Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy—A Syrian Golden Hamster Model
Abstract
1. Introduction
2. Materials and Methods
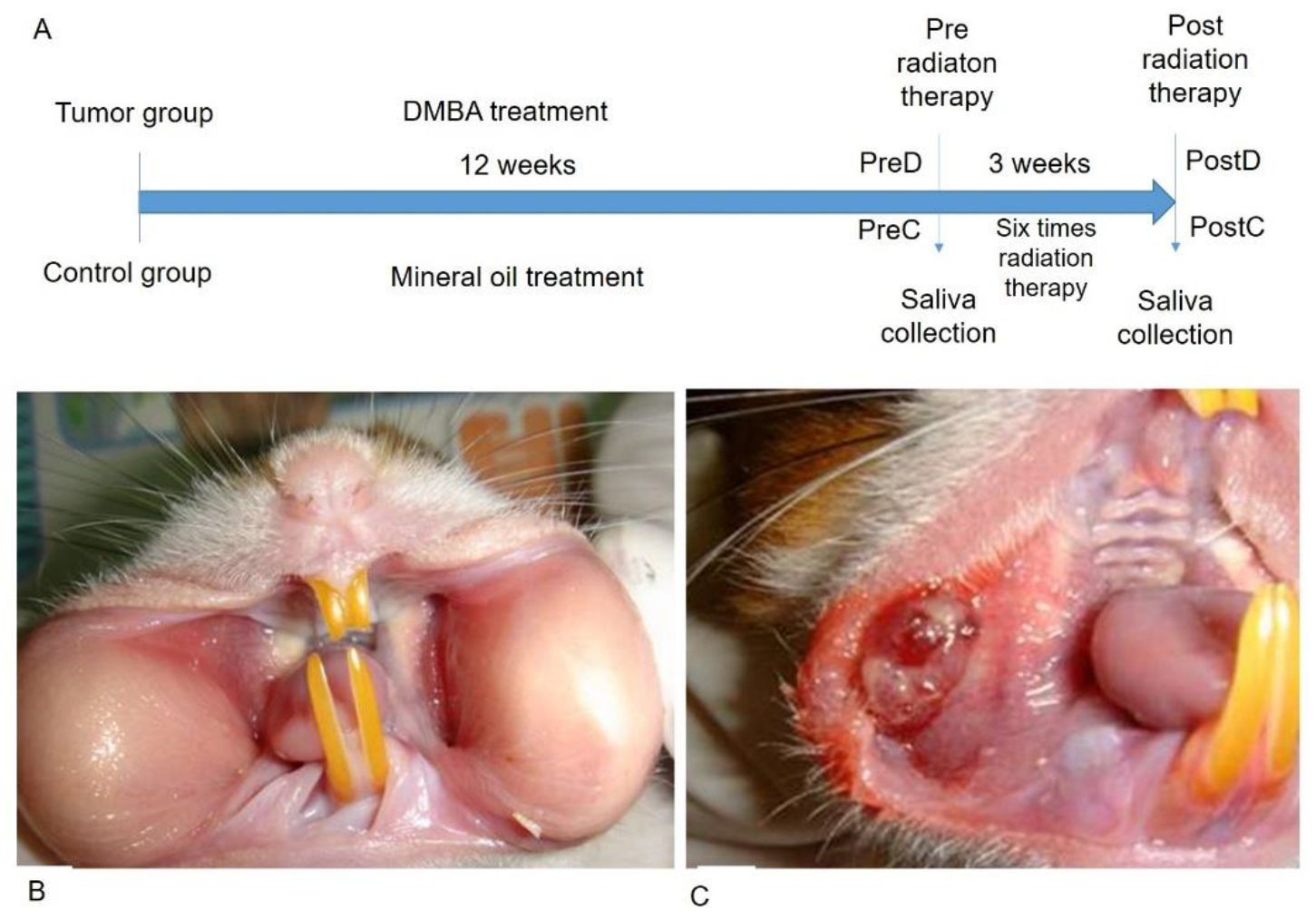
2.1. DMBA-Induced Tumorigenesis
2.2. Radiation Treatment Procedure of the Local Tumor Mass
2.3. Immunohistochemistry Protocol
2.4. Saliva Collection
2.5. Saliva Exosome Isolation and Protein Extraction
2.6. In-Gel Digestion and LC-MS/MS Analysis
2.7. Data Quantification and Statistical Analysis
2.8. Bioinformatics Analysis
3. Results
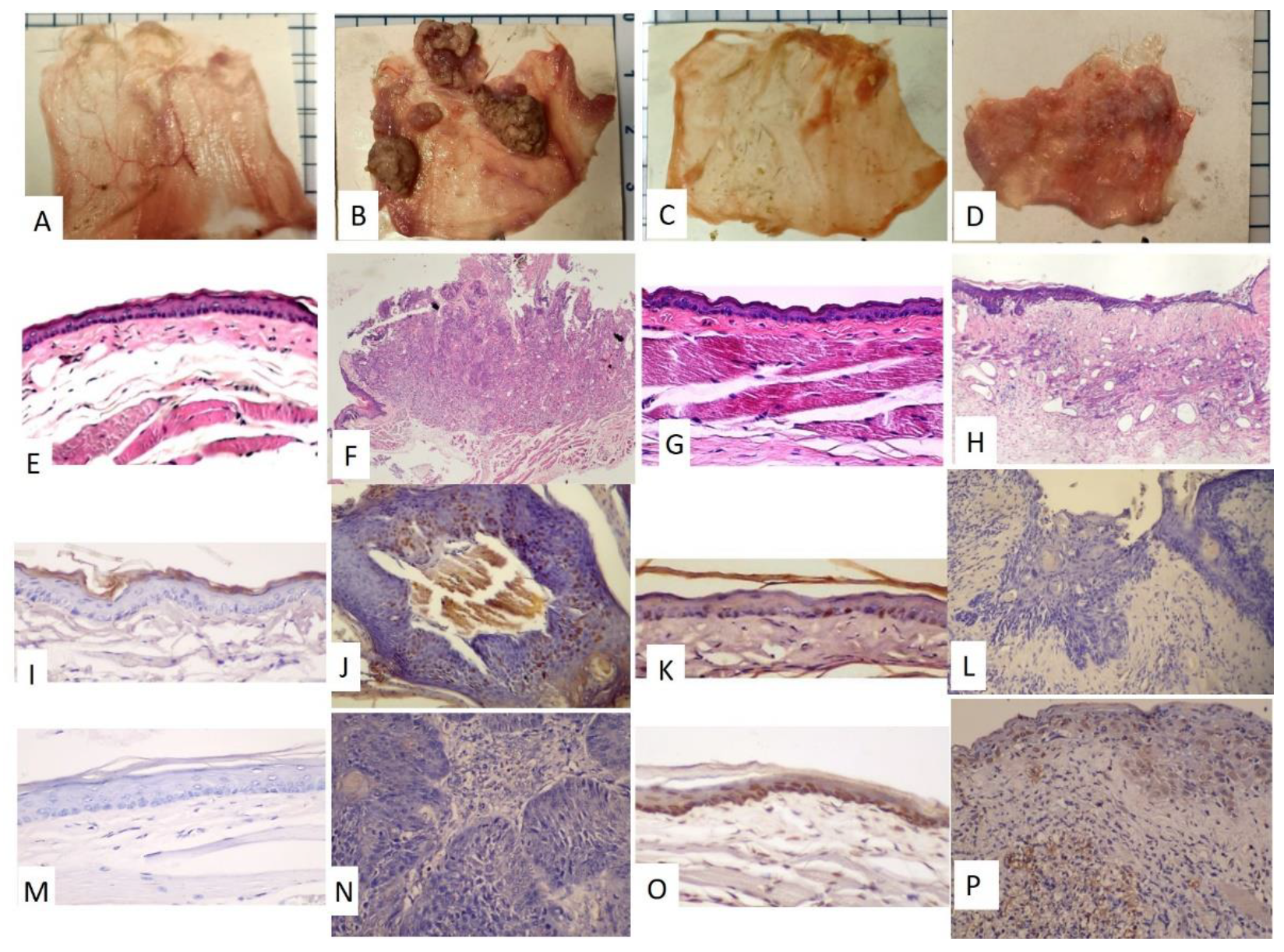
3.1. The Pouches Showed a Tumor Mass after 12 Weeks of DMBA Treatment and the Tumor Mass Showed Apoptosis after Receiving Six Rounds of Radiation Exposure
3.2. The DMBA-Treated PreD Group Expressed a Unique Salivary Exosome Protein, Desmocollin-2
3.3. Five Proteins Related to Biomolecular Breakdown Were Absent in Salivary Exosomes in the PostD Group
3.4. Salivary Exosomes of PreD Lacked the Proteins Associated with Tumorigenesis Signaling Pathways and the p53-Dependent and -Independent DNA Repair Pathway
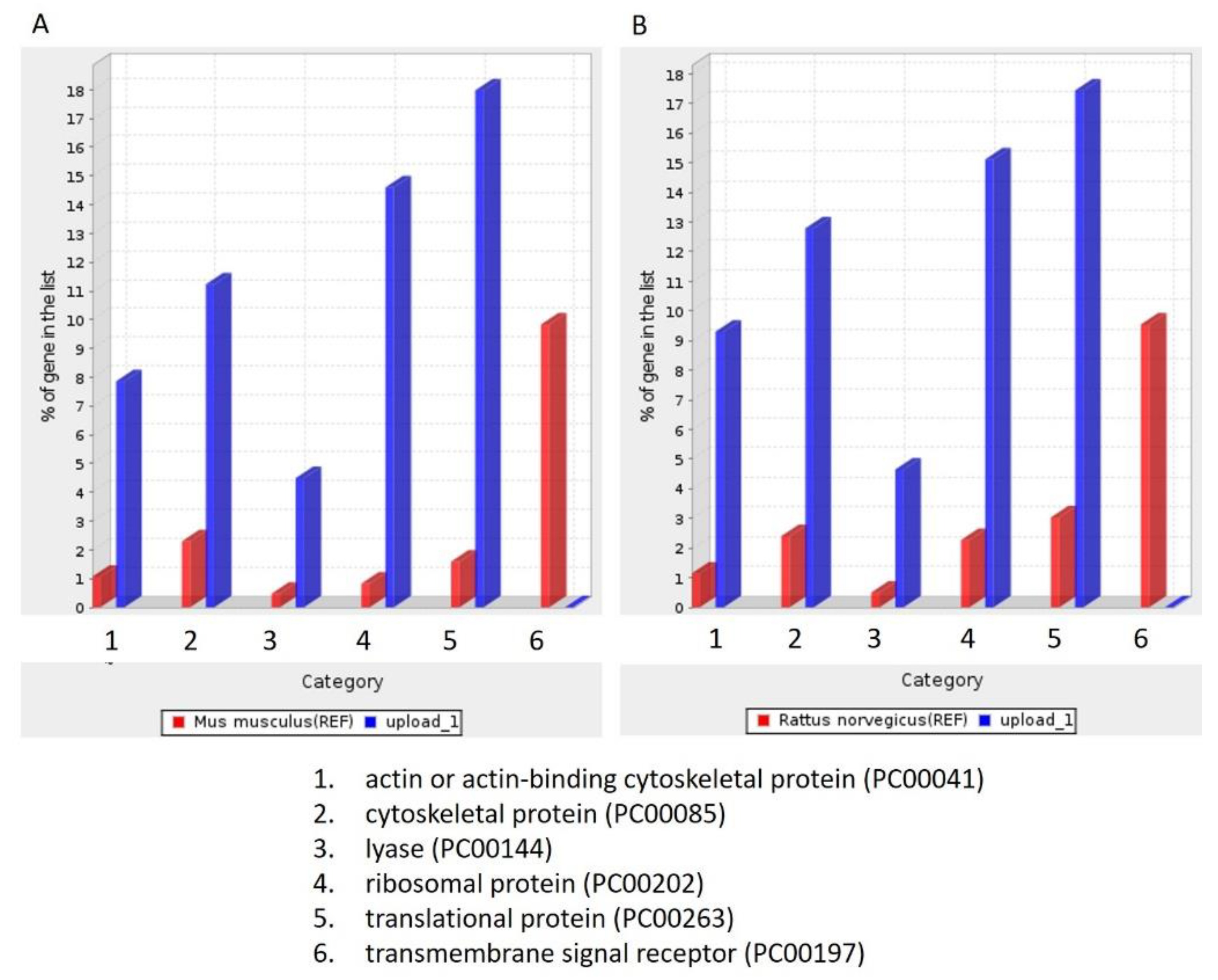
3.5. Molecular Function, Biological Process, and Cellular Component Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Pizzicannella, J.; Diomede, F.; Gugliandolo, A.; Chiricosta, L.; Bramanti, P.; Merciaro, I.; Orsini, T.; Mazzon, E.; Trubiani, O. 3D Printing PLA/Gingival Stem Cells/ EVs Upregulate miR-2861 and -210 during Osteoangiogenesis Commitment. Int. J. Mol. Sci. 2019, 20, 3256. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Tang, K.D.; Kenny, L.; Punyadeera, C. Salivary exosomes as potential biomarkers in cancer. Oral Oncol. 2018, 84, 31–40. [Google Scholar] [CrossRef]
- Zhao, Y.; Yin, L.Y.; Bi, R.R.; Ma, P.; Li, S.B. Current advance in saliva-derived exosome study. Zhonghua Kou Qiang Yi Xue Za Zhi 2016, 51, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Nik Mohamed Kamal, N.N.S.; Awang, R.A.R.; Mohamad, S.; Shahidan, W.N.S. Plasma- and Saliva Exosome Profile Reveals a Distinct MicroRNA Signature in Chronic Periodontitis. Front. Physiol. 2020, 11, 587381. [Google Scholar] [CrossRef]
