Expression Analysis of Five Different Long Non-Coding Ribonucleic Acids in Nonsmall-Cell Lung Carcinoma Tumor and Tumor-Derived Exosomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. GEO Analysis
2.3. Extracellular Vesicle (EV) Isolation from Plasma
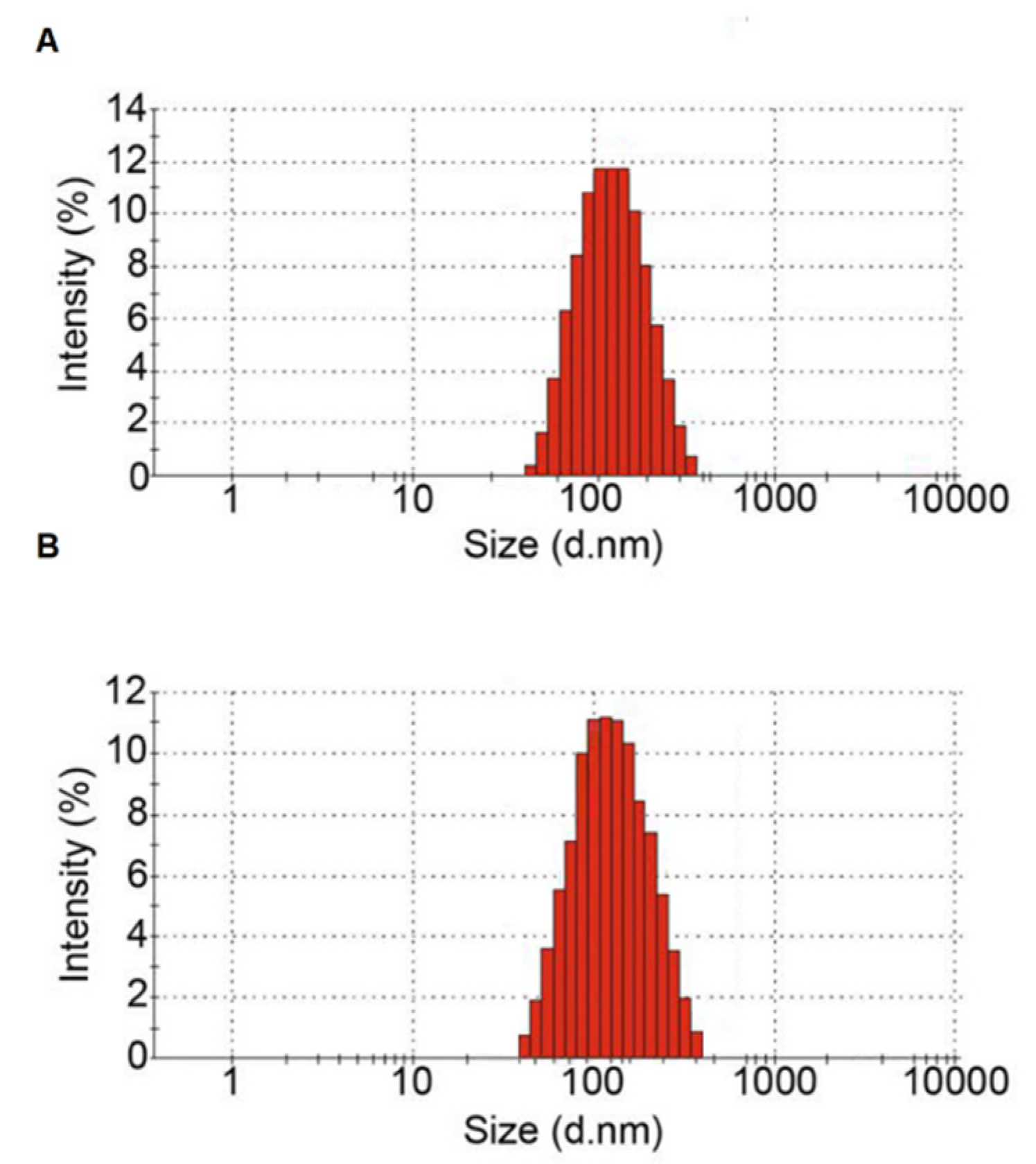
2.4. Measuring Particle Size of Isolated EVs
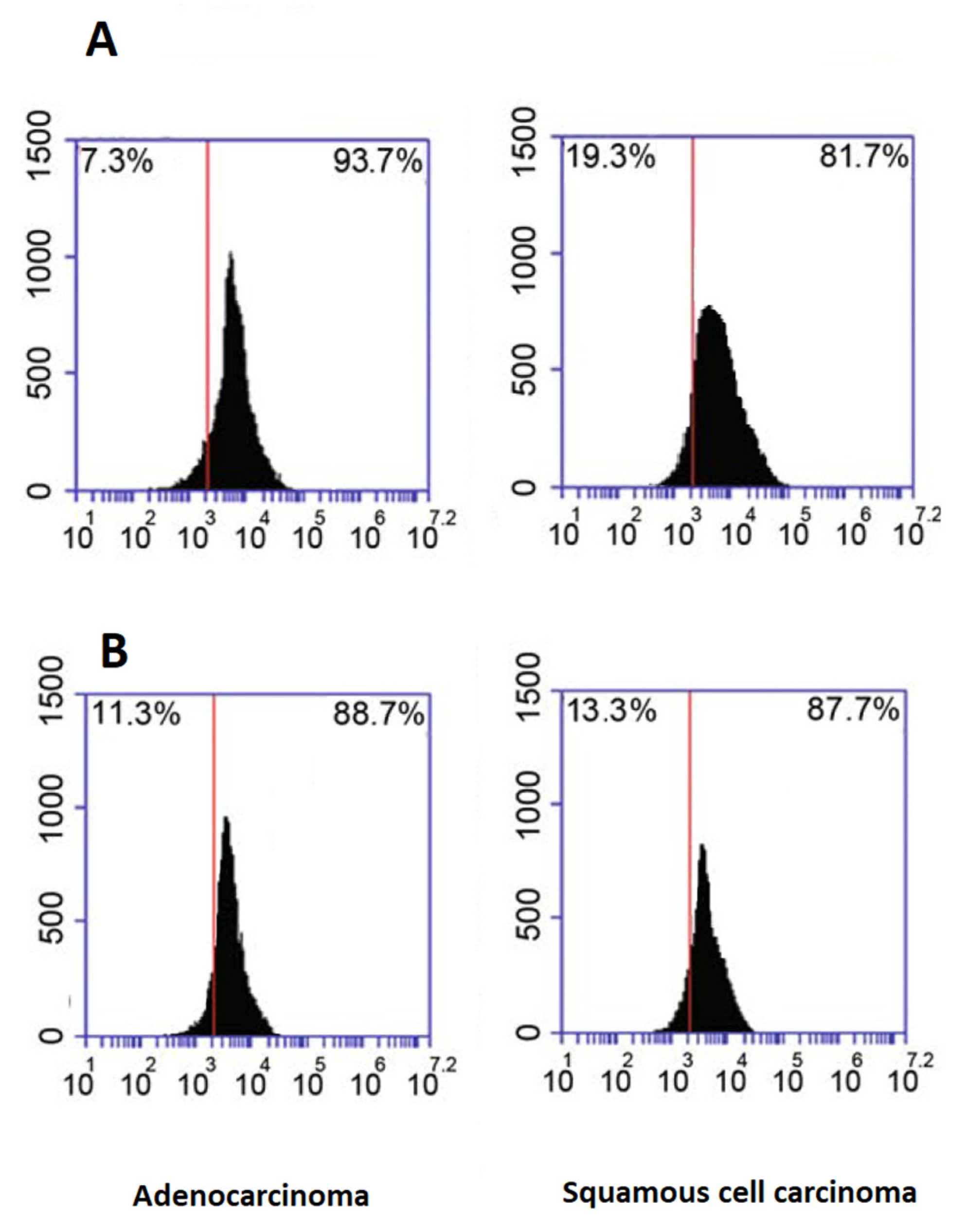
2.5. Flow Cytometry Analysis of Exosome Marker Proteins
2.6. RNA Extraction
2.7. cDNA Synthesis and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
2.8. Statistical Analysis
3. Results
3.1. Characterization of Plasma Isolated EVs
3.2. The Relative Expressions of lncRNAs
3.3. The Correlation between lncRNA Expressions among Different Tissues and the Area under the Curve
3.4. The Association between lncRNA Expression and Clinicopathological Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global epidemiology of lung cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.-H.; Luo, L.; Wampfler, J.A.; Wang, Y.; Liu, D.; Chen, Y.-M.; Adjei, A.A.; Midthun, D.E.; Yang, P. 5-year overall survival in patients with lung cancer eligible or ineligible for screening according to US Preventive Services Task Force criteria: A prospective, observational cohort study. Lancet Oncol. 2019, 20, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Bach, P.B.; Mirkin, J.N.; Oliver, T.K.; Azzoli, C.G.; Berry, D.A.; Brawley, O.W.; Byers, T.; Colditz, G.A.; Gould, M.K.; Jett, J.R. Benefits and harms of CT screening for lung cancer: A systematic review. Jama 2012, 307, 2418–2429. [Google Scholar] [CrossRef] [PubMed]
- Salmaninejad, A.; Estiar, M.A.; Gill, R.K.; Shih, J.H.; Hewitt, S.; Jeon, H.-S.; Fukuoka, J.; Shilo, K.; Shakoori, A.; Jen, J. Expression analysis of p16, c-Myc, and mSin3A in non-small cell lung cancer by computer aided scoring and analysis (CASA). Clin. Lab. 2015, 61, 549–559. [Google Scholar] [CrossRef]
- Jazieh, K.; Khorrami, M.; Saad, A.; Gad, M.; Gupta, A.; Patil, P.; Viswanathan, V.S.; Rajiah, P.; Nock, C.J.; Gilkey, M. Novel imaging biomarkers predict outcomes in stage III unresectable non-small cell lung cancer treated with chemoradiation and durvalumab. J. Immunother. Cancer 2022, 10, e003778. [Google Scholar] [CrossRef]
- Masaoutis, C.; Mihailidou, C.; Tsourouflis, G.; Theocharis, S. Exosomes in lung cancer diagnosis and treatment. From the translating research into future clinical practice. Biochimie 2018, 151, 27–36. [Google Scholar] [CrossRef]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Xiao, H.; Zheng, Y.; Ma, L.; Tian, L.; Sun, Q. Clinically-relevant ABC transporter for anti-cancer drug resistance. Front. Pharmacol. 2021, 12, 648407. [Google Scholar] [CrossRef]
