Deep Learning Supplants Visual Analysis by Experienced Operators for the Diagnosis of Cardiac Amyloidosis by Cine-CMR
Abstract
:1. Introduction
2. Materials and Methods
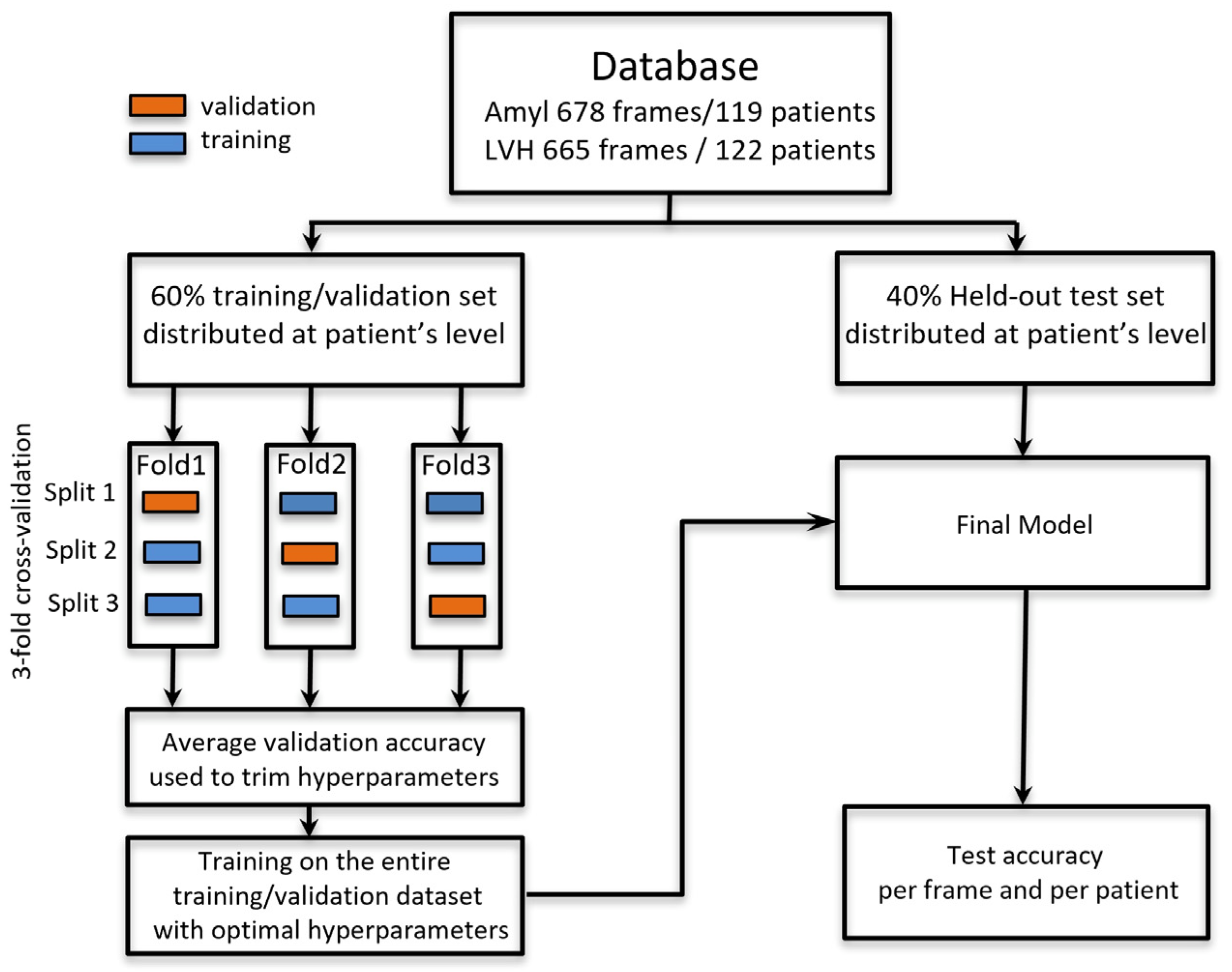
2.1. Study Population
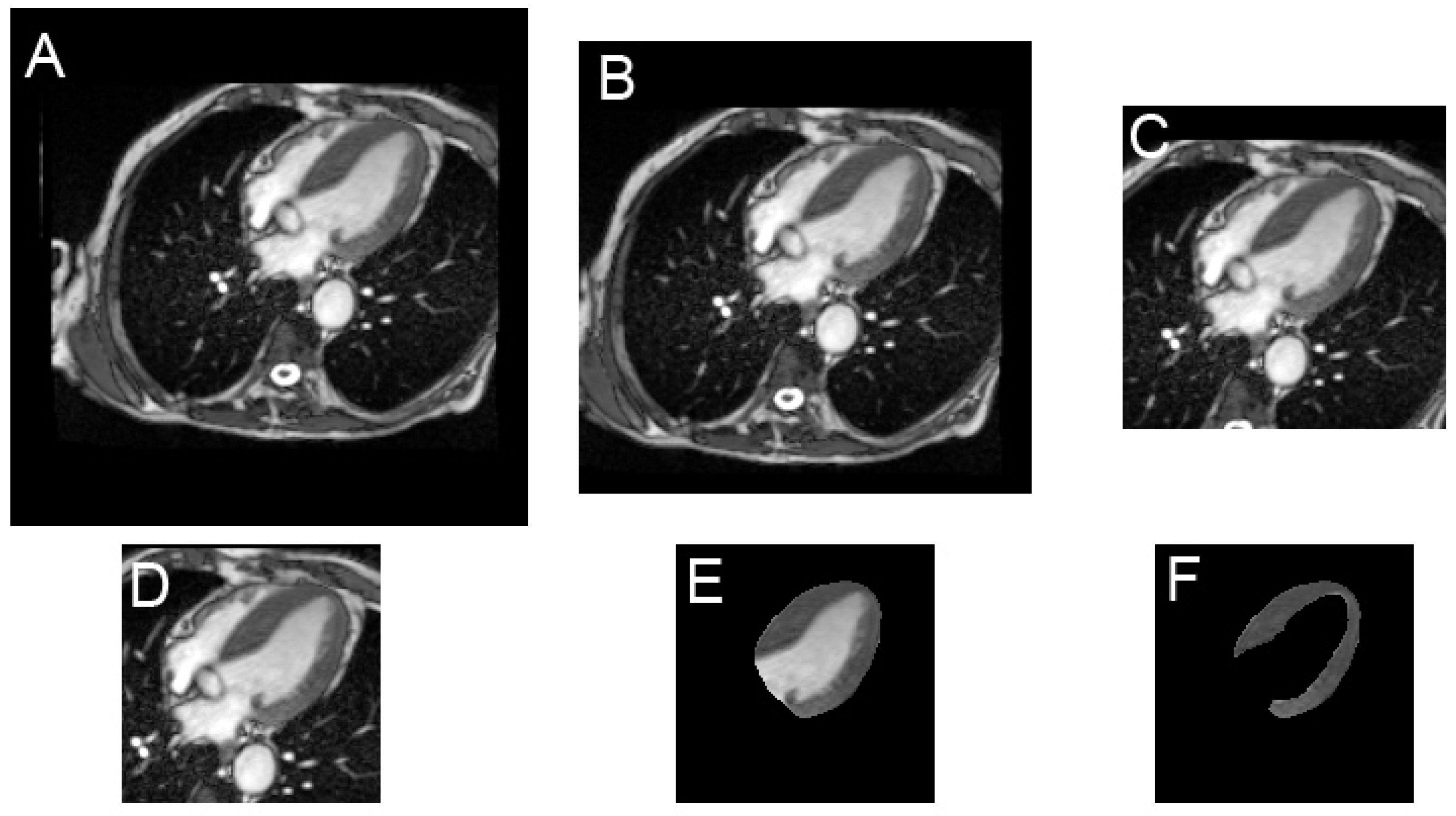
2.2. Image Preparation
2.3. Deep Learning Process
2.4. Experienced Radiologists/Cardiologists Blind Reading
2.5. Evaluation and Statistical Analysis
3. Results
3.1. Amyloidosis vs. LVH Classification Obtained with the Held-Out Test Set According to the Input Shape
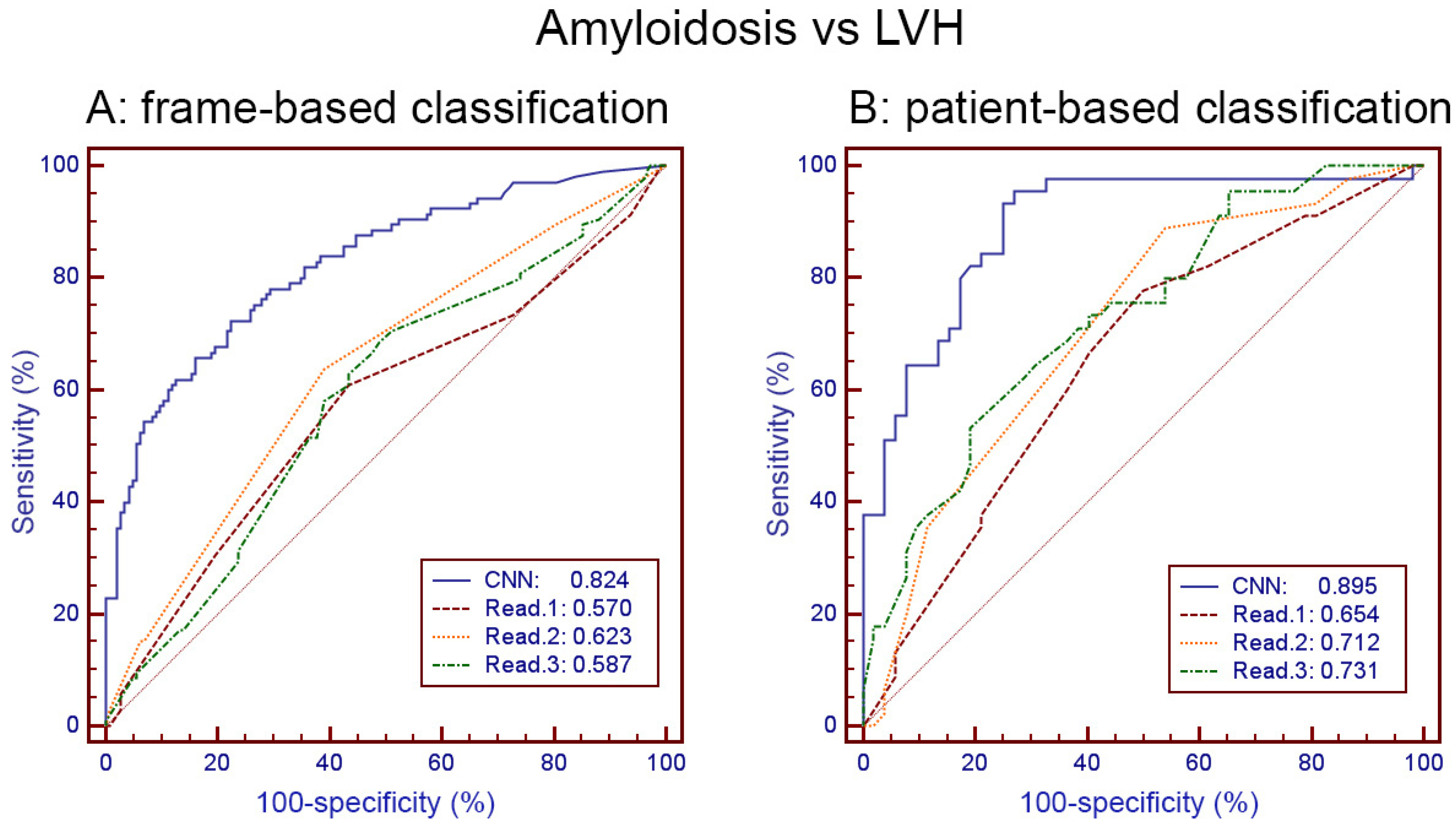
3.2. Amyloidosis vs. LVH Classification Obtained with the Held-Out Test Set by Human Readers and by CNN
3.3. CNN Classification of AL vs. TTR Amyloidosis
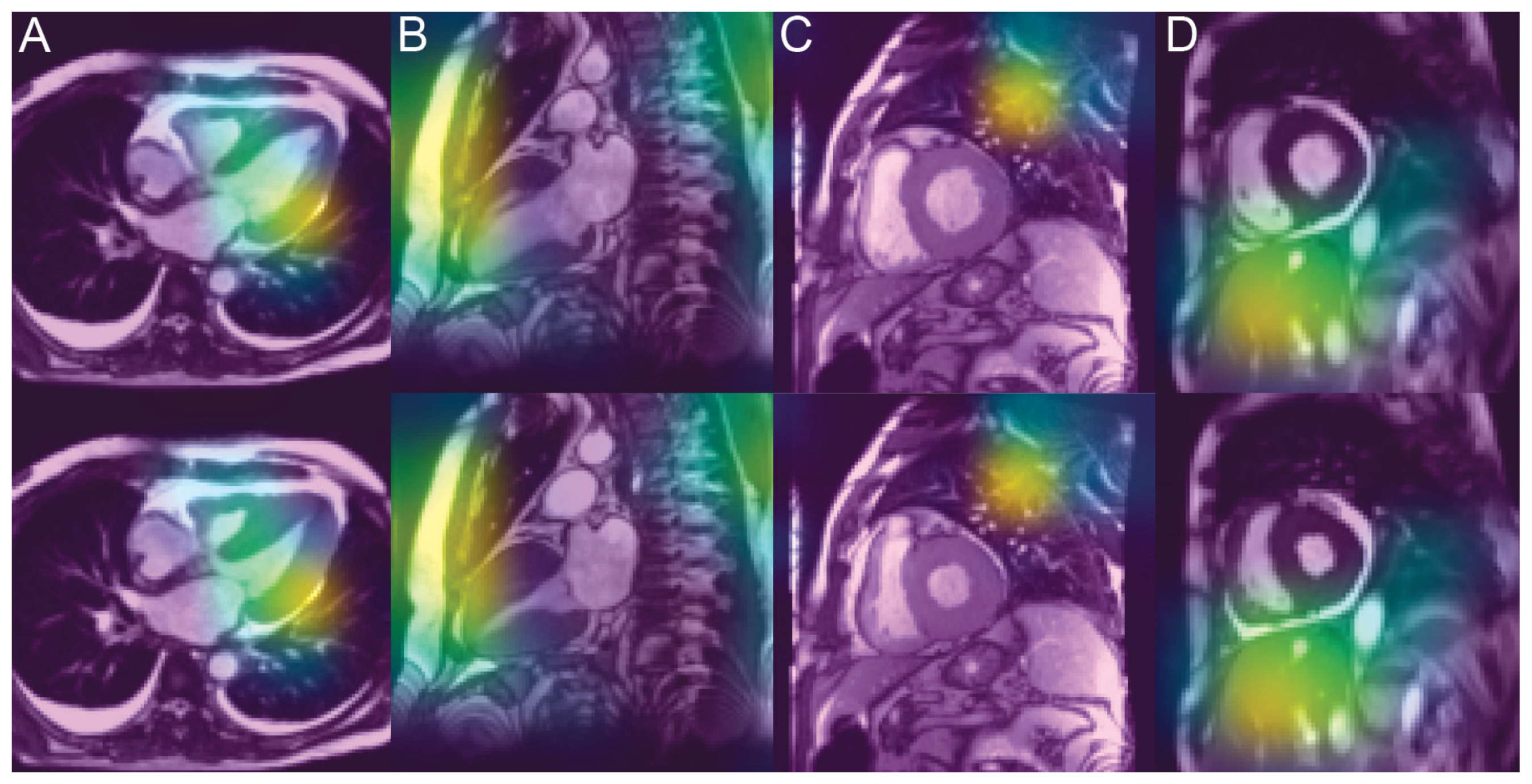
3.4. Analysis of the Saliency Maps
4. Discussion
4.1. Methodological Considerations
4.2. Superiority of CNN Capacities over Human Diagnosis
4.3. Unveiling the Invisible
4.4. Explanation of Classification Remains Unsatisfactory
4.5. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia-Pavia, P.; Rapezzi, C.; Adler, Y.; Arad, M.; Basso, C.; Brucato, A.; Burazor, I.; Caforio, A.L.P.; Damy, T.; Eriksson, U.; et al. Diagnosis and treatment of cardiac amyloidosis. A position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. J. Heart Fail. 2021, 23, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kittleson, M.M.; Maurer, M.S.; Ambardekar, A.V.; Bullock-Palmer, R.P.; Chang, P.P.; Eisen, H.J.; Nair, A.P.; Nativi-Nicolau, J.; Ruberg, F.L. American Heart Association Heart Failure and Transplantation Committee of the Council on Clinical Cardiology. Cardiac Amyloidosis: Evolving Diagnosis and Management: A Scientific Statement From the American Heart Association. Circulation 2020, 142, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Papathanasiou, M.; Carpinteiro, A.; Rischpler, C.; Hagenacker, T.; Rassaf, T.; Luedike, P. Diagnosing cardiac amyloidosis in every-day practice: A practical guide for the cardiologist. Int. J. Cardiol. Heart Vasc. 2020, 28, 100519. [Google Scholar] [CrossRef]
- Maurer, M.S.; Bokhari, S.; Damy, T.; Dorbala, S.; Drachman, B.M.; Fontana, M.; Grogan, M.; Kristen, A.V.; Lousada, I.; Nativi-Nicolau, J.; et al. Expert Consensus Recommendations for the Suspicion and Diagnosis of Transthyretin Cardiac Amyloidosis. Circ. Heart Fail. 2019, 12, e006075. [Google Scholar] [CrossRef] [PubMed]
- Chatzantonis, G.; Bietenbeck, M.; Elsanhoury, A.; Tschöpe, C.; Pieske, B.; Tauscher, G.; Vietheer, J.; Shomanova, Z.; Mahrholdt, H.; Rolf, A.; et al. Diagnostic value of cardiovascular magnetic resonance in comparison to endomyocardial biopsy in cardiac amyloidosis: A multi-centre study. Clin. Res. Cardiol. 2021, 110, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.K.M.; Brizneda, M.V.; Kwon, D.H.; Popovic, Z.B.; Flamm, S.D.; Hanna, M.; Griffin, B.P.; Xu, B. Reference Ranges, Diagnostic and Prognostic Utility of Native T1 Mapping and Extracellular Volume for Cardiac Amyloidosis: A Meta-Analysis. J. Magn. Reason. Imaging 2021, 53, 1458–1468. [Google Scholar] [CrossRef]
- Weinreb, J.C.; Rodby, R.A.; Yee, J.; Wang, C.L.; Fine, D.; McDonald, R.J.; Perazella, M.A.; Dillman, J.R.; Davenport, M.S. Use of Intravenous Gadolinium-based Contrast Media in Patients with Kidney Disease: Consensus Statements from the American College of Radiology and the National Kidney Foundation. Kidney Med. 2021, 3, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Naharro, A.; Treibel, T.A.; Abdel-Gadir, A.; Bulluck, H.; Zumbo, G.; Knight, D.S.; Kotecha, T.; Francis, R.; Hutt, D.F.; Rezk, T.; et al. Magnetic Resonance in Transthyretin Cardiac Amyloidosis. J. Am. Coll. Cardiol. 