- Han, Y.; Jia, L.; Zheng, Y.; Li, W. Salivary Exosomes: Emerging Roles in Systemic Disease. Int. J. Biol. Sci. 2018, 14, 633–643. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef]
- Samavarchi Tehrani, S.; Zaboli, E.; Sadeghi, F.; Khafri, S.; Karimian, A.; Rafie, M.; Parsian, H. MicroRNA-26a-5p as a potential predictive factor for determining the effectiveness of trastuzumab therapy in HER-2 positive breast cancer patients. BioMedicine 2021, 11, 6. [Google Scholar] [CrossRef]
- Lu, Y.; Zheng, Z.; Yuan, Y.; Pathak, J.L.; Yang, X.; Wang, L.; Ye, Z.; Cho, W.C.; Zeng, M.; Wu, L. The Emerging Role of Exosomes in Oral Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 628103. [Google Scholar] [CrossRef]
- Abel, E.L.; Angel, J.M.; Kiguchi, K.; DiGiovanni, J. Multi-stage chemical carcinogenesis in mouse skin: Fundamentals and applications. Nat. Protoc. 2009, 4, 1350–1362. [Google Scholar] [CrossRef]
- Chen, Y.K.; Lin, L.M. DMBA-induced hamster buccal pouch carcinoma and VX2-induced rabbit cancer as a model for human oral carcinogenesis. Expert Rev. Anticancer Ther. 2010, 10, 1485–1496. [Google Scholar] [CrossRef]
- Nagini, S.; Letchoumy, P.V.; Thangavelu, A.; Ramachandran, C.R. Of humans and hamsters: A comparative evaluation of carcinogen activation, DNA damage, cell proliferation, apoptosis, invasion, and angiogenesis in oral cancer patients and hamster buccal pouch carcinomas. Oral Oncol. 2009, 45, e31–e37. [Google Scholar] [CrossRef]
- Wang, W.C.; Liang, S.L.; Chen, Y.K.; Lin, L.M. The therapeutic effect of fractionated radiation on DMBA-induced hamster buccal pouch squamous cell carcinomas. Oral Oncol. 2008, 44, 1160–1166. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Tachikawa, T.; Yamamura, T.; Yoshiki, S. Changes occurring in plasma membranes and intercellular junctions during the process of carcinogenesis and in squamous cell carcinoma. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1984, 47, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Yamaguchi, A.; Sakamoto, K. Aberrant expression and altered cellular localization of desmosomal and hemidesmosomal proteins are associated with aggressive clinicopathological features of oral squamous cell carcinoma. Virchows Arch. 2014, 465, 35–47. [Google Scholar] [CrossRef]