- Entezari, M.; Ghanbarirad, M.; Taheriazam, A.; Sadrkhanloo, M.; Zabolian, A.; Goharrizi, M.A.S.B.; Hushmandi, K.; Aref, A.R.; Ashrafizadeh, M.; Zarrabi, A. Long non-coding RNAs and exosomal lncRNAs: Potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed. Pharmacother. 2022, 150, 112963. [Google Scholar] [CrossRef]
- Chaput, N.; Théry, C. Exosomes: Immune properties and potential clinical implementations. In Seminars in Immunopathology; Springer: Berlin, Germany, 2011. [Google Scholar]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Non-coding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Wang, Y.; Chen, D.; Liu, J.; Jiao, W. Potential clinical application of lncRNAs in non-small cell lung cancer. OncoTargets Ther. 2018, 11, 8045–8052. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ma, H.; Chen, H.; Zhang, Z.; Xue, Q. LncRNA in tumorigenesis of non-small-cell lung cancer: From bench to bedside. Cell Death Discov. 2022, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Nosrati, R.; Salmaninejad, A.; Dehghani, S.; Shahryari, A.; Saberi, A. Organ-specific metastasis of breast cancer: Molecular and cellular mechanisms underlying lung metastasis. Cell. Oncol. 2018, 41, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Temraz, S.; Nasr, R.; Mukherji, D.; Kreidieh, F.; Shamseddine, A. Liquid biopsy derived circulating tumor cells and circulating tumor DNA as novel biomarkers in hepatocellular carcinoma. Expert Rev. Mol. Diagn. 2022, 22, 507–518. [Google Scholar] [CrossRef]
- Xu, N.; Zhu, H.; Shen, D.; Jiang, M.; Zhi, Q.; Ling, C. The emerging lncRNAs as novel regulators in lung cancer. Int. J. Clin. Exp. Med. 2017, 10, 179–203. [Google Scholar]
- Liu, Y.; Zhao, F.; Tan, F.; Tang, L.; Du, Z.; Mou, J.; Zhou, G.; Yuan, C. HNF1A-AS1: A tumor-associated long non-coding RNA. Curr. Pharm. Des. 2022, 28, 1720–1729. [Google Scholar]
- Liu, M.; Zhang, H.; Li, Y.; Wang, R.; Li, Y.; Zhang, H.; Ren, D.; Liu, H.; Kang, C.; Chen, J. HOTAIR, a long noncoding RNA, is a marker of abnormal cell cycle regulation in lung cancer. Cancer Sci. 2018, 109, 2717–2733. [Google Scholar] [CrossRef]
- Li, C.; Qin, F.; Hu, F.; Xu, H.; Sun, G.; Han, G.; Wang, T.; Guo, M. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018, 8, 1–21. [Google Scholar] [CrossRef]