2017, 70, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Binder, C.; Duca, F.; Binder, T.; Rettl, R.; Dachs, T.M.; Seirer, B.; Camuz Ligios, L.; Dusik, F.; Capelle, C.; Qin, H.; et al. Prognostic implications of pericardial and pleural effusion in patients with cardiac amyloidosis. Clin. Res. Cardiol. 2021, 110, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Germain, P.; Vardazaryan, A.; Padoy, N.; Labani, A.; Roy, C.; Schindler, T.H.; El Ghannudi, S. Classification of Cardiomyopathies from MR Cine Images Using Convolutional Neural Network with Transfer Learning. Diagnostics 2021, 11, 1554. [Google Scholar] [CrossRef] [PubMed]
- Martini, N.; Aimo, A.; Barison, A.; Della Latta, D.; Vergaro, G.; Aquaro, G.D.; Ripoli, A.; Emdin, M.; Chiappino, D. Deep learning to diagnose cardiac amyloidosis from cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2020, 22, 84. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef]
- Zhou, H.; Li, L.; Liu, Z.; Zhao, K.; Chen, X.; Lu, M.; Yin, G.; Song, L.; Zhao, S.; Zheng, H.; et al. Deep learning algorithm to improve hypertrophic cardiomyopathy mutation prediction using cardiac cine images. Eur. Radiol. 2021, 31, 3931–3940. [Google Scholar] [CrossRef] [PubMed]
- Mongan, J.; Moy, L.; Kahn, C.E., Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Network for Large Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2019, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Zhang, C.J.P.; Jiang, B.; Chen, J.; Song, J.; Liu, Z.; He, Z.; Wong, S.Y.; Fang, P.H.; Ming, W.K. Artificial Intelligence Versus Clinicians in Disease Diagnosis: Systematic Review. JMIR Med. Inform. 2019, 7, e10010. [Google Scholar] [CrossRef] [Green Version]
