Association of the Timeless Gene with Prognosis and Clinical Characteristics of Human Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Oncomine Database Analysis
2.2. GEPIA Database
2.3. UALCAN Database
2.4. Human Protein Atlas (HPA) Database
2.5. Kaplan–Meier (K-M) Plotter
2.6. cBioPortal for Cancer Genomics
2.7. Database for Annotation, Visualization and Integrated Discovery (DAVID)
2.8. Protein–Protein Interaction Analysis
2.9. A549 and NCI-H226 Cell Culture and Transfection
2.10. Reverse Transcription-Quantitative (RT-q)PCR
2.11. Western Blot Analysis
2.12. Statistical Analysis
3. Results
3.1. Expression Level of the Timeless Gene in Human Lung Cancer
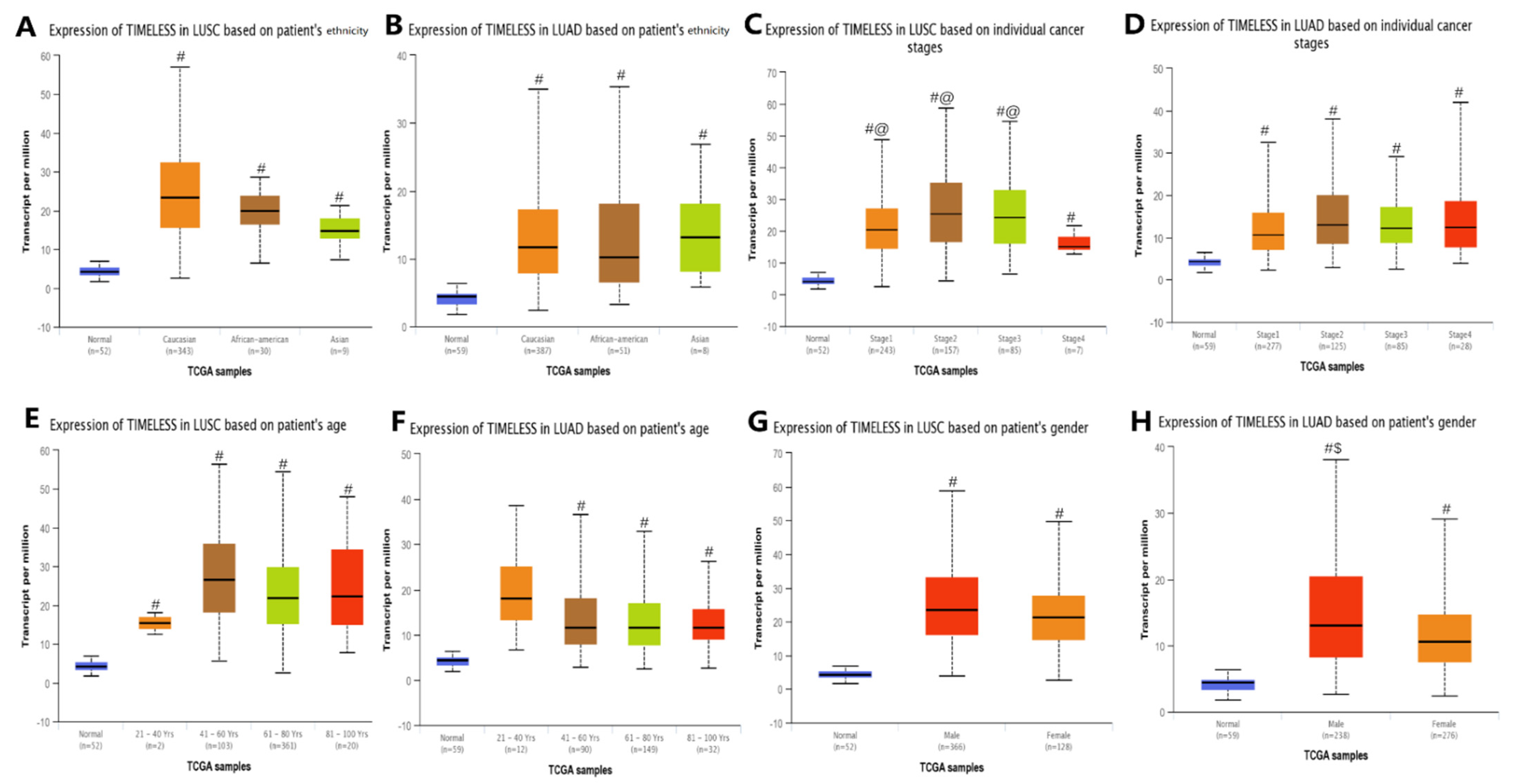
3.2. Expression of Timeless Gene and Clinicopathological Parameters of Patients with Lung Cancer
3.3. Genetic Mutations, Correlations and Networks Analysis of the Timeless Gene in Patients with Lung Cancer
3.4. ESPL1 Expression and Prognostic Potential in Patients with Lung Cancer
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Sangoram, A.M.; Saez, L.; Antoch, M.P.; Gekakis, N.; Staknis, D.; Whiteley, A.; Fruechte, E.M.; Vitaterna, M.H.; Shimomura, K.; King, D.P.; et al. Mammalian circadian autoregulatory loop: A timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 1998, 21, 1101–1113. [Google Scholar] [CrossRef]
- Mao, Y.; Fu, A.; Leaderer, D.; Zheng, T.; Chen, K.; Zhu, Y. Potential cancer-related role of circadian gene TIMELESS suggested by expression profiling and in vitro analyses. BMC Cancer 2013, 13, 498. [Google Scholar] [CrossRef] [PubMed]
- Mazzoccoli, G.; Colangelo, T.; Panza, A.; Rubino, R.; Tiberio, C.; Palumbo, O.; Carella, M.; Trombetta, D.; Gentile, A.; Tavano, F.; et al. Analysis of clock gene-miRNA correlation networks reveals candidate drivers in colorectal cancer. Oncotarget 2016, 7, 45444–45461. [Google Scholar] [CrossRef]
- Xian, H.; Li, Y.; Zou, B.; Chen, Y.; Yin, H.; Li, X.; Pan, Y. Identification of TIMELESS and RORA as key clock molecules of non-small cell lung cancer and the comprehensive analysis. BMC Cancer 2022, 22, 107. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, X.; Yang, H.; Zhao, H.; Xia, B.; You, Y. The expression of the circadian gene TIMELESS in non-small-cell lung cancer and its clinical significance. Int. J. Clin. Exp. Pathol. 2020, 13, 2297–2304. [Google Scholar]
- Bhattacharjee, A.; Richards, W.G.; Staunton, J.; Li, C.; Monti, S.; Vasa, P.; Ladd, C.; Beheshti, J.; Bueno, R.; Gillette, M.; et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA 2001, 98, 13790–13795. [Google Scholar] [CrossRef]
- Hou, J.; Aerts, J.; den Hamer, B.; van Ijcken, W.; den Bakker, M.; Riegman, P.; van der Leest, C.; van der Spek, P.; Foekens, J.A.; Hoogsteden, H.C.; et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef]
- Wachi, S.; Yoneda, K.; Wu, R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics 2005, 21, 4205–4208. [Google Scholar] [CrossRef]