- Weiske, J.; Schoneberg, T.; Schroder, W.; Hatzfeld, M.; Tauber, R.; Huber, O. The fate of desmosomal proteins in apoptotic cells. J. Biol. Chem. 2001, 276, 41175–41181. [Google Scholar] [CrossRef] [PubMed]
- Nava, P.; Laukoetter, M.G.; Hopkins, A.M.; Laur, O.; Gerner-Smidt, K.; Green, K.J.; Parkos, C.A.; Nusrat, A. Desmoglein-2: A novel regulator of apoptosis in the intestinal epithelium. Mol. Biol. Cell 2007, 18, 4565–4578. [Google Scholar] [CrossRef][Green Version]
- Teh, M.T.; Parkinson, E.K.; Thurlow, J.K.; Liu, F.; Fortune, F.; Wan, H. A molecular study of desmosomes identifies a desmoglein isoform switch in head and neck squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 67–76. [Google Scholar] [CrossRef]
- Richardson, J.; Viswanathan, K.; Lucas, A. Serpins, the vasculature, and viral therapeutics. Front. Biosci. 2006, 11, 1042–1056. [Google Scholar] [CrossRef][Green Version]
- Ding, G.; Liu, S.; Ding, Q.; Feng, C. Overexpression of HSD17B4 exerts tumor suppressive function in adrenocortical carcinoma and is not associated with hormone excess. Oncotarget 2017, 8, 114736–114745. [Google Scholar] [CrossRef][Green Version]
- Albakova, Z.; Siam, M.K.S.; Sacitharan, P.K.; Ziganshin, R.H.; Ryazantsev, D.Y.; Sapozhnikov, A.M. Extracellular heat shock proteins and cancer: New perspectives. Transl. Oncol. 2021, 14, 100995. [Google Scholar] [CrossRef]
- Mele, D.A.; Bista, P.; Baez, D.V.; Huber, B.T. Dipeptidyl peptidase 2 is an essential survival factor in the regulation of cell quiescence. Cell Cycle 2009, 8, 2425–2434. [Google Scholar] [CrossRef] [PubMed]
- Nesterova, M.V.; Cho-Chung, Y.S. Chemoprevention with protein kinase A RIalpha antisense in DMBA-mammary carcinogenesis. Ann. N. Y. Acad Sci. 2005, 1058, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Dilman, V.M.; Berstein, L.M.; Zabezhinski, M.A.; Alexandrov, V.A.; Bobrov, J.F.; Pliss, G.B. Inhibition of DMBA-induced carcinogenesis by phenformin in the mammary gland of rats. Arch. Geschwulstforsch 1978, 48, 1–8. [Google Scholar] [PubMed]
| Pre-Radiation | Post-Radiation | |||||
|---|---|---|---|---|---|---|
