Does Anatomical Contextual Information Improve 3D U-Net-Based Brain Tumor Segmentation?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Anatomical Contextual Information
2.3. Model
2.4. Evaluation and Statistical Methods
2.5. Multimodal MR Model Training
2.6. Compensation for Fewer MR Modalities
2.7. Domain Generalization
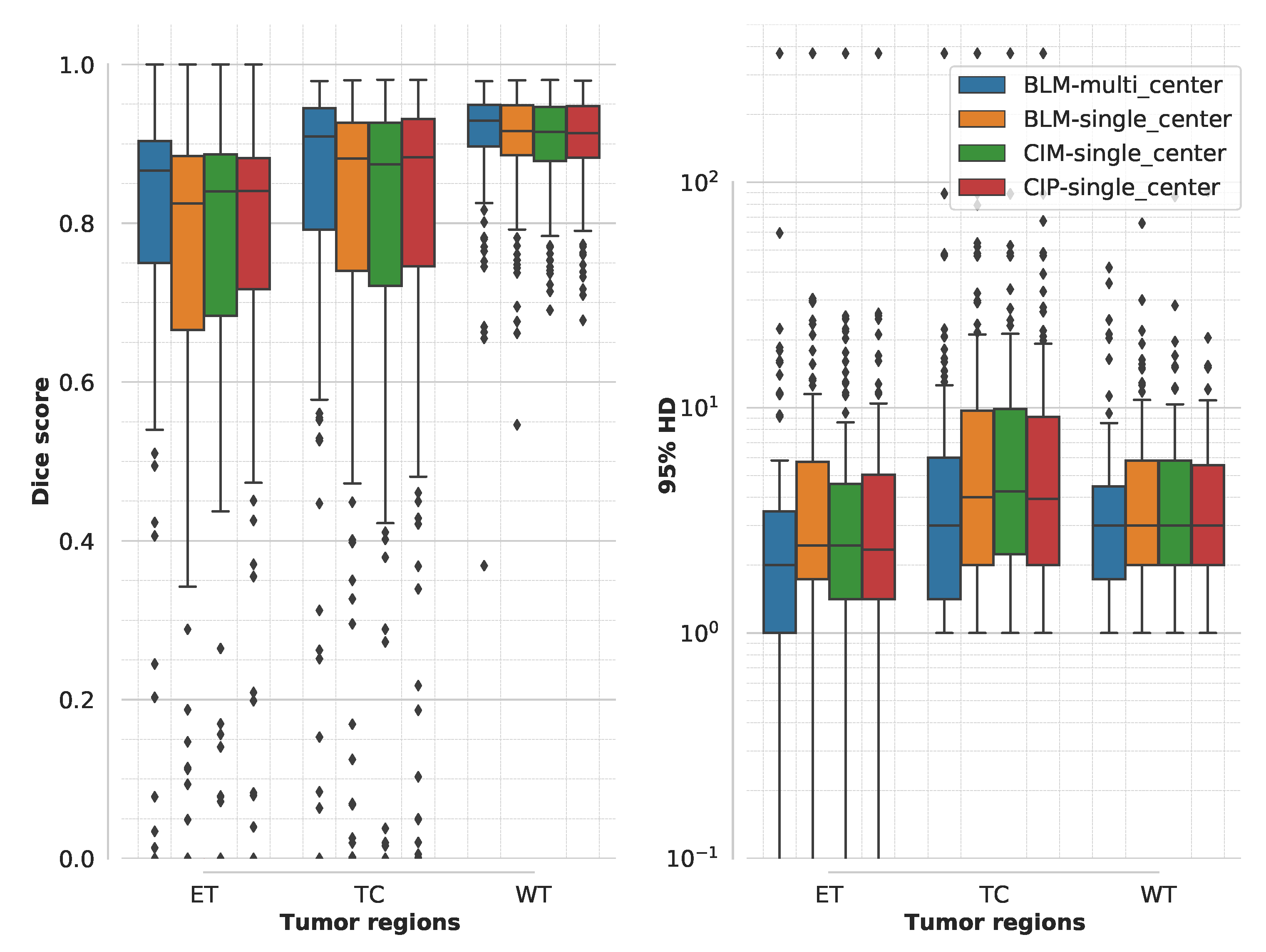
3. Results
3.1. Segmentation Accuracy for Multimodal MR Model Training
3.2. Model Training Time for Multimodal MR Model Training
3.3. Compensation for Fewer MR Modalities
3.4. Domain Generalization
4. Discussion
4.1. Segmentation Accuracy and Training Time for Multimodal MR Model Training
4.2. Quality of the Anatomical Contextual Information
4.3. Compensation for Fewer MR Modalities
4.4. Domain Generalization
4.5. Effect of Contextual Information on LGG and HGG Cases
4.6. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davis, M.E. Glioblastoma: Overview of disease and treatment. Clin. J. Oncol. Nurs. 2016, 20, S2. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, R.S.; Englander, Z.K.; Canoll, P.; Bruce, J.N. Extent of resection in glioma–a review of the cutting edge. World Neurosurg. 2017, 103, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Juratli, T.A.; Tummala, S.S.; Riedl, A.; Daubner, D.; Hennig, S.; Penson, T.; Zolal, A.; Thiede, C.; Schackert, G.; Krex, D.; et al. Radiographic assessment of contrast enhancement and T2/FLAIR mismatch sign in lower grade gliomas: Correlation with molecular groups. J. Neurooncol. 2019, 141, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Müller, D.; van Duijn, R.; Smits, M.; Verburg, N.; Hendriks, E.; Nabuurs, R.; Bot, J.; Eijgelaar, R.; Witte, M.; et al. Inter-rater agreement in glioma segmentations on longitudinal MRI. NeuroImage Clin. 2019, 22, 101727. [Google Scholar] [CrossRef] [PubMed]
- Kubben, P.L.; Postma, A.A.; Kessels, A.G.; van Overbeeke, J.J.; van Santbrink, H. Intraobserver and interobserver agreement in volumetric assessment of glioblastoma multiforme resection. Neurosurgery 2010, 67, 1329–1334. [Google Scholar] [CrossRef]
- Tiwari, A.; Srivastava, S.; Pant, M. Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognit. Lett. 2020, 131, 244–260. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Işın, A.; Direkoğlu, C.; Şah, M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput. Sci. 2016, 102, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Munich, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin, Germany, 2018; pp. 3–11. [Google Scholar]
