Improved Detection of Herpesviruses from Diluted Vitreous Specimens Using Hydrogel Particles
Abstract
1. Introduction
2. Materials and Methods
2.1. Control Viral Panels
2.2. Sampling of Porcine Vitreous
2.3. Comparison of Three Different Nanotrap Particle Types
2.4. Preparation of Simulated Diluted Vitreous Samples
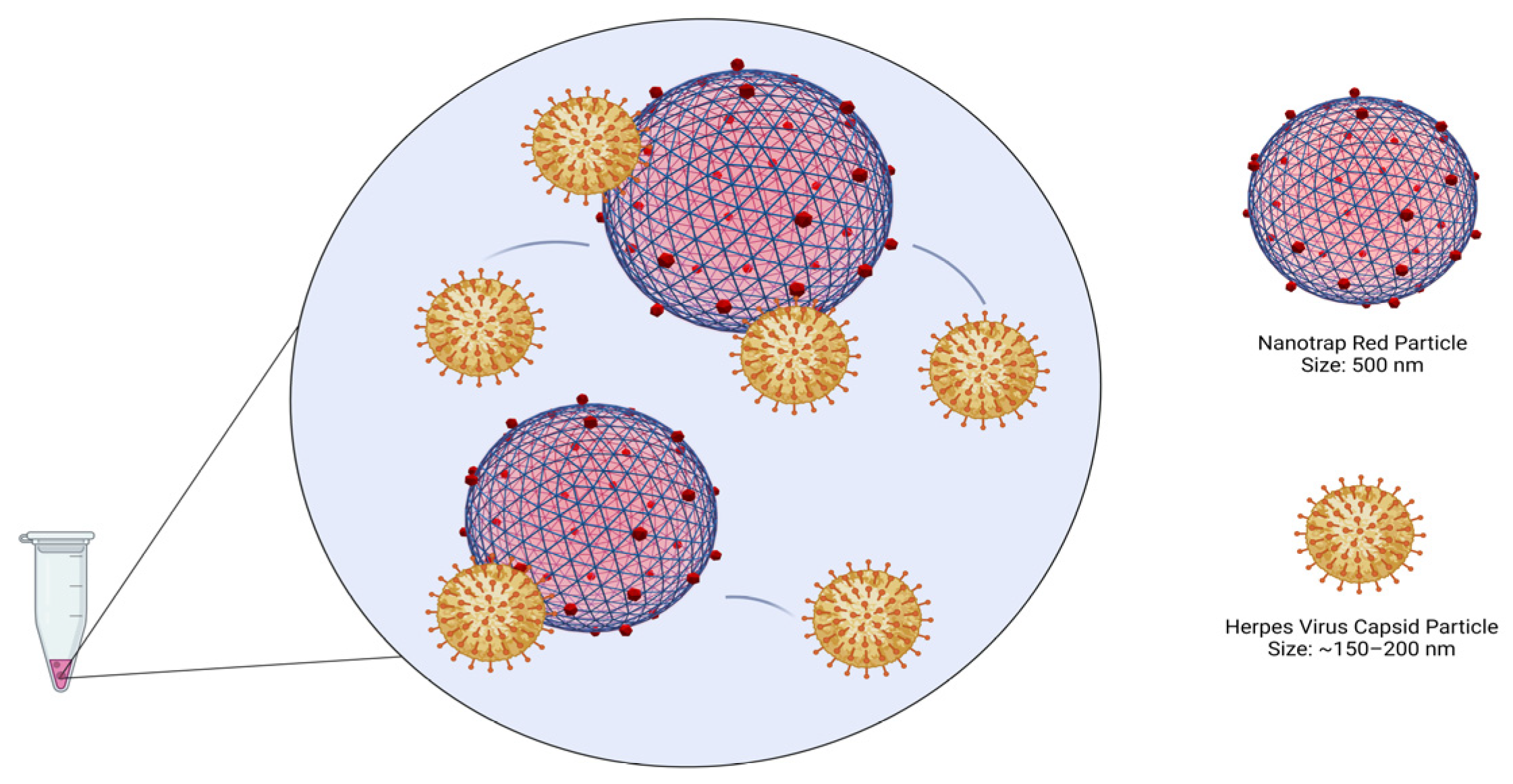
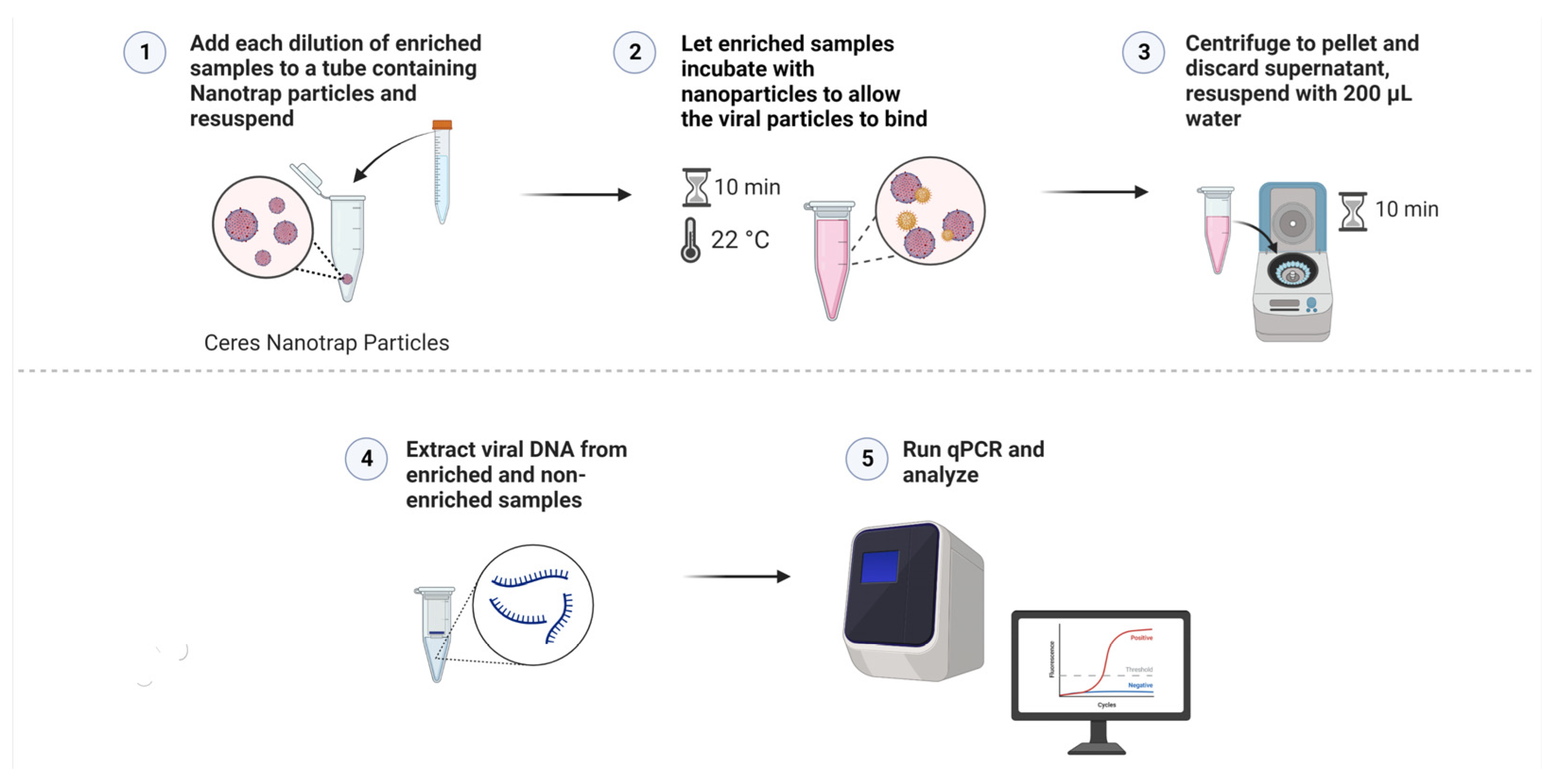
2.5. Viral Capture and Concentration with Nanotrap Red Particles
2.6. Construction of Reference Curves
2.7. Statistical Analyses
3. Results
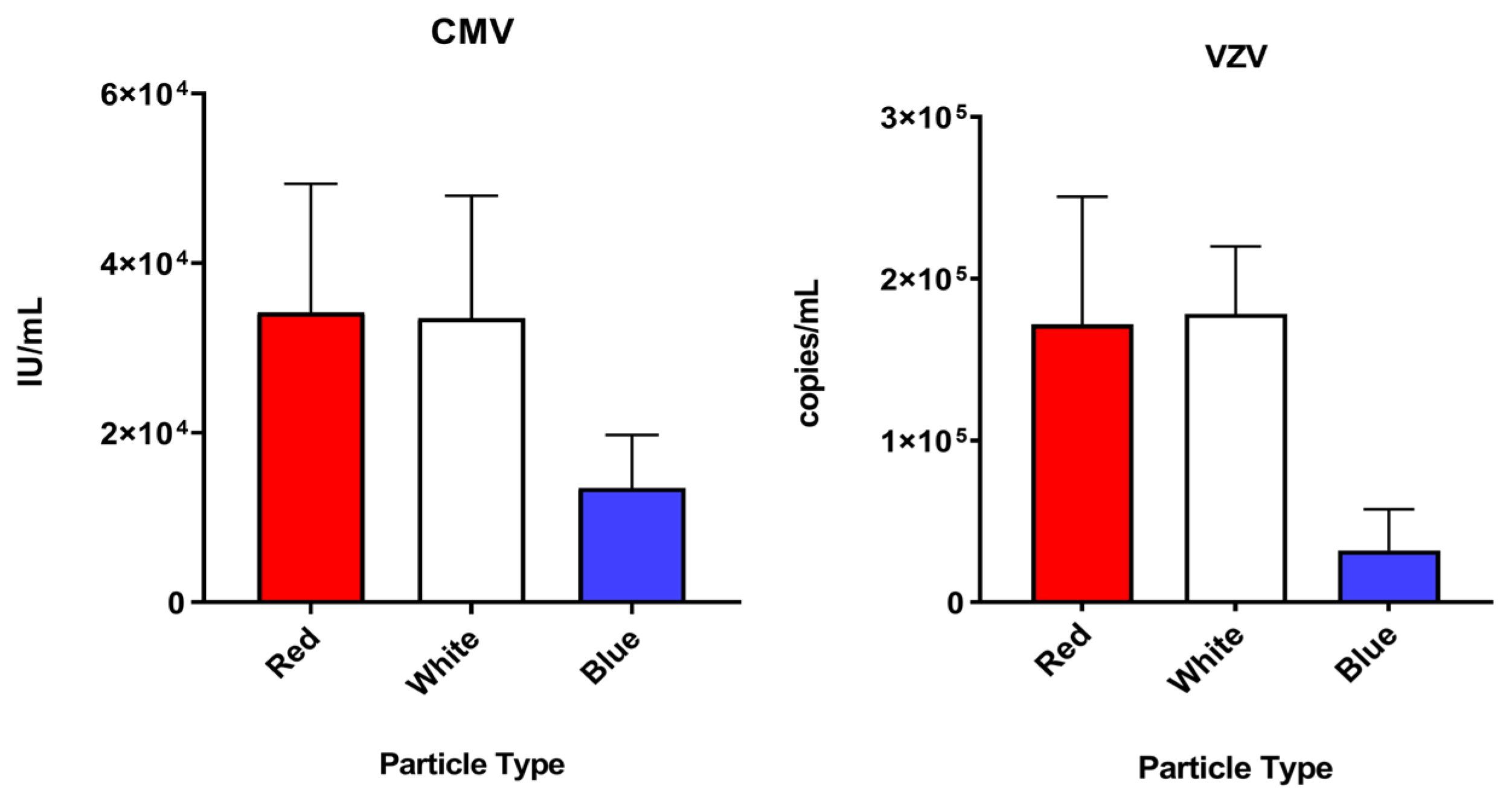
3.1. Performance of Each Particle Type
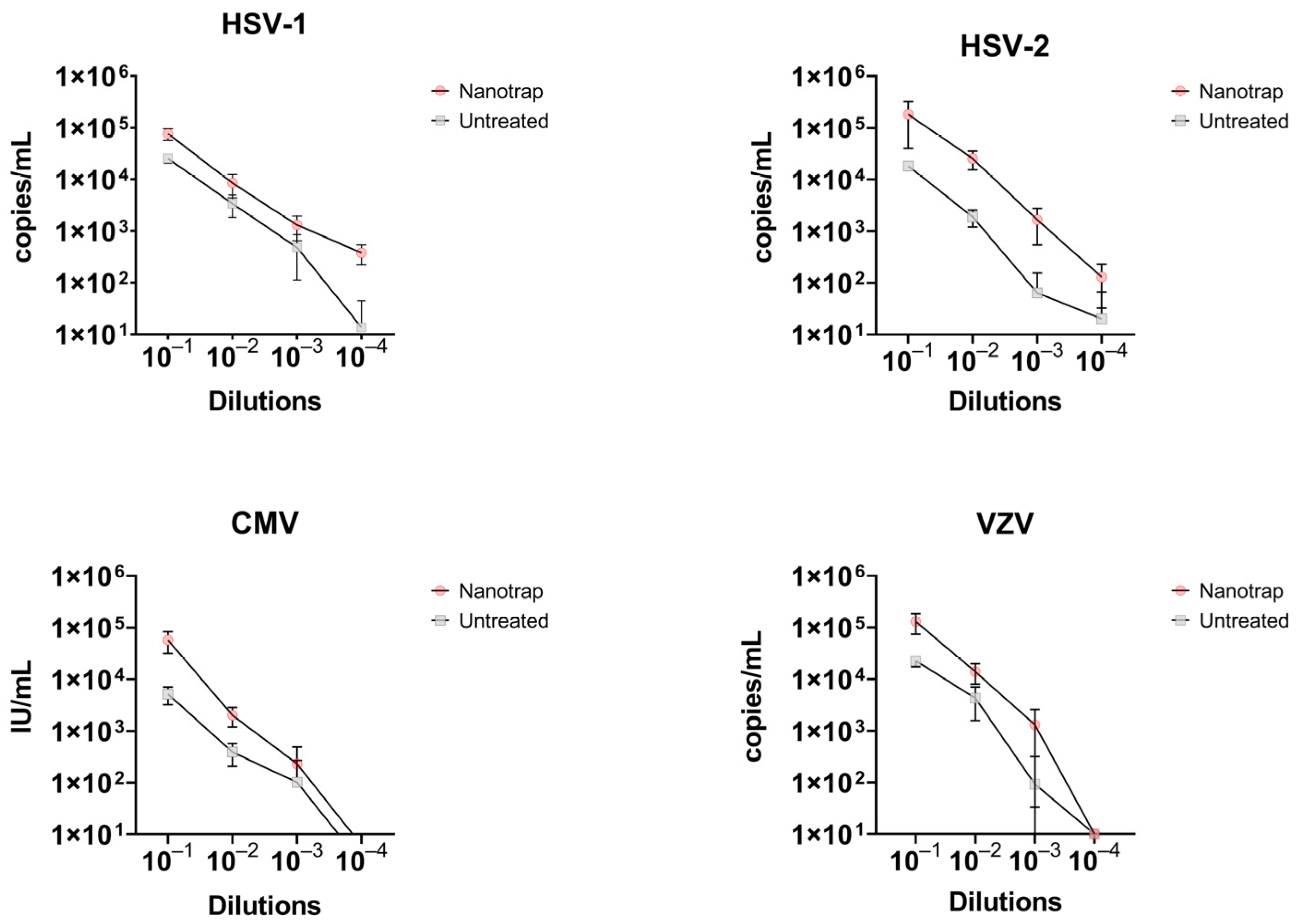
3.2. Comparison of Enriched vs. Non-Enriched Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Suttorp-Schulten, M.S.; Rothova, A. The possible impact of uveitis in blindness: A literature survey. Br. J. Ophthalmol. 1996, 80, 844–848. [Google Scholar] [CrossRef]
- Lin, P. Infectious Uveitis. Curr. Ophthalmol. Rep. 2015, 3, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Agarwal, A.; Mahendradas, P.; Lee, C.S.; Gupta, V.; Pavesio, C.E.; Agrawal, R. Viral posterior uveitis. Surv. Ophthalmol. 2017, 62, 404–445. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.; Konana, V.K.; Ganesh, S.K.; Patnaik, G.; Chan, N.S.W.; Chee, S.P.; Sobolewska, B.; Zierhut, M. Viral anterior uveitis. Indian J. Ophthalmol. 