- Venkadesh, K.V.; Setio, A.A.A.; Schreuder, A.; Scholten, E.T.; Chung, K.; Wille, M.M.W.; Saghir, Z.; van Ginneken, B.; Prokop, M.; Jacobs, C. Deep Learning for Malignancy Risk Estimation of Pulmonary Nodules Detected at Low-Dose Screening CT. Radiology 2021, 300, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki Ben Zadok, O.; Vaturi, M.; Vaxman, I.; Iakobishvili, Z.; Rhurman-Shahar, N.; Kornowski, R.; Hamdan, A. Differences in the characteristics and contemporary cardiac outcomes of patients with light-chain versus transthyretin cardiac amyloidosis. PLoS ONE 2021, 16, e0255487. [Google Scholar] [CrossRef] [PubMed]
- Quarta, C.C.; Solomon, S.D.; Uraizee, I.; Kruger, J.; Longhi, S.; Ferlito, M.; Gagliardi, C.; Milandri, A.; Rapezzi, C.; Falk, R.H. Left ventricular structure and function in transthyretin-related versus light-chain cardiac amyloidosis. Circulation 2014, 129, 1840–1849. [Google Scholar] [CrossRef] [Green Version]
- Dungu, J.N.; Valencia, O.; Pinney, J.H.; Gibbs, S.D.; Rowczenio, D.; Gilbertson, J.A.; Lachmann, H.J.; Wechalekar, A.; Gillmore, J.D.; Whelan, C.J.; et al. CMR-based differentiation of AL and ATTR cardiac amyloidosis. JACC Cardiovasc. Imaging 2014, 7, 133–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlochower, A.; Chow, D.S.; Chang, P.; Khatri, D.; Boockvar, J.A.; Filippi, C.G. Deep Learning AI Applications in the Imaging of Glioma. Top. Magn. Reason. Imaging 2020, 29, 115–121. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Zhu, Q.; Liu, H.; Chang, L.; Duan, S.; Dou, W.; Li, S.; Ye, J. Differentiating Benign from Malignant Renal Tumors Using T2- and Diffusion-Weighted Images: A Comparison of Deep Learning and Radiomics Models Versus Assessment from Radiologists. J. Magn. Reason. Imaging 2021. [Google Scholar] [CrossRef]