- Noori, M.; Bahri, A.; Mohammadi, K. Attention-Guided Version of 2D UNet for Automatic Brain Tumor Segmentation. In Proceedings of the 2019 9th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 24–25 October 2019; pp. 269–275. [Google Scholar]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. No new-net. In International MICCAI Brainlesion Workshop; Springer: Berlin, Germany, 2018; pp. 234–244. [Google Scholar]
- Vercauteren, T.; Unberath, M.; Padoy, N.; Navab, N. Cai4cai: The rise of contextual artificial intelligence in computer-assisted interventions. Proc. IEEE 2019, 108, 198–214. [Google Scholar] [CrossRef] [Green Version]
- Tu, Z.; Bai, X. Auto-context and its application to high-level vision tasks and 3d brain image segmentation. IEEE PAMI 2009, 32, 1744–1757. [Google Scholar]
- Liu, J.; Liu, H.; Tang, Z.; Gui, W.; Ma, T.; Gong, S.; Gao, Q.; Xie, Y.; Niyoyita, J.P. IOUC-3DSFCNN: Segmentation of brain tumors via IOU constraint 3D symmetric full convolution network with multimodal auto-context. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Qamar, S.; Shen, L.; Saeed, A. Context Aware 3D UNet for Brain Tumor Segmentation. arXiv 2020, arXiv:2010.13082. [Google Scholar]
- Chandra, S.; Vakalopoulou, M.; Fidon, L.; Battistella, E.; Estienne, T.; Sun, R.; Robert, C.; Deutsch, E.; Paragios, N. Context aware 3D CNNs for brain tumor segmentation. In International MICCAI Brainlesion Workshop; Springer: Berlin, Germany, 2018; pp. 299–310. [Google Scholar]
- Liu, Z.; Tong, L.; Chen, L.; Zhou, F.; Jiang, Z.; Zhang, Q.; Wang, Y.; Shan, C.; Li, L.; Zhou, H. CANet: Context Aware Network for 3D Brain Tumor Segmentation. arXiv 2020, arXiv:2007.07788. [Google Scholar]
- Pei, L.; Vidyaratne, L.; Rahman, M.M.; Iftekharuddin, K.M. Context aware deep learning for brain tumor segmentation, subtype classification, and survival prediction using radiology images. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Le, N.; Yamazaki, K.; Truong, D.; Quach, K.G.; Savvides, M. A Multi-task Contextual Atrous Residual Network for Brain Tumor Detection & Segmentation. arXiv 2020, arXiv:2012.02073. [Google Scholar]
- Wachinger, C.; Reuter, M.; Klein, T. DeepNAT: Deep convolutional neural network for segmenting neuroanatomy. NeuroImage 2018, 170, 434–445. [Google Scholar] [CrossRef]
- Shen, H.; Wang, R.; Zhang, J.; McKenna, S.J. Boundary-aware fully convolutional network for brain tumor segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; pp. 433–441. [Google Scholar]
