Computer-Aided Diagnosis Improves the Detection of Clinically Significant Prostate Cancer on Multiparametric-MRI: A Multi-Observer Performance Study Involving Inexperienced Readers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients Population and Study Design
2.2. Multiparametric MRI
2.3. Reference Standard
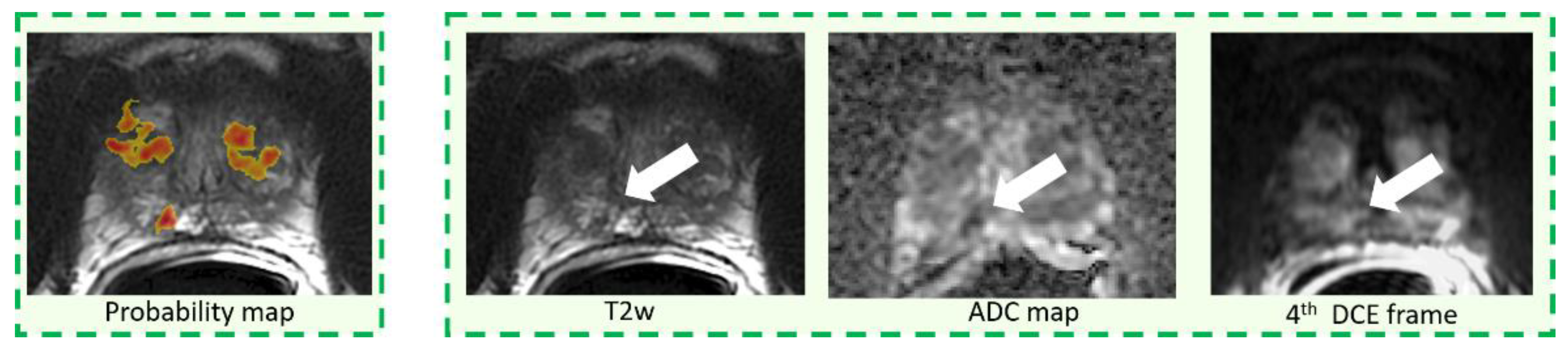
2.4. CAD System and Image Interpretation
2.5. Statistical Analysis
2.6. Power Calculation
3. Results
3.1. Per-Patient Analysis
3.2. Per-Lesion Analysis
3.3. Reading Time and Inter-Reader Agreement
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Bjurlin, M.A.; Carroll, P.R.; Eggener, S.; Fulgham, P.F.; Margolis, D.J.; Pinto, P.A.; Rosenkrantz, A.B.; Rubenstein, J.N.; Rukstalis, D.B.; Taneja, S.S.; et al. Update of the Standard Operating Procedure on the Use of Multiparametric Magnetic Resonance Imaging for the Diagnosis, Staging and Management of Prostate Cancer. J. Urol. 2020, 203, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Loeb, S.; Bjurlin, M.A.; Nicholson, J.; Tammela, T.L.; Penson, D.F.; Carter, H.B.; Carroll, P.; Etzioni, R. Overdiagnosis and overtreatment of prostate cancer. Eur. Urol. 2014, 65, 1046–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monni, F.; Fontanella, P.; Grasso, A.; Wiklund, P.; Ou, Y.C.; Randazzo, M.; Rocco, B.; Montanari, E.; Bianchi, G. Magnetic resonance imaging in prostate cancer detection and management: A systematic review. Minerva Urol. Nefrol. 2017, 69, 567–578. [Google Scholar] [PubMed]
- Russo, F.; Regge, D.; Armando, E.; Giannini, V.; Vignati, A.; Mazzetti, S.; Manfredi, M.; Bollito, E.; Correale, L.; Porpiglia, F. Detection of prostate cancer index lesions with multiparametric magnetic resonance imaging (mp-MRI) using whole-mount histological sections as the reference standard. BJU Int. 2016, 118, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.; Mazzetti, S.; Regge, D.; Ambrosini, I.; Giannini, V.; Manfredi, M.; De Luca, S.; Bollito, E.; Porpiglia, F. Diagnostic Accuracy of Single-plane Biparametric and Multiparametric Magnetic Resonance Imaging in Prostate Cancer: A Randomized Noninferiority Trial in Biopsy-naïve Men. Eur. Urol. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wagaskar, V.G.; Levy, M.; Ratnani, P.; Moody, K.; Garcia, M.; Pedraza, A.M.; Parekh, S.; Pandav, K.; Shukla, B.; Prasad, S.; et al. Clinical Utility of Negative Multiparametric Magnetic Resonance Imaging in the Diagnosis of Prostate Cancer and Clinically Significant Prostate Cancer. Eur. Urol. Open Sci. 2021, 28, 9–16. [Google Scholar] [CrossRef]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Fütterer, J.J.; Briganti, A.; De Visschere, P.; Emberton, M.; Giannarini, G.; Kirkham, A.; Taneja, S.S.; Thoeny, H.; Villeirs, G.; Villers, A. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur. Urol. 2015, 68, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Arumainayagam, N.; Ahmed, H.U.; Moore, C.M.; Freeman, A.; Allen, C.; Sohaib, S.A.; Kirkham, A.; Van Der Meulen, J.; Emberton, M. Multiparametric MR imaging for detection of clinically significant prostate cancer: A validation cohort study with transperineal template prostate mapping as the reference standard. Radiology 2013, 268, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, O.; Weisser, P.; Bodelle, B.; Ackermann, H.; Vogl, T.J. MRI of the prostate: Interobserver agreement compared with histopathologic outcome after radical prostatectomy. Eur. J. Radiol. 2012, 81, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tian, Z.; Zhang, Z.; Fei, B. Computer-aided Detection of Prostate Cancer with MRI: Technology and Applications. Acad. Radiol. 2016, 23, 1024–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. Computer-aided detection of prostate cancer in MRI. IEEE Trans. Med. Imaging 2014, 33, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.R.; Ekberg, J.; Fridell, K. Prostate Cancer Detection in Screening Using Magnetic Resonance Imaging and Artificial Intelligence. Open Artif. Intell. J. 2020. [Google Scholar] [CrossRef]
- Regge, D.; Della Monica, P.; Galatola, G.; Laudi, C.; Zambon, A.; Asnaghi, R.; Correale, L.; Barbaro, B.; Borghi, C.; Campanella, D.; et al. Efficacy of computer-aided detection as a second reader for 6-9-mm lesions at CT colonography: Multicenter prospective trial. Radiology 2013, 266, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Sali, L.; Delsanto, S.; Sacchetto, D.; Correale, L.; Falchini, M.; Ferraris, A.; Gandini, G.; Grazzini, G.; Iafrate, F.; Iussich, G.; et al. Computer-based self-training for CT colonography with and without CAD. Eur. Radiol. 2018, 28, 4783–4791. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Browning, P.; Wahi-Anwar, M.W.; Murphy, M.; Delgado, J.; Greenspan, H.; Abtin, F.; Ghahremani, S.; Yaghmai, N.; da Costa, I.; et al. Integration of Chest CT CAD into the Clinical Workflow and Impact on Radiologist Efficiency. Acad. Radiol. 2019, 26, 626–631. [Google Scholar] [CrossRef]
- Greer, M.D.; Lay, N.; Shih, J.H.; Barrett, T.; Bittencourt, L.K.; Borofsky, S.; Kabakus, I.; Law, Y.M.; Marko, J.; Shebel, H.; et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: An international multi-reader study. Eur. Radiol. 2018, 28, 4407–4417. [Google Scholar] [CrossRef] [PubMed]
- Hambrock, T.; Vos, P.C.; Hulsbergen-Van De Kaa, C.A.; Barentsz, J.O.; Huisman, H.J. Prostate cancer: Computer-aided diagnosis with multiparametric 3-T MR imaging-Effect on observer performance. Radiology 2013, 266, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Giannini, V.; Mazzetti, S.; Armando, E.; Carabalona, S.; Russo, F.; Giacobbe, A.; Muto, G.; Regge, D. Multiparametric magnetic resonance imaging of the prostate with computer-aided detection: Experienced observer performance study. Eur. Radiol. 2017, 27, 4200–4208. [Google Scholar] [CrossRef] [PubMed]