- Selamat, S.A.; Chung, B.S.; Girard, L.; Zhang, W.; Zhang, Y.; Campan, M.; Siegmund, K.D.; Koss, M.N.; Hagen, J.A.; Lam, W.L.; et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012, 22, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y.; Shiraishi, K.; Iwakawa, R.; Furuta, K.; Tsuta, K.; Shibata, T.; Yamamoto, S.; et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012, 72, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Stearman, R.S.; Dwyer-Nield, L.; Zerbe, L.; Blaine, S.A.; Chan, Z.; Bunn, P.A., Jr.; Johnson, G.L.; Hirsch, F.R.; Merrick, D.T.; Franklin, W.A.; et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 2005, 167, 1763–1775. [Google Scholar] [CrossRef]
- Liu, J.Y.; Qian, C.Y.; Gao, Y.F.; Chen, J.; Zhou, H.H.; Yin, J.Y. Association between DNA mismatch repair gene polymorphisms and platinum-based chemotherapy toxicity in non-small cell lung cancer patients. Chin. J. Cancer 2017, 36, 12. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Heiden, M.G.V.; Jacks, T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016, 24, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Banach, E.; Pawlak, J.; Kapelski, P.; Szczepankiewicz, A.; Rajewska-Rager, A.; Skibinska, M.; Czerski, P.; Twarowska-Hauser, J.; Dmitrzak-Weglarz, M. Clock genes polymorphisms in male bipolar patients with comorbid alcohol abuse. J. Affect. Disord. 2018, 241, 142–146. [Google Scholar] [CrossRef]
- Jiang, Y.D.; Yuan, X.; Bai, Y.L.; Wang, G.Y.; Zhou, W.W.; Zhu, Z.R. Knockdown of timeless Disrupts the Circadian Behavioral Rhythms in Laodelphax striatellus (Hemiptera: Delphacidae). Environ Entomol. 2018, 47, 1216–1225. [Google Scholar] [CrossRef]
- Lam, V.H.; Li, Y.H.; Liu, X.; Murphy, K.A.; Diehl, J.S.; Kwok, R.S.; Chiu, J.C. CK1α Collaborates with DOUBLETIME to Regulate PERIOD Function in the Drosophila Circadian Clock. J. Neurosci. 2018, 38, 10631–10643. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Wu, X.; Ma, D.; Wu, J.; Wang, L.; Jiang, Y.; Fei, Y.; Zhu, C.; Tan, R.; et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018, 563, 131–136. [Google Scholar] [CrossRef]
- Young, L.M.; Marzio, A.; Perez-Duran, P.; Reid, D.A.; Meredith, D.N.; Roberti, D.; Star, A.; Rothenberg, E.; Ueberheide, B.; Pagano, M. TIMELESS Forms a Complex with PARP1 Distinct from Its Complex with TIPIN and Plays a Role in the DNA Damage Response. Cell Rep. 2015, 13, 451–459. [Google Scholar] [CrossRef]
- Ao, Y.; Zhao, Q.; Yang, K.; Zheng, G.; Lv, X.; Su, X. A role for the clock period circadian regulator 2 gene in regulating the clock gene network in human oral squamous cell carcinoma cells. Oncol. Lett. 2018, 15, 4185–4192. [Google Scholar] [CrossRef]
- Chi, L.; Zou, Y.; Qin, L.; Ma, W.; Hao, Y.; Tang, Y.; Luo, R.; Wu, Z. TIMELESS contributes to the progression of breast cancer through activation of MYC. Breast Cancer Res. 2017, 19, 53. [Google Scholar] [CrossRef]
- Reszka, E.; Przybek, M. Circadian Genes in Breast Cancer. Adv. Clin. Chem. 2016, 75, 53–70. [Google Scholar]