- Neisius, U.; El-Rewaidy, H.; Nakamori, S.; Rodriguez, J.; Manning, W.J.; Nezafat, R. Radiomic Analysis of Myocardial Native T1 Imaging Discriminates Between Hypertensive Heart Disease and Hypertrophic Cardiomyopathy. JACC Cardiovasc. Imaging 2019, 12, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Larroza, A.; Materka, A.; López-Lereu, M.P.; Monmeneu, J.V.; Bodí, V.; Moratal, D. Differentiation between acute and chronic myocardial infarction by means of texture analysis of late gadolinium enhancement and cine cardiac magnetic resonance imaging. Eur. J. Radiol. 2017, 92, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, D.; Liu, X.; Fan, X.; Wang, K.; Li, S.; Zhang, Z.; Ma, K.; Qian, T.; Jiang, T.; et al. Molecular subtyping of diffuse gliomas using magnetic resonance imaging: Comparison and correlation between radiomics and deep learning. Eur. Radiol. 2021, 52. [Google Scholar] [CrossRef] [PubMed]
- Truhn, D.; Schrading, S.; Haarburger, C.; Schneider, H.; Merhof, D.; Kuhl, C. Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-enhancing Lesions at Multiparametric Breast MRI. Radiology 2019, 290, 290–297. [Google Scholar] [CrossRef]
- Sun, Q.; Lin, X.; Zhao, Y.; Li, L.; Yan, K.; Liang, D.; Sun, D.; Li, Z.C. Deep Learning vs. Radiomics for Predicting Axillary Lymph Node Metastasis of Breast Cancer Using Ultrasound Images: Don’t Forget the Peritumoral Region. Front. Oncol. 2020, 10, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Chang, M.; Wang, R.; Xi, I.L.; Chang, K.; Huang, R.Y.; Vallières, M.; Habibollahi, P.; Dagli, M.S.; Palmer, M.; et al. Deep Learning Based on MRI for Differentiation of Low- and High-Grade in Low-Stage Renal Cell Carcinoma. J. Magn. Reason. Imaging 2020, 52, 1542–1549. [Google Scholar] [CrossRef]