- Szucs-Farkas, Z.; Patak, M.A.; Yuksel-Hatz, S.; Ruder, T.; Vock, P. Improved detection of pulmonary nodules on energy-subtracted chest radiographs with a commercial computer-aided diagnosis software: Comparison with human observers. Eur. Radiol. 2010, 20, 1289–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boone, D.; Mallett, S.; McQuillan, J.; Taylor, S.A.; Altman, D.G.; Halligan, S. Assessment of the incremental benefit of computer-aided detection (CAD) for interpretation of CT colonography by experienced and inexperienced readers. PLoS ONE 2015, 10, e0136624. [Google Scholar] [CrossRef] [Green Version]
- Barentsz, J.O.; Weinreb, J.C.; Verma, S.; Thoeny, H.C.; Tempany, C.M.; Shtern, F.; Padhani, A.R.; Margolis, D.; Macura, K.J.; Haider, M.A.; et al. Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use. Eur. Urol. 2016, 69, 41. [Google Scholar] [CrossRef] [PubMed]
- Giannini, V.; Mazzetti, S.; Vignati, A.; Russo, F.; Bollito, E.; Porpiglia, F.; Stasi, M.; Regge, D. A fully automatic computer aided diagnosis system for peripheral zone prostate cancer detection using multi-parametric magnetic resonance imaging. Comput. Med. Imaging Graph. 2015, 46, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Mazzetti, S.; Giannini, V.; Russo, F.; Regge, D. Computer-aided diagnosis of prostate cancer using multi-parametric MRI: Comparison between PUN and Tofts models. Phys. Med. Biol. 2018, 63, 095004. [Google Scholar] [CrossRef] [PubMed]
- Dhand, N.K.; Khatkar, M.S. Statulator: An Online Statistical Calculator. Sample Size Calculator for Comparing Two Paired Proportions. Available online: http//statulator.com/SampleSize/ss2PP.html (accessed on 21 November 2017).
- Petrick, N.; Haider, M.; Summers, R.M.; Yeshwant, S.C.; Brown, L.; Iuliano, E.M.; Louie, A.; Choi, J.R.; Pickhardt, P.J. CT colonography with computer-aided detection as a second reader: Observer performance study. Radiology 2008, 246, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Iussich, G.; Correale, L.; Senore, C.; Segnan, N.; Laghi, A.; Iafrate, F.; Campanella, D.; Neri, E.; Cerri, F.; Hassan, C.; et al. CT colonography: Preliminary assessment of a double-read paradigm that uses computer-aided detection as the first reader. Radiology 2013, 268, 743–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barinov, L.; Jairaj, A.; Becker, M.; Seymour, S.; Lee, E.; Schram, A.; Lane, E.; Goldszal, A.; Quigley, D.; Paster, L. Impact of Data Presentation on Physician Performance Utilizing Artificial Intelligence-Based Computer-Aided Diagnosis and Decision Support Systems. J. Digit. Imaging 2019, 32, 408–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Gao, X.; Liu, L.; Shu, R.; Yan, J.; Zhang, G.; Xiao, Y.; Ju, Y.; Zhao, N.; Song, H. Performance and reading time of automated breast us with or without computer-aided detection. Radiology 2019, 292, 540–549. [Google Scholar] [CrossRef] [PubMed]