| Protein Name | Gene Name | Control | Tumor | Control | Tumor | Protein Function |
| desmocollin-2 isoform X2 | Dsc2 | UD | 400 | UD | UD | Component of intercellular desmosome junctions Contribute to epidermal cell positioning (stratification). |
| desmocollin-2 isoform X1 | Dsc2 | UD | 400 | UD | UD | |
| desmocollin-2 isoform X3 | Dsc2 | UD | 400 | UD | UD | |
| serpin A9 | Serpina9 | UD | 108.2 | 291.8 | UD | Protease inhibitor that inhibits trypsin and trypsin-like serine proteases. |
| peroxisomal multifunctional enzyme type 2 | Hsd17b4 | UD | 39.9 | 360.1 | UD | Bifunctional enzyme acting on the peroxisomal beta-oxidation pathway for fatty acids. |
| alpha-mannosidase | Man2b2 | UD | 295.9 | 104.1 | UD | Necessary for the catabolism of N-linked carbohydrates released during glycoprotein turnover. |
| heat shock 70 kDa protein 13 | Hspa13 | UD | 333.4 | 66.6 | UD | Plays a pivotal role in the protein quality control system, ensuring the correct folding of proteins, re-folding of misfolded proteins and controlling, and targeting of proteins for subsequent degradation |
| dipeptidyl peptidase 2 | Dpp7 | UD | 331.5 | 68.5 | UD | Plays an important role in the degradation of some oligopeptides |
| Reference | Reactome Pathways | Fold Enrichment | Raw p-Value | FDR |
|---|---|---|---|---|
| Rattus norvegicus | CREB1 phosphorylation through the activation of adenylate cyclase (R-RNO-442720) | >100 | 3.23 × 10−4 | 1.20 × 10−2 |
| PKA activation in glucagon signalling (R-RNO-164378) | >100 | 3.23 × 10−4 | 1.18 × 10−2 | |
| Glucagon signaling in metabolic regulation (R-RNO-163359) | 71.71 | 5.50 × 10−4 | 1.74 × 10−2 | |
| p53-independent G1/S DNA damage checkpoint (R-RNO-69613) | 15.06 | 1.22 × 10−3 | 2.87 × 10−2 | |
| p53-independent DNA damage response (R-RNO-69610) | 15.06 | 1.22 × 10−3 | 2.83 × 10−2 | |
| p53-dependent G1/S DNA damage checkpoint (R-RNO-69580) | 11.95 | 2.31 × 10−3 | 3.64 × 10−2 | |
| p53-dependent G1 DNA damage response (R-RNO-69563) | 11.95 | 2.31 × 10−3 | 3.60 × 10−2 | |
| Mus musculus | CREB1 phosphorylation through the activation of adenylate cyclase (R-MMU-442720) | 98.82 | 3.33 × 10−4 | 1.25 × 10−2 |