2020, 68, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Port, A.D.; Orlin, A.; Kiss, S.; Patel, S.; D’Amico, D.J.; Gupta, M.P. Cytomegalovirus Retinitis: A Review. J. Ocul. Pharmacol. Ther. 2017, 33, 224–234. [Google Scholar] [CrossRef]
- De Groot-Mijnes, J.D.F.; Rothova, A.; Van Loon, A.M.; Schuller, M.; Ten Dam-Van Loon, N.H.; De Boer, J.H.; Schuurman, R.; Weersink, A.J.L. Polymerase Chain Reaction and Goldmann-Witmer Coefficient Analysis Are Complimentary for the Diagnosis of Infectious Uveitis. Am. J. Ophthalmol. 2006, 141, 313–318. [Google Scholar] [CrossRef]
- Shoughy, S.S.; Alkatan, H.M.; Al-Abdullah, A.A.; El-Khani, A.; de Groot-Mijnes, J.D.; Tabbara, K.F. Polymerase chain reaction in unilateral cases of presumed viral anterior uveitis. Clin. Ophthalmol. 2015, 9, 2325–2328. [Google Scholar] [CrossRef]
- Relvas, L.J.M.; Antoun, J.; de Groot-Mijnes, J.D.F.; Motulsky, E.; Ten Dam-Van Loon, N.H.; Makhoul, D.; Willermain, F.; Caspers, L. Diagnosis of Cytomegalovirus Anterior Uveitis in Two European Referral Centers. Ocul. Immunol. Inflamm. 2018, 26, 116–121. [Google Scholar] [CrossRef]
- Scheepers, M.A.; Lecuona, K.A.; Rogers, G.; Bunce, C.; Corcoran, C.; Michaelides, M. The value of routine polymerase chain reaction analysis of intraocular fluid specimens in the diagnosis of infectious posterior uveitis. Sci. World J. 2013, 2013, 545149. [Google Scholar] [CrossRef]