| Total | Positive | Negative | p-Value | |
|---|---|---|---|---|
| Demographic | ||||
| Number of patients, n (%) | 90 | 45 | 45 | |
| Age, y (IQR) | 66.7 (63.3–74.7) | 68.5 (65.0–75.2) | 65.7 (62.0–72.0) | 0.059 |
| PSA, ng/mL (IQR) | 7.3 (6.0–10.9) | 6.9 (5.9–11.9) | 7.6 (6.4–10.7) | 0.422 |
| Prostate volume, mL (IQR) | 52.2 (36.5–80.2) | 40.9 (29.6–55.8) | 70.2 (48.2–91.0) | <0.001 |
| PSAD, ng/mL/mL (IQR) | 0.14 (0.11–0.24) | 0.18 (0.13–0.30) | 0.13 (0.09–0.17) | <0.001 |
| Imaging | ||||
| Longest lesion diameter, mm (IQR) | - | 12 (7.8–18) | - | |
| PI-RADS v2 assessment, n (%) | ||||
| 1 | 24 (27%) | - | 24 (53%) | |
| 2 | 21 (23%) | - | 21 (47%) | |
| 3 | 4 (5%) | 4 (9%) | - | |
| 4 | 20 (22%) | 20 (44%) | - | |
| 5 | 21 (23%) | 21 (47%) | - | |
| Gleason Score | ||||
| 3 + 3 | 13 (29%) | 13 | - | |
| 3 + 4 | 16 (36%) | 16 | - | |
| 4 + 3 | 10 (22%) | 10 | - | |
| 4 + 4 | 3 (7%) | 3 | - | |
| 4 + 5 | 1 (2%) | 1 | - | |
| 5 + 4 | 1 (2%) | 1 | - | |
| 5 + 5 | 1 (2%) | 1 | - |
| Unassisted Reading (%) | Assisted Reading (%) | p Value | |
|---|---|---|---|
| Sensitivity | |||
| Reader 1 | 64.4 (29/45) [48.8–78.1] | 64.4 (29/45) [48.8–78.1] | 0.500 |
| Reader 2 | 60.0 (27/45) [44.3–74.3] | 64.4 (29/45) [48.8–78.1] | 0.387 |
| Reader 3 | 77.8 (35/45) [62.9–88.8] | 82.2 (37/45) [67.9–92.0] | 0.344 |
| Average | 67.4 (91/135) [58.8–75.2] | 70.4 (95/135) [61.9–77.9] | 0.298 |
| CAD standalone | - | 95.6 (43/45) [84.8–99.5] | <0.001 |
| Sensitivity for GS = 6 | |||
| Reader 1 | 61.5 (8/13) [31.6–86.1] | 46.2 (6/13) [19.2–74.9] | 0.363 |
| Reader 2 | 53.8 (7/13) [25.1–80.8] | 38.5 (5/13) [13.9–68.4] | 0.344 |
| Reader 3 | 76.9 (10/13) [46.2–95.0] | 69.2 (9/13) [38.6–90.9] | 0.500 |
| Average | 64.1 (25/39) [47.2–78.8] | 51.3 (20/39) [34.8–67.6] | 0.166 |
| CAD standalone | - | 92.3 (12/13) [64.0–99.8] | 0.005 |
| Sensitivity for GS > 6 | |||
| Reader 1 | 65.6 (21/32) [46.8–81.4] | 71.9 (23/32) [53.2–86.2] | 0.344 |
| Reader 2 | 62.5 (20/32) [43.7–78.9] | 75.0 (24/32) [56.6–88.5] | 0.109 |
| Reader 3 | 78.1 (25/32) [60.0–90.7] | 87.5 (28/32) [71.0–96.5] | 0.125 |
| Average | 68.7 (66/96) [58.5–77.8] | 78.1 (75/96) [68.5–85.9] | 0.018 |
| CAD standalone | - | 95.6 (31/32) [78.1–99.9] | 0.012 |
| Sensitivity for max diameter 4–9 mm | |||
| Reader 1 | 41.1 (7/17) [18.4–67.1] | 52.9 (9/17) [27.8–77.0] | 0.344 |
| Reader 2 | 41.1 (7/17) [18.4–67.1] | 47.1 (8/17) [23.0–72.2] | 0.500 |