- Yuan, Y.; Qin, W.; Buyyounouski, M.; Ibragimov, B.; Hancock, S.; Han, B.; Xing, L. Prostate cancer classification with multiparametric MRI transfer learning model. Med. Phys. 2019, 46, 756–765. [Google Scholar] [CrossRef]
- Chacko, L.; Boldrini, M.; Martone, R.; Law, S.; Martinez-Naharrro, A.; Hutt, D.F.; Kotecha, T.; Patel, R.K.; Razvi, Y.; Rezk, T.; et al. Cardiac Magnetic Resonance-Derived Extracellular Volume Mapping for the Quantification of Hepatic and Splenic Amyloid. Circ. Cardiovasc. Imaging 2021, 14, e012506. [Google Scholar] [CrossRef] [PubMed]
- Misumi, Y.; Ueda, M.; Yamashita, T.; Masuda, T.; Kinoshita, Y.; Tasaki, M.; Nagase, T.; Ando, Y. Novel screening for transthyretin amyloidosis by using fat ultrasonography. Ann. Neurol. 2017, 81, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Slivnick, J.A.; Tong, M.S.; Nagaraja, H.N.; Elamin, M.B.; Wallner, A.; O’Brien, A.; Raman, S.V.; Zareba, K.M. Novel Cardiovascular Magnetic Resonance Model to Distinguish Immunoglobulin Light Chain From Transthyretin Cardiac Amyloidosis. JACC Cardiovasc. Imaging 2021, 14, 302–304. [Google Scholar] [CrossRef]
| Amyloidosis | LVH | p | |
|---|---|---|---|
| N patients | 119 | 122 | |
| Age (years) | 74.65 ± 9.53 | 59.50 ± 14.34 | 0.0001 |
| Sex (F/M) | 31/88 | 39/83 | 0.31 |
| Weight (kg) | 70.80 ± 15.16 | 82.95 ± 20.50 | 0.0001 |
| Height (m) | 169.9 ± 8.84 | 170.36 ± 10.05 | 0.78 |
| BSA (m2) | 1.84 ± 0.22 | 2.00 ± 0.27 | <0.0001 |
| IVS (mm) | 18.11 ± 3.54 | 18.38 ± 3.54 | 0.56 |
| LVMI (g/m2) | 115.96 ± 29.08 | 116.58 ± 31.43 | 0.88 |
| LVDVI (mL/m2) | 69.88 ± 22.21 | 74.51 ± 20.82 | 0.36 |
| LVEF (%) | 58.96 ± 10.93 | 67.33 ± 12.18 | <0.0001 |
| LA surface (cm2) | 31.55 ± 5.23 | 25.47 ± 5.96 | 0.0002 |