| PKA activation in glucagon signalling (R-MMU-164378) | 98.82 | 3.33 × 10−4 | 1.22 × 10−2 | |
| Glucagon signaling in metabolic regulation (R-MMU-163359) | 70.59 | 5.68 × 10−4 | 1.87 × 10−2 | |
| p53-independent G1/S DNA damage checkpoint (R-MMU-69613) | 14.53 | 1.35 × 10−3 | 3.10 × 10−2 | |
| p53-independent DNA damage response (R-MMU-69610) | 14.53 | 1.35 × 10−3 | 3.06 × 10−2 | |
| p53-dependent G1/S DNA damage checkpoint (R-MMU-69580) | 11.58 | 2.52 × 10−3 | 3.95 × 10−2 | |
| p53-dependent G1 DNA damage response (R-MMU-69563) | 11.58 | 2.52 × 10−3 | 3.92 × 10−2 |
| Reference List | Reference List | ||
|---|---|---|---|
| Rattus norvegicus | Fold Enrichment | Mus musculus | Fold Enrichment |
| signal sequence binding (GO:0005048) | 23.53 | signal sequence binding (GO:0005048) | 24.71 |
| ubiquitin-like protein conjugating enzyme activity (GO:0061650) | 17.51 | structural constituent of ribosome (GO:0003735) | 22.06 |
| actin filament binding (GO:0051015) | 12.45 | actin filament binding (GO:0051015) | 12.56 |
| lyase activity (GO:0016829) | 8.88 | structural molecule activity (GO:0005198) | 10.47 |
| actin binding (GO:0003779) | 8.66 | lyase activity (GO:0016829) | 9.24 |
| structural constituent of ribosome (GO:0003735) | 7.21 | actin binding (GO:0003779) | 8.77 |
| mRNA binding (GO:0003729) | 6.34 | molecular adaptor activity (GO:0060090) | 8.75 |
| RNA binding (GO:0003723) | 5.81 | mRNA binding (GO:0003729) | 8.72 |
| GTPase activity (GO:0003924) | 5.4 | RNA binding (GO:0003723) | 7.72 |
| structural molecule activity (GO:0005198) | 5.27 | GTPase activity (GO:0003924) | 5.43 |
| protein-containing complex binding (GO:0044877) | 5.24 | protein-containing complex binding (GO:0044877) | 5.15 |
| cytoskeletal protein binding (GO:0008092) | 4.64 | cytoskeletal protein binding (GO:0008092) | 4.61 |
| binding (GO:0005488) | 1.61 | nucleic acid binding (GO:0003676) | 2.18 |
| molecular_function (GO:0003674) | 1.47 | heterocyclic compound binding (GO:1901363) | 2.05 |
| organic cyclic compound binding (GO:0097159) | 2.01 | ||
| binding (GO:0005488) | 1.67 | ||
| molecular_function (GO:0003674) | 1.5 |
| Reference List | Reference List | ||
|---|---|---|---|