| Reader 3 | 64.7 (11/17) [38.3–85.8] | 76.5 (13/17) [50.1–93.2] | 0.313 |
| Average | 49.0 (25/51) [34.7–63.4] | 58.8 (30/51) [44.2–72.4] | 0.151 |
| CAD standalone | - | 94.1 (16/17) [71.3–99.8] | 0.004 |
| Sensitivity for max diameter ≥ 10 mm | |||
| Reader 1 | 78.6 (22/28) [59.0–91.7] | 71.4 (20/28) [51.3–86.8] | 0.363 |
| Reader 2 | 71.4 (20/28) [51.3–86.8] | 75.0 (21/28) [55.1–89.3] | 0.500 |
| Reader 3 | 85.7 (24/28) [67.3–96.0] | 85.7 (24/28) [67.3–96.0] | 0.500 |
| Average | 78.6 (66/84) [68.3–86.8] | 77.4 (65/84) [66.9–85.8] | 0.500 |
| CAD standalone | - | 96.4 (27/28) [81.6–99.9] | 0.012 |
| Specificity | |||
| Reader 1 | 95.6 (43/45) [84.9–99.5] | 100 (45/45) [92.1–100.0] | 0.250 |
| Reader 2 | 97.8 (44/45) [88.2–99.9] | 80.0 (36/45) [65.4–90.4] | 0.004 |
| Reader 3 | 91.1 (41/45) [78.8–97.5] | 88.9 (40/45) [75.9–96.3] | 0.500 |
| Average | 94.8 (128/135) [89.6–97.9] | 89.6 (121/135) [83.2–94.2] | 0.072 |
| Unassisted Reading | Assisted Reading | p Value | |
|---|---|---|---|
| Sensitivity | |||
| Reader 1 | 60.8 (31/51) [46.1–74.2] | 54.9 (28/51) [40.3–68.9] | 0.304 |
| Reader 2 | 52.9 (27/51) [38.5–67.1] | 56.9 (29/51) [42.2–70.6] | 0.387 |
| Reader 3 | 70.6 (36/51) [56.2–82.5] | 72.6 (37/51) [58.3–84.1] | 0.500 |
| Average | 61.4 (94/153) [53.2–69.2] | 61.4 (94/153) [53.2–69.2] | 0.500 |
| CAD standalone | - | 84.6 (45/51) [76.1–95.6] | 0.001 |
| Sensitivity for GS = 6 | |||
| Reader 1 | 57.1 (8/14) [28.9–82.3] | 42.9 (6/14) [17.7–71.1] | 0.363 |
| Reader 2 | 50.0 (7/14) [23.0–77.0] | 35.7 (5/14) [12.8–64.9] | 0.344 |
| Reader 3 | 71.4 (10/14) [41.9–91.6] | 71.4 (10/14) [41.9–91.6] | 0.500 |
| Average | 59.6 (25/42) [43.3–74.4] | 50.0 (21/42) [34.2–65.8] | 0.240 |
| CAD standalone | - | 92.9 (13/14) [66.1–99.8] | 0.002 |
| Sensitivity for GS > 6 | |||
| Reader 1 | 62.2 (23/37) [44.8–77.5] | 59.5 (22/37) [42.1–75.2] | 0.500 |
| Reader 2 | 54.1 (20/37) [36.9–70.5] | 64.9 (24/37) [47.5–79.8] | 0.109 |
| Reader 3 | 70.3 (26/37) [53.0–84.1] | 73.0 (27/37) [55.9–86.2] | 0.500 |
| Average | 62.2 (69/111) [52.4–71.2] | 65.8 (73/111) [56.2–74.5] | 0.240 |
| CAD standalone | - | 86.5 (32/37) [71.2–95.5] | 0.008 |
| Sensitivity for max diameter 4–9 mm | |||
| Reader 1 | 39.1 (9/23) [19.7–61.5] | 39.1 (9/23) [19.7–61.5] | 0.500 |
| Reader 2 | 34.8 (8/23) [16.4–57.3] | 39.1 (9/23) [19.7–61.5] | 0.500 |
| Reader 3 | 56.6 (13/23) [34.5–76.8] | 60.9 (14/23) [38.5–80.3] | 0.500 |
| Average | 43.5 (30/69) [31.6–56.0] | 46.4 (32/69) [34.3–58.8] | 0.407 |
| CAD standalone | - | 78.2 (18/23) [56.3–92.5] | 0.004 |
| Sensitivity for max diameter ≥ 10 mm | |||
| Reader 1 | 78.6 (22/28) [59.0–91.7] | 67.9 (19/28) [47.6–84.1] | 0.254 |
| Reader 2 | 67.9 (19/28) [47.6–84.1] | 71.4 (20/28) [51.3–86.8] | 0.500 |