- Shen, H.; Zhang, J.; Zheng, W. Efficient symmetry-driven fully convolutional network for multimodal brain tumor segmentation. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 3864–3868. [Google Scholar]
- Kao, P.Y.; Ngo, T.; Zhang, A.; Chen, J.W.; Manjunath, B. Brain tumor segmentation and tractographic feature extraction from structural MR images for overall survival prediction. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018; pp. 128–141. [Google Scholar]
- Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006, 31, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Brady, M.; Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 2001, 20, 45–57. [Google Scholar] [CrossRef]
- Tudorascu, D.L.; Karim, H.T.; Maronge, J.M.; Alhilali, L.; Fakhran, S.; Aizenstein, H.J.; Muschelli, J.; Crainiceanu, C.M. Reproducibility and bias in healthy brain segmentation: Comparison of two popular neuroimaging platforms. Front. Neurosci. 2016, 10, 503. [Google Scholar] [CrossRef]
- Ashburner, J.; Barnes, G.; Chen, C.C.; Daunizeau, J.; Flandin, G.; Friston, K.; Kiebel, S.; Kilner, J.; Litvak, V.; Moran, R.; et al. SPM12 Manual; Wellcome Trust Centre for Neuroimaging: London, UK, 2014; Volume 2464. [Google Scholar]
- Bruce, F. FreeSurfer. NeuroImage 2012, 62, 774–781. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Full, P.M.; Vollmuth, P.; Maier-Hein, K.H. nnU-Net for Brain Tumor Segmentation. arXiv 2020, arXiv:2011.00848. [Google Scholar]
- Tofts, P. Quantitative MRI of the Brain: Measuring Changes Caused by Disease; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Hollingsworth, K.G. Reducing acquisition time in clinical MRI by data undersampling and compressed sensing reconstruction. Phys. Med. Biol. 2015, 60, R297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Liu, Z.; Qiao, Y.; Xiang, T.; Loy, C.C. Domain generalization: A survey. arXiv 2021, arXiv:2103.02503. [Google Scholar]





| Grade | Model | Median Dice Score | Median 95% HD | ||||
|---|---|---|---|---|---|---|---|
| [Min, Max] | [Min, Max] | ||||||
| TC | ET | WT | TC | ET | WT | ||
| LGG (18 cases) | BLM | 79.96 | 36.72 | 93.28 | 7.21 | 39.66 | 3.00 |
| [0.00, 98.39] | [0.00, 100.00] | [0.00, 97.09] | [1.00, 373.13] | [0.00, 373.13] | [1.00, 373.13] | ||
| CIM | 81.95 | 44.87 | 93.51 | 6.98 | 25.18 | 3.00 | |
| [0.00, 98.38] | [0.00, 100.00] | [0.00, 96.98] | [1.00, 373.13] | [0.00, 373.13] | [1.41, 373.13] | ||
| CIP | 82.27 | 51.27 | 93.22 | 7.64 | 23.17 | 3.08 | |
| [0.00, 80.00] | [0.00, 100.00] | [10.08, 97.18] | [1.00, 373.13] | [0.00, 373.13] | [1.00, 18.49] | ||