- Santos, H.; Ferracioli-Oda, E.; Barbosa, T.S.; Otani, C.S.V.; Tanaka, T.; Silva, L.; Lopes, G.O.; Doi, A.; Hirata, C.E.; Yamamoto, J.H. Usefulness of aqueous and vitreous humor analysis in infectious uveitis. Clinics 2020, 75, e1498. [Google Scholar] [CrossRef]
- Luchini, A.; Geho, D.H.; Bishop, B.; Tran, D.; Xia, C.; Dufour, R.L.; Jones, C.D.; Espina, V.; Patanarut, A.; Zhou, W.; et al. Smart hydrogel particles: Biomarker harvesting: One-step affinity purification, size exclusion, and protection against degradation. Nano Lett. 2008, 8, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Shafagati, N.; Fite, K.; Patanarut, A.; Baer, A.; Pinkham, C.; An, S.; Foote, B.; Lepene, B.; Kehn-Hall, K. Enhanced detection of respiratory pathogens with nanotrap particles. Virulence 2016, 7, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Carey, B.D.; Callahan, V.; Lee, J.H.; Bracci, N.; Patnaik, A.; Smith, A.K.; Narayanan, A.; Lepene, B.; Kehn-Hall, K. Use of Nanotrap particles for the capture and enrichment of Zika, chikungunya and dengue viruses in urine. PLoS ONE 2020, 15, e0227058. [Google Scholar] [CrossRef] [PubMed]
- Shafagati, N.; Narayanan, A.; Baer, A.; Fite, K.; Pinkham, C.; Bailey, C.; Kashanchi, F.; Lepene, B.; Kehn-Hall, K. The use of NanoTrap particles as a sample enrichment method to enhance the detection of Rift Valley Fever Virus. PLoS Negl. Trop. Dis. 2013, 7, e2296. [Google Scholar] [CrossRef]