| Reader 3 | 82.1 (23/28) [63.1–93.9] | 82.1 (23/28) [63.1–93.9] | 0.500 |
| Average | 76.2 (64/84) [65.6–84.8] | 73.8 (62/84) [63.1–82.8] | 0.407 |
| CAD standalone | - | 96.4 (27/28) [81.6–99.9] | 0.005 |
| Unassisted Reading | p Value | Assisted Reading | p Value | |||
|---|---|---|---|---|---|---|
| Reader 1 | 146 (91–199) | 35 (25–77) | <0.001 | |||
| Reader 2 | 120 (84–210) | 70 (37–99) | <0.001 | |||
| Reader 3 | 255 (180–403) | 100 (56–180) | <0.001 | |||
| Average | 170 (101–270) | 66 (33–108) | <0.001 | |||
| Biopsy + | Biopsy − | Biopsy + | Biopsy − | |||
| Reader 1 | 187 (135–281) | 122 (90–158) | <0.001 | 70 (47–99) | 25 (20–35) | <0.001 |
| Reader 2 | 180 (117–233) | 90 (60–129) | <0.001 | 80 (49–106) | 50 (30–90) | 0.007 |
| Reader 3 | 331 (224–473) | 200 (127–291) | <0.001 | 120 (70–205) | 98 (40–170) | 0.051 |
| Average | 210 (143–325) | 125 (90–200) | <0.001 | 90 (50–120) | 40 (25–90) | <0.001 |
| Per-Patient Analysis | Per-Lesion Analysis | |||
|---|---|---|---|---|
| CAD-Assisted | Reader 1 | Reader 3 | Reader 1 | Reader 3 |
| Reader 2 | 0.647 (0.488–0.806) | 0.641 (0.482–0.799) | 0.722 (0.531–0.913) | 0.631 (0.427–0.834) |
| Reader 3 | 0.704 (0.561–0.846) | - | 0.582 (0.364–0.800) | - |
| Overall | 0.662 (0.542–0.781) | 0.641 (0.483–0.780) | ||
| Unassisted | Reader 1 | Reader 3 | Reader 1 | Reader 3 |
| Reader 2 | 0.746 (0.598–0.893) | 0.602 (0.438–0.766) | 0.603 (0.385–0.821) | 0.441 (0.188–0.693) |
| Reader 3 | 0.605 (0.440–0.770) | - | 0.397 (0.159–0.635) | - |
| Overall | 0.646 (0.527–0.765) | 0.476 (0.317–0.634) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannini, V.; Mazzetti, S.; Cappello, G.; Doronzio, V.M.; Vassallo, L.; Russo, F.; Giacobbe, A.; Muto, G.; Regge, D. Computer-Aided Diagnosis Improves the Detection of Clinically Significant Prostate Cancer on Multiparametric-MRI: A Multi-Observer Performance Study Involving Inexperienced Readers. Diagnostics 2021, 11, 973. https://doi.org/10.3390/diagnostics11060973
Giannini V, Mazzetti S, Cappello G, Doronzio VM, Vassallo L, Russo F, Giacobbe A, Muto G, Regge D. Computer-Aided Diagnosis Improves the Detection of Clinically Significant Prostate Cancer on Multiparametric-MRI: A Multi-Observer Performance Study Involving Inexperienced Readers. Diagnostics. 2021; 11(6):973. https://doi.org/10.3390/diagnostics11060973
Chicago/Turabian StyleGiannini, Valentina, Simone Mazzetti, Giovanni Cappello, Valeria Maria Doronzio, Lorenzo Vassallo, Filippo Russo, Alessandro Giacobbe, Giovanni Muto, and Daniele Regge. 2021. "Computer-Aided Diagnosis Improves the Detection of Clinically Significant Prostate Cancer on Multiparametric-MRI: A Multi-Observer Performance Study Involving Inexperienced Readers" Diagnostics 11, no. 6: 973. https://doi.org/10.3390/diagnostics11060973