- Abedpoor, N.; Taghian, F.; Hajibabaie, F. Cross Brain–Gut Analysis Highlighted Hub Genes and LncRNA Networks Differentially Modified During Leucine Consumption and Endurance Exercise in Mice with Depression-Like Behaviors. Mol. Neurobiol. 2022, 59, 4106–4123. [Google Scholar] [CrossRef]
- Yousefi, M.; Bahrami, T.; Salmaninejad, A.; Nosrati, R.; Ghaffari, P.; Ghaffari, S.H. Lung cancer-associated brain metastasis: Molecular mechanisms and therapeutic options. Cell. Oncol. 2017, 40, 419–441. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.L. Lung Cancer: Epidemiology and Screening. Surg. Clin. 2022, 102, 335–344. [Google Scholar]
- Yousefi, M.; Ghaffari, P.; Nosrati, R.; Dehghani, S.; Salmaninejad, A.; Abarghan, Y.J.; Ghaffari, S.H. Prognostic and therapeutic significance of circulating tumor cells in patients with lung cancer. Cell. Oncol. 2020, 43, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, F.; Chen, A.; Hua, Q.; Gao, W. Costimulatory molecule-related lncRNA model as a potential prognostic biomarker in non-small cell lung cancer. Cancer Med. 2022. [Google Scholar] [CrossRef]
- Whiteside, E.J.; Seim, I.; Pauli, J.P.; O’Keeffe, A.J.; Thomas, P.B.; Carter, S.L.; Walpole, C.M.; Fung, J.N.; Josh, P.; Herington, A.C. Identification of a long non-coding RNA gene, growth hormone secretagogue receptor opposite strand, which stimulates cell migration in non-small cell lung cancer cell lines. Int. J. Oncol. 2013, 43, 566–574. [Google Scholar] [CrossRef]
- Thomas, P.B. Characterisation of the Long Noncoding RNA GHSROS in Prostate and Breast Cancer. Ph.D. Thesis, Queensland University of Technology, Brisbane City, Australia, 2018. [Google Scholar]
- Chen, S.; Li, P.; Xiao, B.; Guo, J. Long Noncoding RNA HMlincRNA717 and AC130710 Have Been Officially Named as Gastric Cancer Associated Transcript 2 (GACAT2) and GACAT3, Respectively; Springer: Berlin, Germany, 2014; Volume 35, pp. 8351–8352. [Google Scholar]
- Sun, X.; Cao, G.; Cao, Y.; Jiang, X.; Li, X.; Ye, X.; Wang, D.; Yan, S. Association of LncRNA HMlincRNA717 with prognosis in pancreatic cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2230–2234. [Google Scholar]
- Xie, X.; Liu, H.-T.; Mei, J.; Ding, F.-B.; Xiao, H.-B.; Hu, F.-Q.; Hu, R.; Wang, M.-S. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. Int. J. Clin. Exp. Pathol. 2014, 7, 8881. [Google Scholar]