- Jaworski, E.; Saifuddin, M.; Sampey, G.; Shafagati, N.; Van Duyne, R.; Iordanskiy, S.; Kehn-Hall, K.; Liotta, L.; Petricoin, E., 3rd; Young, M.; et al. The use of Nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS ONE 2014, 9, e96778. [Google Scholar] [CrossRef]
- Bispo, P.J.M.; Davoudi, S.; Sahm, M.L.; Ren, A.; Miller, J.; Romano, J.; Sobrin, L.; Gilmore, M.S. Rapid Detection and Identification of Uveitis Pathogens by Qualitative Multiplex Real-Time PCR. Investig. Ophthalmol. Vis. Sci. 2018, 59, 582–589. [Google Scholar] [CrossRef]
- Harper, T.W.; Miller, D.; Schiffman, J.C.; Davis, J.L. Polymerase chain reaction analysis of aqueous and vitreous specimens in the diagnosis of posterior segment infectious uveitis. Am. J. Ophthalmol. 2009, 147, 140–147.e2. [Google Scholar] [CrossRef]
- de Groot-Mijnes, J.D.; de Visser, L.; Zuurveen, S.; Martinus, R.A.; Völker, R.; ten Dam-van Loon, N.H.; de Boer, J.H.; Postma, G.; de Groot, R.J.; van Loon, A.M.; et al. Identification of new pathogens in the intraocular fluid of patients with uveitis. Am. J. Ophthalmol. 2010, 150, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, L.L.; Gibler, T.M.; Van Gelder, R.N. Real-time quantitative polymerase chain reaction diagnosis of infectious posterior uveitis. Arch. Ophthalmol. 2002, 120, 1534–1539. [Google Scholar] [CrossRef]