| Rattus norvegicus | Fold Enrichment | Mus musculus | Fold Enrichment |
| protein K48-linked ubiquitination (GO:0070936) | 47.06 | regulation of endocytosis (GO:0030100) | 27.45 |
| regulation of endocytosis (GO:0030100) | 26.89 | actin filament organization (GO:0007015) | 7.97 |
| actin filament organization (GO:0007015) | 7.93 | translational elongation (GO:0006414) | 6.56 |
| ubiquitin-dependent protein catabolic process (GO:0006511) | 5.88 | translation (GO:0006412) | 6.56 |
| modification-dependent protein catabolic process (GO:0019941) | 5.67 | ribonucleoprotein complex biogenesis (GO:0022613) | 6.53 |
| modification-dependent macromolecule catabolic process (GO:0043632) | 5.58 | peptide biosynthetic process (GO:0043043) | 6.47 |
| regulation of cellular component organization (GO:0051128) | 4.76 | cellular protein-containing complex assembly (GO:0034622) | 5.19 |
| cellular protein-containing complex assembly (GO:0034622) | 4.57 | regulation of cellular component organization (GO:0051128) | 4.87 |
| intracellular protein transport (GO:0006886) | 4.29 | protein-containing complex assembly (GO:0065003) | 4.87 |
| protein transport (GO:0015031) | 4.14 | intracellular transport (GO:0046907) | 4.47 |
| peptide transport (GO:0015833) | 4.09 | intracellular protein transport (GO:0006886) | 4.36 |
| intracellular transport (GO:0046907) | 4.08 | protein transport (GO:0015031) | 4.19 |
| establishment of protein localization (GO:0045184) | 4.05 | peptide transport (GO:0015833) | 4.16 |
| amide transport (GO:0042886) | 4.01 | establishment of protein localization (GO:0045184) | 4.11 |
| establishment of localization in cell (GO:0051649) | 3.74 | establishment of localization in cell (GO:0051649) | 4.07 |
| nitrogen compound transport (GO:0071705) | 3.72 | amide transport (GO:0042886) | 4.06 |
| cellular component assembly (GO:0022607) | 3.55 | nitrogen compound transport (GO:0071705) | 3.77 |
| cellular component biogenesis (GO:0044085) | 3.43 | cellular component biogenesis (GO:0044085) | 3.77 |
| protein localization (GO:0008104) | 3.4 | cellular component assembly (GO:0022607) | 3.76 |
| cellular localization (GO:0051641) | 3.18 | cellular localization (GO:0051641) | 3.47 |