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Ji, C.; Zhang, L.; Di, Y.; Lou, W.; Zhang, X.; Xu, J. Novel Implications of Exosomes and lncRNAs in the Diagnosis and Treatment of Pancreatic Cancer. In Novel Implications of Exosomes in Diagnosis and Treatment of Cancer and Infectious Diseases; IntechOpen: London, UK, 2017; pp. 3–33. [Google Scholar]
- Xie, Z.; Chen, X.; Li, J.; Guo, Y.; Li, H.; Pan, X.; Jiang, J.; Liu, H.; Wu, B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016, 7, 25408–25419. [Google Scholar] [CrossRef]
- Wang, Y.-L.; Liu, L.-C.; Hung, Y.; Chen, C.-J.; Lin, Y.-Z.; Wu, W.-R.; Wang, S.-C. Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast 2019, 46, 64–69. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, L.; Deng, G.; Ding, Y.; Bi, K.; Jin, H.; Shu, J.; Yang, J.; Deng, H.; Wang, Z. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci. China Life Sci. 2020, 63, 1265–1268. [Google Scholar] [CrossRef] [PubMed]
- Castellano, J.J.; Navarro, A.; Viñolas, N.; Marrades, R.M.; Moises, J.; Cordeiro, A.; Saco, A.; Muñoz, C.; Fuster, D.; Molins, L. LincRNA-p21 impacts prognosis in resected non–small cell lung Cancer patients through angiogenesis regulation. J. Thorac. Oncol. 2016, 11, 2173–2182. [Google Scholar] [CrossRef] [PubMed]
- Ao, X.; Jiang, M.; Zhou, J.; Liang, H.; Xia, H.; Chen, G. lincRNA-p21 inhibits the progression of non-small cell lung cancer via targeting miR-17-5p. Oncol. Rep. 2019, 41, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Işın, M.; Uysaler, E.; Özgür, E.; Köseoğlu, H.; Şanlı, Ö.; Yücel, Ö.B.; Gezer, U.; Dalay, N. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front. Genet. 2015, 6, 168. [Google Scholar] [PubMed]
- Gezer, U.; Özgür, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, Y.; Li, Q.; Duan, P. lncRNA HNF1A-AS1 modulates non–small cell lung cancer progression by targeting miR-149-5p/Cdk6. J. Cell. Biochem. 2019, 120, 18736–18750. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liang, T.; Li, C.; Li, Y.; Jin, S.; Liu, Y. Long non-coding RNA HNF1A-AS1 up-regulation in non-small cell lung cancer correlates to poor survival. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4858–4863. [Google Scholar]
- Luo, X.; Wei, J.; Yang, F.-L.; Pang, X.-X.; Shi, F.; Wei, Y.-X.; Liao, B.-Y.; Wang, J.-L. Exosomal lncRNA HNF1A-AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA-34b/TUFT1 axis. Cancer Cell Int. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Słomka, A.; Kornek, M.; Cho, W.C. Small Extracellular Vesicles and Their Involvement in Cancer Resistance: An Up-to-Date Review. Cells 2022, 11, 2913. [Google Scholar] [CrossRef]


