M.Y. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad. Radiol. 2008, 15, 1513–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truhn, D.; Schrading, S.; Haarburger, C.; Schneider, H.; Merhof, D.; Kuhl, C. Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-enhancing Lesions at Multiparametric Breast MRI. Radiology 2019, 290, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, P.; Onishi, N.; Sadinski, M.; Gallagher, K.M.; Hughes, M.; Martinez, D.F.; Morris, E.A.; Sutton, E.J. Characterization of Sub-1 cm Breast Lesions Using Radiomics Analysis. J. Magn. Reson. Imaging 2019, 50, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Gullo, R.L.; Daimiel, I.; Rossi Saccarelli, C.; Bitencourt, A.; Gibbs, P.; Fox, M.J.; Thakur, S.B.; Martinez, D.F.; Jochelson, M.S.; Morris, E.A.; et al. Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers. Eur. Radiol. 2020, 30, 6721–6731. [Google Scholar] [CrossRef]
- Zhang, Q.; Peng, Y.; Liu, W.; Bai, J.; Zheng, J.; Yang, X.; Zhou, L. Radiomics Based on Multimodal MRI for the Differential Diagnosis of Benign and Malignant Breast Lesions. J. Magn. Reson. Imaging 2020, 52, 596–607. [Google Scholar] [CrossRef]
- Verburg, E.; van Gils, C.H.; Bakker, M.F.; Viergever, M.A.; Pijnappel, R.M.; Veldhuis, W.B.; Gilhuijs, K.G.A. Computer-Aided Diagnosis in Multiparametric Magnetic Resonance Imaging Screening of Women With Extremely Dense Breasts to Reduce False-Positive Diagnoses. Investig. Radiol. 2020, 55, 438–444. [Google Scholar] [CrossRef]


| Patient Characteristic | Number (Percentage) |
|---|---|
| Mean age (years; SD) | 49 years ± 12 years |
| Menopausal status | |
| Fertile | 55 (59.1%) |
| Menopause | 38 (40.9%) |
| Breast Findings | |
| Benign | 58 (55.8%) |
| Malignant | 46 (44.2%) |
| Benign Lesions | Malignant Lesions | ||||
|---|---|---|---|---|---|
| Mass | 50 (86.2%) | Mass | 35 (76%) | ||
| NMLE | 8 (13.8%) | NMLE | 11(24%) | ||
| Histopathology | |||||
| Fibroadenoma or fibroadenomatoid change | 30 (51.8%) | IDC | Histological Grade 1: 4 (8.6%) | ||
| Phyllodes tumor | 1 (1.7%) | Histological Grade 2: 18 (39.2%) | |||
| Adenosis, stromal fibrosis, ductal ectasia, or normal breast parenchyma | 10 (17.3%) | Histological Grade 3: 20 (43.6%) | |||
| FCC | 5 (8.6%) | ILC | 2 (4.3%) | ||
| ADH or ALH | 4 (6.9%) | ||||
| PASH | 3 (5.2%) | ||||
| Papilloma | 2 (3.4%) | ||||
| Hamartoma | 1 (1.7%) | DCIS | 2 (4.3%) | ||
| Fat necrosis | 1 (1.7%) | ||||
| Epithelial intraductal proliferation without atypia | 1 (1.7%) | ||||
| Dataset | Sensitivity | Specificity | PPV | NPV | Accuracy | AUC |
|---|---|---|---|---|---|---|
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | |
| DWI | 68.1 (52.8–80.9) | 77.2 (64.2–87.3) | 71.1 (59.5–80.5) | 74.6 (65.4–82.0) | 73.1 (63.5–81.3) | 0.79 (0.70–0.88) |
| DCE | 76.6 (62.0–87.7) | 77.2 (64.2–87.3) | 73.5 (62.6–82.1) | 80 (70.1–87.2) | 76.9 (67.6–84.6) | 0.83 (0.75–0.91) |
| Multiparametric | 85.1 (71.7–93.8) | 79 (66.1–88.6) | 76.9 (66.5–84.8) | 86.5 (76.2–92.8) | 81.7 (73.0–88.6) | 0.85 (0.7–0.92) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daimiel Naranjo, I.; Gibbs, P.; Reiner, J.S.; Lo Gullo, R.; Sooknanan, C.; Thakur, S.B.; Jochelson, M.S.; Sevilimedu, V.; Morris, E.A.; Baltzer, P.A.T.; et al. Radiomics and Machine Learning with Multiparametric Breast MRI for Improved Diagnostic Accuracy in Breast Cancer Diagnosis. Diagnostics 2021, 11, 919. https://doi.org/10.3390/diagnostics11060919
Daimiel Naranjo I, Gibbs P, Reiner JS, Lo Gullo R, Sooknanan C, Thakur SB, Jochelson MS, Sevilimedu V, Morris EA, Baltzer PAT, et al. Radiomics and Machine Learning with Multiparametric Breast MRI for Improved Diagnostic Accuracy in Breast Cancer Diagnosis. Diagnostics. 2021; 11(6):919. https://doi.org/10.3390/diagnostics11060919
Chicago/Turabian StyleDaimiel Naranjo, Isaac, Peter Gibbs, Jeffrey S. Reiner, Roberto Lo Gullo, Caleb Sooknanan, Sunitha B. Thakur, Maxine S. Jochelson, Varadan Sevilimedu, Elizabeth A. Morris, Pascal A. T. Baltzer, and et al. 2021. "Radiomics and Machine Learning with Multiparametric Breast MRI for Improved Diagnostic Accuracy in Breast Cancer Diagnosis" Diagnostics 11, no. 6: 919. https://doi.org/10.3390/diagnostics11060919