| HGG (18 cases) | BLM | 94.83 | 89.22 | 92.79 | 1.41 | 1.41 | 2.00 |
| [65.76, 97.56] | [75.54, 96.57] | [86.64, 97.19] | [1.00, 21.47] | [1.00, 3.00] | [1.00, 7.87] | ||
| CIM | 94.80 | 89.16 | 92.94 | 1.57 | 1.41 | 1.87 | |
| [64.22, 97.37] | [74.74, 96.17] | [86.90, 97.17] | [1.00, 22.03] | [1.00, 3.00] | [1.00, 7.87] | ||
| CIP | 94.54 | 89.12 | 92.74 | 1.57 | 1.41 | 1.87 | |
| [67.68, 97.48] | [74.21, 96.55] | [86.96, 97.27] | [1.00, 14.87] | [1.00, 3.00] | [1.00, 7.55] | ||
| Model | Dataset | Mean Dice Score | Claimed Improvement Using Context Awareness | ||
|---|---|---|---|---|---|
| TC | ET | WT | |||
| Isensee et al. [33] * | BraTS2020 1 | 85.95 | 82.03 | 88.95 | no context-aware mechanism used |
| Liu et al. [15] | BraTS2017 3 | 84.00 | 78.00 | 89.00 | comparison between models with and without context-aware mechanism not available |
| Liu et al. [18] | BraTS2019 2 | 85.10 | 75.90 | 88.50 | |
| Ahmad et al. [16] | BraTS2020 1 | 84.67 | 79.10 | 89.12 | |
| Chandra et al. [17] | BraTS2018 1 | 73.33 | 61.82 | 82.99 | |
| Pei et al. [19] | BraTS2019/20 3 | 83.50 | 82.10 | 89.50 | |
| Shen et al. [23] | BraTS2013 3 | 71.80 | 72.50 | 88.70 | 2–3% (no p-value) |
| Shen et al. [22] | BraTS2015 1 | 82.00 | 75.00 | 87.00 | 1.3% (p-value < 0.01) |
| Kao et al. [24] | BraTS2018 1 | 79.30 | 74.90 | 87.50 | 1–2% (no p-value) |
| Le et al. [20] | BraTS2018 2 | 88.88 | 81.41 | 90.95 | 2% (no p-value) |
| BLM | BraTS2020 3 (36 cases) | 81.80 | 67.20 | 90.80 | none |
| CIM | 81.90 | 77.00 | 90.10 | ||
| CIP | 81.80 | 70.40 | 90.50 | ||
| BLM | BraTS2020 2 (125 cases) | 81.60 | 73.41 | 90.54 | none |
| CIM | 81.61 | 73.43 | 90.58 | ||
| CIP | 81.44 | 74.05 | 90.69 | ||
| Model | Average Training Time [hh:mm:ss] | Epochs [250 Mini-Batches Each] |
|---|---|---|
| BLM | 8:15:11 | 79 |
| CIM | 20:02:39 | 140 |
| CIP | 12:58:57 | 103 |
| Grade | Model | Median Dice Score | Median 95% HD | ||||
|---|---|---|---|---|---|---|---|
| [Min, Max] | [Min, Max] | ||||||
| TC | ET | WT | TC | ET | WT | ||
| LGG (18 cases) | BLM | 79.99 | 61.08 | 86.65 | 9.25 | 176.67 | 86.01 |
| [0.00, 98.06] | [0.00, 100.00] | [0.00, 95,34] | [1.00, 373.13] | [0.00, 373.13] | [0.00, 373.13] | ||
| CIM | 78.21 | 37.93 | 83.85 | 6.42 | 37.85 | 9.26 | |
| [0.00, 98.04] | [0.00, 100.00] | [0.00, 95.16] | [1.00, 373.13] | [0.00, 373.13] | [2.00, 373.13] | ||
| CIP | 79.58 | 52.22 | 83.27 | 7.87 | 23.43 | 10.37 | |
| [0.00, 98.21] | [0.00, 100.00] | [0.00, 94.87] | [1.00, 373.13] | [0.00, 373.13] | [2.24, 373.13] | ||