| Variable | Number | |
|---|---|---|
| Gender N (%) | Female | 54 (32.1) |
| Male | 114 (67.9) | |
| Mean age ± SD | 62.22 ± 6.59 | |
| Subtype N (%) | Adenocarcinoma | 116(69) |
| Squamous cell carcinoma | 52 (31) | |
| Stage N (%) | 1 | 47 (28) |
| 2 | 71 (42.3) | |
| 3 | 50 (29.8) | |
| Body mass index N (%) | <30 | 110 (65.5) |
| >30 | 58 (34.5) | |
| Smoking | 147 (87.5) | |
| Sample | ADC | SQCC |
|---|---|---|
| Average of particle diameter (nm) | 121.2 | 118.6 |
| Polydispersity index (PDI) a | 0.247 | 0.231 |
| Major peak of particle diameter (nm) | 171.4 | 159.8 |
| Percentage of 20–200 nm diameter (%) | 81.4 | 79.8 |
| Genes | Target Sample | Compared To | Relative Expression | Std. Error | p Value | 95% Confidence Interval | |
|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||
| GHSROS | tumor | normal | 3.41071 * | 0.43620 | 0.000 | 2.3629 | 4.4585 |
| exosome | 0.08333 | 0.43620 | 1.000 | −0.9644 | 1.1311 | ||
| exosome | normal | 3.32738 * | 0.43620 | 0.000 | 2.2796 | 4.3752 | |
| tumor | −0.08333 | 0.43620 | 1.000 | −1.1311 | 0.9644 | ||
| HMlincRNA717 | tumor | normal | −5.07143 * | 0.43958 | 0.000 | −6.1273 | −4.0155 |
| exosome | −0.00595 | 0.43958 | 1.000 | −1.0618 | 1.0499 | ||
| exosome | normal | −5.06548 * | 0.43958 | 0.000 | −6.1214 | −4.0096 | |
| tumor | 0.00595 | 0.43958 | 1.000 | −1.0499 | 1.0618 | ||
| HNF1A-AS1 | tumor | normal | 2.79167 * | 0.44683 | 0.000 | 1.7184 | 3.8650 |
| exosome | 0.23810 | 0.44683 | 1.000 | −0.8352 | 1.3114 | ||
| exosome | normal | 2.55357 * | 0.44683 | 0.000 | 1.4803 | 3.6269 | |
| tumor | −0.23810 | 0.44683 | 1.000 | −1.3114 | 0.8352 | ||
| HOTAIR | tumor | normal | 3.02381 * | 0.68063 | 0.000 | 1.3889 | 4.6587 |
| exosome | 0.01190 | 0.68063 | 1.000 | −1.6230 | 1.6468 | ||
| exosome | normal | 3.01190 * | 0.68063 | 0.000 | 1.3770 | 4.6468 | |
| tumor | −0.01190 | 0.68063 | 1.000 | −1.6468 | 1.6230 | ||
| LINCRNA-p21 | tumor | normal | −5.16667 * | 0.42132 | 0.000 | −6.1787 | −4.1546 |
| exosome | 0.42262 | 0.42132 | 0.949 | −0.5894 | 1.4347 | ||
| exosome | normal | −5.58929 * | 0.42132 | 0.000 | −6.6013 | −4.5772 | |
| tumor | −0.42262 | 0.42132 | 0.949 | −1.4347 | 0.5894 | ||
| Tumor vs. ANCT | Exosome vs. ANCT | |||||
|---|---|---|---|---|---|---|
| Gene | Standard Error | Odds Ratio | p Value | Standard Error | Odds Ratio | p Value |
| GHSROS | 0.050 | 1.308 | 0.000 | 0.048 | 1.234 | 0.000 |
| HMlincRNA717 | 0.045 | 0.771 | 0.000 | 0.048 | 0.759 | 0.000 |