| cellular component organization or biogenesis (GO:0071840) | 2.34 | protein localization (GO:0008104) | 3.41 |
| cellular component organization (GO:0016043) | 2.3 | cellular component organization or biogenesis (GO:0071840) | 2.36 |
| cellular component organization (GO:0016043) | 2.28 |
| Reference List | Reference List | ||
|---|---|---|---|
| Rattus norvegicus | Fold Enrichment | Mus musculus | Fold Enrichment |
| proteasome regulatory particle, lid subcomplex (GO:0008541) | 71.71 | proteasome regulatory particle, lid subcomplex (GO:0008541) | 61.76 |
| proteasome accessory complex (GO:0022624) | 50.2 | proteasome accessory complex (GO:0022624) | 46.32 |
| proteasome regulatory particle (GO:0005838) | 50.2 | proteasome regulatory particle (GO:0005838) | 46.32 |
| eukaryotic translation initiation factor 3 complex (GO:0005852) | 33.47 | cytosolic small ribosomal subunit (GO:0022627) | 36.33 |
| endocytic vesicle (GO:0030139) | 24.29 | eukaryotic translation initiation factor 3 complex (GO:0005852) | 35.29 |
| proteasome complex (GO:0000502) | 18.82 | small ribosomal subunit (GO:0015935) | 29.65 |
| early endosome (GO:0005769) | 17.93 | cytosolic ribosome (GO:0022626) | 29.54 |
| endopeptidase complex (GO:1905369) | 17.11 | cytoplasmic stress granule (GO:0010494) | 27.45 |
| peptidase complex (GO:1905368) | 12.98 | cytosolic large ribosomal subunit (GO:0022625) | 26.01 |
| small ribosomal subunit (GO:0015935) | 10.32 | endocytic vesicle (GO:0030139) | 25.56 |
| cytosolic small ribosomal subunit (GO:0022627) | 10.2 | ribosomal subunit (GO:0044391) | 22.98 |
| actin cytoskeleton (GO:0015629) | 7.84 | ribosome (GO:0005840) | 21.18 |
| cytosolic ribosome (GO:0022626) | 7.67 | proteasome complex (GO:0000502) | 19.5 |
| ribosomal subunit (GO:0044391) | 7.38 | large ribosomal subunit (GO:0015934) | 18.76 |
| ribosome (GO:0005840) | 7.09 | early endosome (GO:0005769) | 17.97 |
| cytosolic large ribosomal subunit (GO:0022625) | 6.38 | endopeptidase complex (GO:1905369) | 17.65 |
| large ribosomal subunit (GO:0015934) | 5.75 | peptidase complex (GO:1905368) | 12.56 |
| ribonucleoprotein complex (GO:1990904) | 5.42 | ribonucleoprotein complex (GO:1990904) | 9.34 |