- Sugita, S.; Shimizu, N.; Watanabe, K.; Mizukami, M.; Morio, T.; Sugamoto, Y.; Mochizuki, M. Use of multiplex PCR and real-time PCR to detect human herpes virus genome in ocular fluids of patients with uveitis. Br. J. Ophthalmol. 2008, 92, 928–932. [Google Scholar] [CrossRef]
- Nakano, S.; Tomaru, Y.; Kubota, T.; Takase, H.; Mochizuki, M.; Shimizu, N.; Sugita, S. Evaluation of a Multiplex Strip PCR Test for Infectious Uveitis: A Prospective Multicenter Study. Am. J. Ophthalmol. 2020, 213, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, M.P.; Bansal, R.; Gupta, A.; Ram, J.; Ratho, R.K. Development and evaluation of multiplex real-time PCR for diagnosis of HSV-1, VZV, CMV, and Toxoplasma gondii in patients with infectious uveitis. Diagn. Microbiol. Infect. Dis. 2017, 89, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Matos, K.; Muccioli, C.; Belfort Junior, R.; Rizzo, L.V. Correlation between clinical diagnosis and PCR analysis of serum, aqueous, and vitreous samples in patients with inflammatory eye disease. Arq. Bras. Oftalmol. 2007, 70, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.M.; Chandler, D.; Short, G.A.; Margolis, T.P. Polymerase chain reaction—Based assays of vitreous samples for the diagnosis of viral retinitis: Use in diagnostic dilemmas. Ophthalmology 1998, 105, 37–45. [Google Scholar] [CrossRef] [PubMed]