| HNF1A-AS1 | 0.045 | 1.151 | 0.000 | 0.045 | 1.169 | 0.000 |
| HOTAIR | 0.029 | 1.085 | 0.000 | 0.029 | 1.101 | 0.000 |
| LINCRNA-p21 | 0.059 | 0.689 | 0.000 | 0.055 | 0.672 | 0.000 |
| Specificity (%) | Sensitivity (%) | Negative Predictive Value (%) | Positive Predictive Value (%) | AUC (%) | Cut off Point | Gene | |
|---|---|---|---|---|---|---|---|
| 76.4 | 71.5 | 68.4 | 76.1 | 73.4 | >3.51 | GHSROS | Tumor compared to ANCT |
| 71.3 | 72.3 | 70.1 | 70.4 | 67.7 | >2.59 | HNF1A-AS1 | |
| 89.8 | 91.6 | 87.7 | 91.1 | 85.4 | >2.71 | HOTAIR | |
| 75.6 | 76.6 | 77.6 | 84.3 | 73.9 | >−4.42 | HMlincRNA717 | |
| 82 | 81.4 | 80.4 | 79.2 | 68.1 | >−5.02 | LINCRNA-p21 | |
| 94.4 | 93.7 | 91.3 | 93.8 | 93.7 | Combined LncRNAs | ||
| 77.3 | 74.1 | 71.1 | 76.2 | 75.1 | >3.12 | GHSROS | Exosome compared to ANCT |
| 70.8 | 73.4 | 70.2 | 69.8 | 68.1 | >2.09 | HNF1A-AS1 | |
| 88.6 | 92.2 | 88.4 | 92.1 | 87.2 | >1.87 | HOTAIR | |
| 74.7 | 78.7 | 75.6 | 86.7 | 72.2 | >−4.86 | HMlincRNA717 | |
| 83.4 | 82.1 | 81.3 | 76.4 | 68.5 | >−6.67 | LINCRNA-p21 | |
| 95.7 | 94.2 | 94.1 | 93.6 | 94.7 | Combined LncRNAs |
| GHSROS | |||||||||
| Tumoral: ANCT (N) | Tumoral: Exosome (N) | Exosome: ANCT (N) | |||||||
| Down | Up | p Value | Down | Up | p Value | Down | Up | p Value | |
| Age | |||||||||
| <60 years | 15 | 45 | 0.893 | 27 | 33 | 0.612 | 15 | 45 | 0.794 |
| >60 years | 26 | 82 | 53 | 55 | 29 | 79 | |||
| Gender | |||||||||
| Female | 12 | 42 | 0.65 | 25 | 29 | 0.813 | 16 | 38 | 0.485 |
| Male | 29 | 85 | 55 | 59 | 28 | 86 | |||
| Subtype | |||||||||
| SCC | 13 | 39 | 0.904 | 22 | 30 | 0.356 | 13 | 39 | 0.814 |
| Adeno | 28 | 88 | 58 | 58 | 31 | 85 | |||
| Stage | |||||||||
| 1 | 14 | 33 | 20 | 27 | 18 | 29 | |||
| 2 | 15 | 56 | 0.561 | 33 | 38 | 0.513 | 14 | 57 | 0.073 |
| 3 | 12 | 38 | 27 | 23 | 12 | 38 | |||
| Smoking | |||||||||
| Yes | 40 | 107 | 0.005 | 75 | 72 | 0.599 | 75 | 72 | 0.484 |
| No | 14 | 7 | 12 | 9 | 9 | 12 | |||
| BMI | |||||||||
| <30 | 28 | 82 | 0.663 | 56 | 54 | 0.24 | 30 | 80 | 0.66 |
| >30 | 13 | 45 | 24 | 34 | 14 | 44 | |||
| HMlincRNA717 | |||||||||
| Tumoral: ANCT (N) | Tumoral: Exosome (N) | Exosome: ANCT (N) | |||||||
| Down | Up | p Value | Down | Up | p Value | Down | Up | p Value | |
| Age | |||||||||
| <60 years | 46 | 14 | 0.978 | 29 | 31 | 0.258 | 50 | 10 | 0.657 |
| >60 years | 83 | 25 | 62 | 46 | 87 | 21 | |||
| Gender | |||||||||
| Female | 45 | 9 | 0.167 | 33 | 21 | 0.214 | 61 | 13 | 0.196 |