| cytosol (GO:0005829) | 4.17 | actin cytoskeleton (GO:0015629) | 7.84 |
| cytoskeleton (GO:0005856) | 3.39 | cytosol (GO:0005829) | 5.53 |
| intracellular non-membrane-bounded organelle (GO:0043232) | 2.68 | cytoplasmic vesicle (GO:0031410) | 3.69 |
| non-membrane-bounded organelle (GO:0043228) | 2.68 | intracellular vesicle (GO:0097708) | 3.67 |
| protein-containing complex (GO:0032991) | 2.54 | cytoskeleton (GO:0005856) | 3.38 |
| cytoplasm (GO:0005737) | 2.4 | intracellular non-membrane-bounded organelle (GO:0043232) | 2.95 |
| intracellular organelle (GO:0043229) | 1.91 | non-membrane-bounded organelle (GO:0043228) | 2.95 |
| organelle (GO:0043226) | 1.87 | protein-containing complex (GO:0032991) | 2.81 |
| intracellular (GO:0005622) | 1.86 | cytoplasm (GO:0005737) | 2.68 |
| cellular_component (GO:0005575) | 1.38 | intracellular organelle (GO:0043229) | 2 |
| cellular anatomical entity (GO:0110165) | 1.32 | organelle (GO:0043226) | 1.96 |
| intracellular (GO:0005622) | 1.92 | ||
| cellular_component (GO:0005575) | 1.38 | ||
| cellular anatomical entity (GO:0110165) | 1.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-C.; Huang, M.-Y.; Chen, Y.-K.; Lan, W.-C.; Shieh, T.-M.; Shih, Y.-H. Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy—A Syrian Golden Hamster Model. Diagnostics 2022, 12, 65. https://doi.org/10.3390/diagnostics12010065
Wang W-C, Huang M-Y, Chen Y-K, Lan W-C, Shieh T-M, Shih Y-H. Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy—A Syrian Golden Hamster Model. Diagnostics. 2022; 12(1):65. https://doi.org/10.3390/diagnostics12010065
Chicago/Turabian StyleWang, Wen-Chen, Ming-Yii Huang, Yuk-Kwan Chen, Wan-Chen Lan, Tzong-Ming Shieh, and Yin-Hwa Shih. 2022. "Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy—A Syrian Golden Hamster Model" Diagnostics 12, no. 1: 65. https://doi.org/10.3390/diagnostics12010065
APA StyleWang, W.-C., Huang, M.-Y., Chen, Y.-K., Lan, W.-C., Shieh, T.-M., & Shih, Y.-H. (2022). Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy—A Syrian Golden Hamster Model. Diagnostics, 12(1), 65. https://doi.org/10.3390/diagnostics12010065