| Systolic time (ms) | 321 ± 39 | 332 ± 40 | 0.095 |
| T1 (ms) | 1138.5 ± 48.1 | 1038.0 ± 56.2 | <0.0001 |
| ECV (%) | 53.97 ± 11.17 | 26.89 ± 4.00 | <0.0001 |
| N long axis frames/patient | 2.24 ± 0.93 | 2.22 ± 0.94 | 0.93 |
| N short axis frames/patient | 3.41 ± 1.45 | 3.59 ± 1.27 | 0.49 |
| N frames/patient | 5.68 ± 1.85 | 5.47 ± 1.81 | 0.58 |
| N frame post-gadolinium | 171/676 | 167/667 | 0.96 |
| N patient with pericard. | 54 (45%) | 27 (22%) | 0.00013 |
| N patients with pleural. | 45 (38%) | 10 (8%) | 0.00001 |
| N patients with both. | 24 (20%) | 3 (2.5%) | 0.00001 |
| Frame-Based | Patient-Based | |||
|---|---|---|---|---|
| Input Shape | Accuracy | ROC AUC | Accuracy | ROC AUC |
| 160 × 160/D + S | 0.759 | 0.836 [0.786–0.878] | 0.812 | 0.937 [0.828–0.987] |
| 160 × 160/D | 0.760 (ns) | 0.820 (ns) [0.769–0.864] | 0.833 (ns) | 0.918 (ns) [0.802–0.978] |
| 160 × 160/S | 0.733 (ns) | 0.801 (0.04) [0.749–0.848] | 0.833 (ns) | 0.890 (ns) [0.767–0.962] |
| 256 × 256/D + S | 0.710 (ns) | 0.790 (0.03) [0.735–0.836] | 0.771 (ns) | 0.803 (0.02) [0.663–0.904] |
| 224 × 224/D + S | 0.728 (ns) | 0.823 (ns) [0.772–0.867] | 0.812 (ns) | 0.852 (ns) [0.720–0.938] |
| 128 × 128/D + S | 0.740 (ns) | 0.808 (ns) [0.756–0.853] | 0.812 (ns) | 0.922 (ns) [0.807–0.979] |
| Epicardial ROI | 0.722 (ns) | 0.787 (0.01) [0.762–0.810] | 0.791 (ns) | 0.888 (ns) [0.839–0.927] |
| Myocard. ROI | 0.662 (0.05) | 0.719 (0.01) [0.693–0.745] | 0.714 (ns) | 0.814 (0.03) [0.756–0.863] |
| Frame-Based | Patient-Based | |||||
|---|---|---|---|---|---|---|
| Metric | Accur. | Sensitiv. Specific. | ROC AUC | Accur. | Sensitiv. Specific. | ROC AUC |
| CNN | 0.746 | 77.0 71.0 | 0.824 [0.770–0.869] | 0.825 | 85.7 77.6 | 0.895 [0.816–0.948] |