| Male | 84 | 30 | 58 | 56 | 96 | 18 | |||
| Subtype | |||||||||
| SCC | 35 | 17 | 0.051 | 23 | 29 | 0.084 | 43 | 9 | 0.798 |
| Adeno | 94 | 22 | 68 | 48 | 94 | 22 | |||
| Stage | |||||||||
| 1 | 38 | 9 | 25 | 22 | 39 | 8 | |||
| 2 | 56 | 15 | 0.387 | 42 | 29 | 0.473 | 62 | 9 | 0.097 |
| 3 | 35 | 15 | 24 | 26 | 36 | 14 | |||
| Smoking | |||||||||
| Yes | 64 | 83 | 0.637 | 73 | 74 | 0.861 | 70 | 77 | 0.102 |
| No | 8 | 13 | 10 | 11 | 14 | 7 | |||
| BMI | |||||||||
| <30 | 85 | 25 | 0.837 | 30 | 50 | 0.892 | 87 | 23 | 0.258 |
| >30 | 44 | 14 | 31 | 27 | 50 | 8 | |||
| HNF1A-AS1 | |||||||||
| Tumoral: ANCT (N) | Tumoral: Exosome (N) | Exosome: ANCT (N) | |||||||
| Down | Up | p Value | Down | Up | p Value | Down | Up | p Value | |
| Age | |||||||||
| <60 years | 23 | 37 | 0.07 | 26 | 34 | 0.981 | 22 | 38 | 0.943 |
| >60 years | 27 | 81 | 47 | 61 | 39 | 69 | |||
| Gender | |||||||||
| Female | 12 | 42 | 0.141 | 22 | 32 | 0.626 | 16 | 28 | 0.215 |
| Male | 38 | 76 | 51 | 63 | 45 | 69 | |||
| Subtype | |||||||||
| SCC | 13 | 39 | 0.366 | 20 | 32 | 0.382 | 16 | 36 | 0.317 |
| Adeno | 37 | 79 | 53 | 63 | 45 | 71 | |||
| Stage | |||||||||
| 1 | 16 | 31 | 14 | 33 | 19 | 28 | 0.764 | ||
| 2 | 16 | 55 | 0.21 | 32 | 39 | 0.052 | 24 | 47 | |
| 3 | 18 | 32 | 27 | 23 | 18 | 32 | |||
| Smoking | |||||||||
| Yes | 70 | 77 | 0.1 | 61 | 86 | 0.475 | 66 | 81 | 0.067 |
| No | 6 | 15 | 7 | 14 | 5 | 16 | |||
| BMI | |||||||||
| <30 | 31 | 79 | 0.537 | 47 | 63 | 0.794 | 38 | 72 | 0.513 |
| >30 | 19 | 39 | 26 | 32 | 23 | 35 | |||
| HOTAIR | |||||||||
| Tumoral: ANCT (N) | Tumoral: Exosome (N) | Exosome: ANCT (N) | |||||||
| Down | Up | p Value | Down | Up | p Value | Down | Up | p Value | |
| Age | |||||||||
| <60 years | 27 | 33 | 0.105 | 29 | 31 | 0.473 | 21 | 39 | 0.827 |
| >60 years | 35 | 73 | 46 | 62 | 36 | 72 | |||
| Gender | |||||||||
| Female | 21 | 33 | 0.714 | 25 | 29 | 0.767 | 21 | 33 | 0.35 |
| Male | 41 | 73 | 50 | 64 | 36 | 78 | |||
| Subtype | |||||||||
| SCC | 24 | 28 | 0.096 | 24 | 28 | 0.792 | 21 | 31 | 0.237 |
| Adeno | 38 | 78 | 51 | 65 | 36 | 80 | |||
| Stage | |||||||||
| 1 | 17 | 30 | 23 | 24 | 16 | 31 | |||
| 2 | 32 | 39 | 0.1 | 31 | 40 | 0.771 | 29 | 42 | 0.156 |
| 3 | 13 | 37 | 21 | 29 | 12 | 38 | |||
| Smoking | |||||||||
| Yes | 58 | 89 | 0.475 | 57 | 90 | 0.044 | 62 | 85 | 0.234 |
| No | 10 | 11 | 13 | 8 | 6 | 15 | |||
| BMI | |||||||||
| <30 | 38 | 72 | 0.383 | 48 | 62 | 0.718 | 36 | 74 | 0.651 |
| >30 | 24 | 34 | 27 | 31 | 21 | 37 | |||