| HGG (18 cases) | BLM | 94.41 | 89.70 | 90.35 | 1.87 | 1.41 | 3.86 |
| [87.55, 97.10] | [69.71, 96.29] | [71.90, 94.51] | [1.00, 4.90] | [1.00, 2.83] | [1.73, 15.17] | ||
| CIM | 93.90 | 89.86 | 89.58 | 1.87 | 1.41 | 3.93 | |
| [66.94, 97.06] | [70.93, 96.41] | [77.27, 94.49] | [1.00, 13.49] | [1.00, 3.00] | [2.00, 13.00] | ||
| CIP | 94.10 | 89.75 | 89.96 | 1.87 | 1.41 | 3.86 | |
| [63.08, 97.34] | [72.14, 96.45] | [79.54, 94.61] | [1.00, 22.38] | [1.00, 3.00] | [1.73, 10.72] | ||
| Grade | Model | Median Dice Score | Median 95% HD | ||||
|---|---|---|---|---|---|---|---|
| [Min, Max] | [Min, Max] | ||||||
| TC | ET | WT | TC | ET | WT | ||
| LGG (18 cases) | BLM | 78.47 | 60.81 | 92.77 | 6.40 | 7.64 | 2.73 |
| [0.00, 97.31] | [0.00, 100.00] | [76.48, 97.11] | [0.00, 97.31] | [1.00, 373.13] | [0.00, 373.13] | ||
| CIM | 75.99 | 56.71 | 93.02 | 6.71 | 13.42 | 2.53 | |
| [0.00, 96.85] | [0.00, 100.00] | [84.68, 97.21] | [1.00, 373.13] | [0.00, 373.13] | [1.41, 12.69] | ||
| CIP | 73.53 | 60.19 | 93.29 | 6.54 | 13.98 | 2.45 | |
| [0.00, 97.05] | [0.00, 100.00] | [14.81, 96.94] | [1.00, 373.13] | [0.00, 373.13] | [1.00, 14.78] | ||
| HGG (18 cases) | BLM | 91.68 | 85.66 | 89.30 | 2.24 | 2.00 | 3.23 |
| [63.67, 96.15] | [53.57, 94.62] | [76.57, 96,62] | [1.00, 12.37] | [1.00, 6.63] | [1.00, 75.21] | ||
| CIM | 90.97 | 86.37 | 89.13 | 2.34 | 2.00 | 3.67 | |
| [67.18, 96.62] | [53.79, 94.29] | [80.83, 96.84] | [1.14, 10.05] | [1.00, 6.48] | [1.00, 13.64] | ||
| CIP | 92.16 | 86.33 | 89.41 | 2.27 | 1.23 | 3.38 | |
| [63.55, 96.36] | [54.59, 94.25] | [81.16, 96.63] | [1.41, 13.19] | [1.00, 6.40] | [1.00, 10.20] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tampu, I.E.; Haj-Hosseini, N.; Eklund, A. Does Anatomical Contextual Information Improve 3D U-Net-Based Brain Tumor Segmentation? Diagnostics 2021, 11, 1159. https://doi.org/10.3390/diagnostics11071159
Tampu IE, Haj-Hosseini N, Eklund A. Does Anatomical Contextual Information Improve 3D U-Net-Based Brain Tumor Segmentation? Diagnostics. 2021; 11(7):1159. https://doi.org/10.3390/diagnostics11071159
Chicago/Turabian StyleTampu, Iulian Emil, Neda Haj-Hosseini, and Anders Eklund. 2021. "Does Anatomical Contextual Information Improve 3D U-Net-Based Brain Tumor Segmentation?" Diagnostics 11, no. 7: 1159. https://doi.org/10.3390/diagnostics11071159
APA StyleTampu, I. E., Haj-Hosseini, N., & Eklund, A. (2021). Does Anatomical Contextual Information Improve 3D U-Net-Based Brain Tumor Segmentation? Diagnostics, 11(7), 1159. https://doi.org/10.3390/diagnostics11071159