| Read 1 | 0.585 (0.001) | 66.4 50.85 | 0.570 [0.506–0.632] (0.0001) | 0.629 (0.004) | 67.4 58.8 | 0.654 [0.550–0.747] (0.001) |
| Read 2 | 0.623 (0.005) | 69.6 54.5 | 0.623 [0.560–0.684] (0.0001) | 0.649 (0.009) | 69.6 60.8 | 0.712 [0.611–0.799] (0.0002) |
| Read 3 | 0.585 (0.001) | 66.4 50.9 | 0.587 [0.523–0.649] (0.0001) | 0.660 (0.013) | 71.1 61.5 | 0.731 [0.631–0.816] (0.002) |
| Read (avg) | 0.605 (0.0008) | 69.2 52.7 | 0.630 [0.567–0.690] (0.0001) | 0.660 (0.008) | 72.1 61.1 | 0.727 [0.627–0.813] (0.002) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Germain, P.; Vardazaryan, A.; Padoy, N.; Labani, A.; Roy, C.; Schindler, T.H.; El Ghannudi, S. Deep Learning Supplants Visual Analysis by Experienced Operators for the Diagnosis of Cardiac Amyloidosis by Cine-CMR. Diagnostics 2022, 12, 69. https://doi.org/10.3390/diagnostics12010069
Germain P, Vardazaryan A, Padoy N, Labani A, Roy C, Schindler TH, El Ghannudi S. Deep Learning Supplants Visual Analysis by Experienced Operators for the Diagnosis of Cardiac Amyloidosis by Cine-CMR. Diagnostics. 2022; 12(1):69. https://doi.org/10.3390/diagnostics12010069
Chicago/Turabian StyleGermain, Philippe, Armine Vardazaryan, Nicolas Padoy, Aissam Labani, Catherine Roy, Thomas Hellmut Schindler, and Soraya El Ghannudi. 2022. "Deep Learning Supplants Visual Analysis by Experienced Operators for the Diagnosis of Cardiac Amyloidosis by Cine-CMR" Diagnostics 12, no. 1: 69. https://doi.org/10.3390/diagnostics12010069
APA StyleGermain, P., Vardazaryan, A., Padoy, N., Labani, A., Roy, C., Schindler, T. H., & El Ghannudi, S. (2022). Deep Learning Supplants Visual Analysis by Experienced Operators for the Diagnosis of Cardiac Amyloidosis by Cine-CMR. Diagnostics, 12(1), 69. https://doi.org/10.3390/diagnostics12010069