| LINCRNA-p21 | |||||||||
| Tumoral: ANCT (N) | Tumoral: Exosome (N) | Exosome: ANCT (N) | |||||||
| Down | Up | p Value | Down | Up | p Value | Down | Up | p Value | |
| Age | |||||||||
| <60 years | 52 | 8 | 0.808 | 29 | 31 | 0.369 | 53 | 7 | 0.57 |
| >60 years | 95 | 13 | 60 | 48 | 92 | 16 | |||
| Gender | |||||||||
| Female | 45 | 9 | 0.261 | 27 | 27 | 0.595 | 47 | 7 | 0.85 |
| Male | 102 | 12 | 62 | 52 | 98 | 16 | |||
| Subtype | |||||||||
| SCC | 42 | 10 | 0.077 | 25 | 27 | 0.394 | 44 | 8 | 0.669 |
| Adeno | 105 | 11 | 64 | 52 | 101 | 15 | |||
| Stage | |||||||||
| 1 | 40 | 7 | 24 | 23 | 38 | 9 | |||
| 2 | 60 | 11 | 0.252 | 37 | 34 | 0.872 | 63 | 8 | 0.436 |
| 3 | 47 | 3 | 28 | 22 | 44 | 6 | |||
| Smoking | |||||||||
| Yes | 90 | 57 | 0.63 | 92 | 55 | 0.223 | 89 | 58 | 0.765 |
| No | 14 | 7 | 16 | 5 | 12 | 9 | |||
| BMI | |||||||||
| <30 | 94 | 16 | 0.27 | 57 | 53 | 0.679 | 94 | 16 | 0.657 |
| >30 | 53 | 5 | 32 | 26 | 51 | 7 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talebi, S.; Abadi, A.J.; Kazemioula, G.; Hosseini, N.; Taheri, F.; Pourali, S.; Mahdloo, T.; Rezaei, M.; Mirinezhad, M.; Ajami, N.; et al. Expression Analysis of Five Different Long Non-Coding Ribonucleic Acids in Nonsmall-Cell Lung Carcinoma Tumor and Tumor-Derived Exosomes. Diagnostics 2022, 12, 3209. https://doi.org/10.3390/diagnostics12123209
Talebi S, Abadi AJ, Kazemioula G, Hosseini N, Taheri F, Pourali S, Mahdloo T, Rezaei M, Mirinezhad M, Ajami N, et al. Expression Analysis of Five Different Long Non-Coding Ribonucleic Acids in Nonsmall-Cell Lung Carcinoma Tumor and Tumor-Derived Exosomes. Diagnostics. 2022; 12(12):3209. https://doi.org/10.3390/diagnostics12123209
Chicago/Turabian StyleTalebi, Samaneh, Asal Jalal Abadi, Golnesa Kazemioula, Nayyerehalsadat Hosseini, Forough Taheri, Saba Pourali, Touba Mahdloo, Marzieh Rezaei, Mohammadreza Mirinezhad, Naser Ajami, and et al. 2022. "Expression Analysis of Five Different Long Non-Coding Ribonucleic Acids in Nonsmall-Cell Lung Carcinoma Tumor and Tumor-Derived Exosomes" Diagnostics 12, no. 12: 3209. https://doi.org/10.3390/diagnostics12123209
APA StyleTalebi, S., Abadi, A. J., Kazemioula, G., Hosseini, N., Taheri, F., Pourali, S., Mahdloo, T., Rezaei, M., Mirinezhad, M., Ajami, N., & Salmaninejad, A. (2022). Expression Analysis of Five Different Long Non-Coding Ribonucleic Acids in Nonsmall-Cell Lung Carcinoma Tumor and Tumor-Derived Exosomes. Diagnostics, 12(12), 3209. https://doi.org/10.3390/diagnostics12123209