- Zhang, J.S.; Yuan, P.; Yan, Z.Y.; Lu, R.; Li, B.; E Geng, X.; Mu, J.; Zhang, H.X. Timeless promotes the proliferation of hepatocellular carcinoma cell by reprogramming of glucose metabolism. Zhonghua Zhong Liu Za Zhi 2018, 40, 499–505. [Google Scholar]
- Zhang, W.; He, W.; Shi, Y.; Zhao, J.; Liu, S.; Zhang, F.; Yang, J.; Xie, C.; Zhang, Y. Aberrant TIMELESS expression is associated with poor clinical survival and lymph node metastasis in early-stage cervical carcinoma. Int. J. Oncol. 2017, 50, 173–184. [Google Scholar] [CrossRef]
- Fu, A.; Leaderer, D.; Zheng, T.; Hoffman, A.E.; Stevens, R.G.; Zhu, Y. Genetic and epigenetic associations of circadian gene TIMELESS and breast cancer risk. Mol. Carcinog. 2012, 51, 923–929. [Google Scholar] [CrossRef]
- Lesicka, M.; Jablonska, E.; Wieczorek, E.; Seroczyńska, B.; Siekierzycka, A.; Skokowski, J.; Kalinowski, L.; Wasowicz, W.; Reszka, E. Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS ONE 2018, 13, e0199622. [Google Scholar] [CrossRef]
- Reszka, E.; Przybek, M.; Muurlink, O.; Pepłonska, B. Circadian gene variants and breast cancer. Cancer Lett. 2017, 390, 137–145. [Google Scholar] [CrossRef]
- Yoshida, K.; Sato, M.; Hase, T.; Elshazley, M.; Yamashita, R.; Usami, N.; Taniguchi, T.; Yokoi, K.; Nakamura, S.; Kondo, M.; et al. TIMELESS is overexpressed in lung cancer and its expression correlates with poor patient survival. Cancer Sci. 2013, 104, 171–177. [Google Scholar] [CrossRef]
- Sun, Y.; Kucej, M.; Fan, H.Y.; Yu, H.; Sun, Q.Y.; Zou, H. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell 2009, 137, 123–132. [Google Scholar] [CrossRef]
- Finetti, P.; Guille, A.; Adelaide, J.; Birnbaum, D.; Chaffanet, M.; Bertucci, F. ESPL1 is a candidate oncogene of luminal B breast cancers. Breast Cancer Res. Treat. 2014, 147, 51–59. [Google Scholar] [CrossRef]
- Zhang, C.; Min, L.; Zhang, L.; Ma, Y.; Yang, Y.; Shou, C. Combined analysis identifies six genes correlated with augmented malignancy from non-small cell to small cell lung cancer. Tumour Biol. 2016, 37, 2193–2207. [Google Scholar] [CrossRef] [PubMed]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, J.; Chen, J.; Wang, J.; Xia, Z.; Jia, Y. Association of the Timeless Gene with Prognosis and Clinical Characteristics of Human Lung Cancer. Diagnostics 2022, 12, 2681. https://doi.org/10.3390/diagnostics12112681
Ye J, Chen J, Wang J, Xia Z, Jia Y. Association of the Timeless Gene with Prognosis and Clinical Characteristics of Human Lung Cancer. Diagnostics. 2022; 12(11):2681. https://doi.org/10.3390/diagnostics12112681
Chicago/Turabian StyleYe, Jishi, Jingli Chen, Juan Wang, Zhongyuan Xia, and Yifan Jia. 2022. "Association of the Timeless Gene with Prognosis and Clinical Characteristics of Human Lung Cancer" Diagnostics 12, no. 11: 2681. https://doi.org/10.3390/diagnostics12112681
APA StyleYe, J., Chen, J., Wang, J., Xia, Z., & Jia, Y. (2022). Association of the Timeless Gene with Prognosis and Clinical Characteristics of Human Lung Cancer. Diagnostics, 12(11), 2681. https://doi.org/10.3390/diagnostics12112681