| Nanotrap Red | Nanotrap White | Nanotrap Blue | ||||
|---|---|---|---|---|---|---|
| Mean * | SD | Mean | SD | Mean | SD | |
| CMV | 34,157.8 | 15,194.1 | 33,502.2 | 144,512 | 13,454.8 | 6271.9 |
| HSV-1 | 110,954.9 | 76,777.7 | 81,236.4 | 14,932 | 57,353.6 | 13,377.8 |
| HSV-2 | 26,255.2 | 7395.3 | 14,253.5 | 7496.8 | 12,822.3 | 9859.6 |
| VZV | 171,683.8 | 79,047.8 | 178,146.5 | 41,784.4 | 31,991.6 | 25,461.7 |
| Enriched | Non-Enriched | Fold Increase | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean * | SD | Mean Ct | Mean | SD | Mean Ct | |||
| HSV-1 | 104 | 76,252.5 | 19,527.4 | 23.96 | 25,351.6 | 4671.6 | 25.58 | 3.0 |
| 103 | 8484.1 | 4075.9 | 27.31 | 3397.3 | 1559.0 | 28.68 | 2.5 | |
| 102 | 1303.1 | 657.7 | 30.16 | 480.5 | 367.3 | 32.25 | 2.7 | |
| 101 | 380.2 | 160.3 | 31.95 | 0 § | - | >40 | - | |
| HSV-2 | 104 | 276,249.6 | 22,581.5 | 22.67 | 18,241.9 | 1953.7 | 26.60 | 15.1 |
| 103 | 25,437.6 | 10,008.6 | 26.20 | 1863.7 | 679.14 | 29.98 | 13.6 | |
| 102 | 1638.9 | 1105.1 | 30.62 | 61.9 | 89.6 | 37.46 | 26.5 | |
| 101 | 126.9 | 95.5 | 34.20 | 0 | - | >40 | - | |
| CMV | 104 | 57,465.4 | 25,857.7 | 23.71 | 5162.7 | 1948.1 | 27.42 | 11.1 |
| 103 | 2022.9 | 825.4 | 28.86 | 390.5 | 182.3 | 31.41 | 5.2 | |
| 102 | 230.9 | 258.13 | 33.76 | 101 | 170.6 | 37.12 | 2.3 | |
| 101 | 0 | - | >40 | 0 | - | >40 | ||
| VZV | 104 | 130,149.9 | 56,026.1 | 29.06 | 22,669.5 | 5395.1 | 31.48 | 5.7 |
| 103 | 13,950.6 | 5956.7 | 32.28 | 4313.8 | 2735.0 | 34.34 | 3.2 | |
| 102 | 1303.9 | 1270.9 | 36.91 | 0 | - | >40 | - | |
| 101 | 0 | - | >40 | 0 | - | >40 | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belanger, N.L.; Barbero, R.; Barclay, R.; Lepene, B.; Sobrin, L.; Bispo, P.J.M. Improved Detection of Herpesviruses from Diluted Vitreous Specimens Using Hydrogel Particles. Diagnostics 2022, 12, 3016. https://doi.org/10.3390/diagnostics12123016
Belanger NL, Barbero R, Barclay R, Lepene B, Sobrin L, Bispo PJM. Improved Detection of Herpesviruses from Diluted Vitreous Specimens Using Hydrogel Particles. Diagnostics. 2022; 12(12):3016. https://doi.org/10.3390/diagnostics12123016
Chicago/Turabian StyleBelanger, Nicole L., Robbie Barbero, Robert Barclay, Benjamin Lepene, Lucia Sobrin, and Paulo J. M. Bispo. 2022. "Improved Detection of Herpesviruses from Diluted Vitreous Specimens Using Hydrogel Particles" Diagnostics 12, no. 12: 3016. https://doi.org/10.3390/diagnostics12123016
APA StyleBelanger, N. L., Barbero, R., Barclay, R., Lepene, B., Sobrin, L., & Bispo, P. J. M. (2022). Improved Detection of Herpesviruses from Diluted Vitreous Specimens Using Hydrogel Particles. Diagnostics, 12(12), 3016. https://doi.org/10.3390/diagnostics12123016







