Relevance of ARID1A Mutations in Endometrial Carcinomas
Abstract
:1. Introduction
2. Materials and Methods
2.1. NGS
2.2. IHC
2.3. ARID1A RNA Expression
2.4. Structure Analysis
3. Results
3.1. ARID1A Mutational Status
3.2. RNA Expression
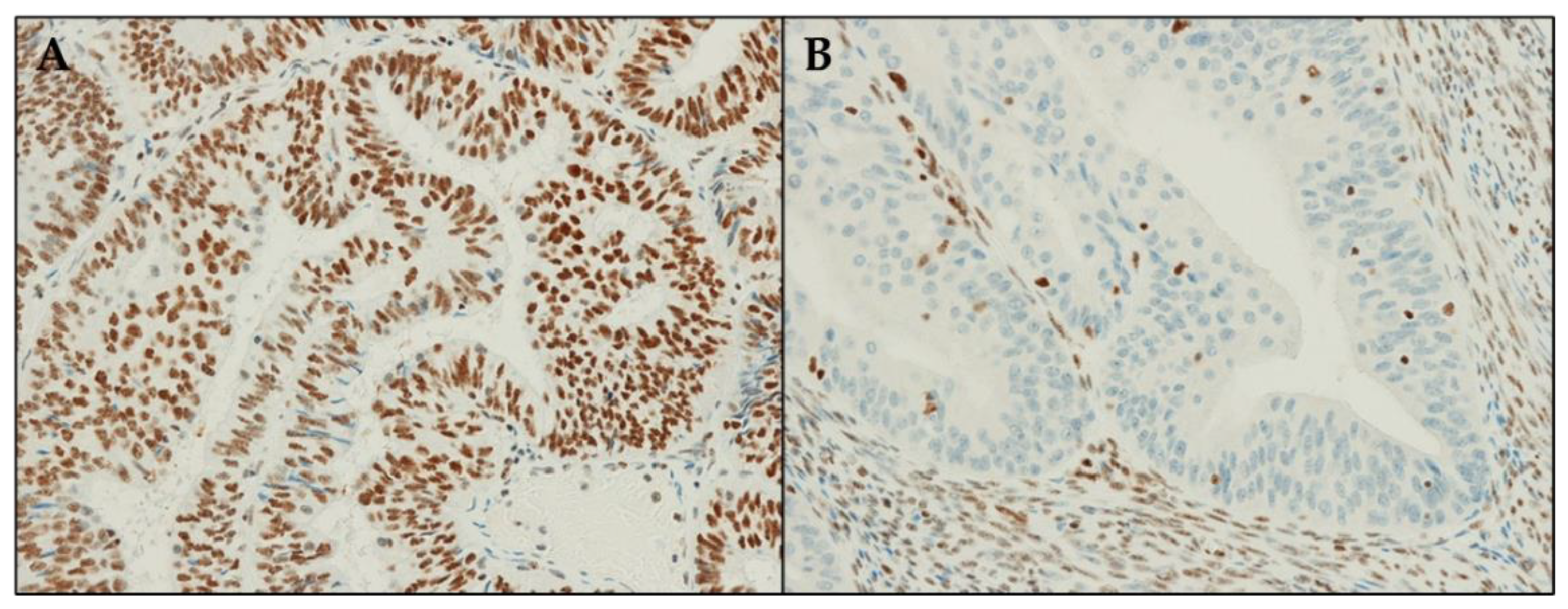
3.3. ARID1A Protein Expression
3.4. ARID1A Structure Data
- p.Leu2195Arg (6LTH/6LTJ) (Case #5). This residue is located at the buried edge of a partially buried α-helix. Leu2195 forms hydrophobic contacts with ARID1A Val2168 and Leu2241. The observed mutation can cause mild to severe effects on the latter interactions considering the positive charge and the H-bond donor capability of the arginine side chain tail, even if it is difficult to predict their extent.
- p.Leu1100Phe (1RYU) (Case #12). Leu1100 is located in the ARID1A hydrophobic core and is partially accessible to the solvent. The vicinity of several ARID1A aromatic residues (Tyr 1055, Tyr1096, Tyr1101, Phe1103) can cause the formation of new interactions when Leu1100 is mutated in phenylalanine. On the other hand, it is difficult to predict the effect of this mutation on the ARID1A folding.
- p.Asn1705Ser (6LTH/6LTJ) (Case #14). This residue is located on a solvent-exposed α-helix. Asn1705 is H-bonded to ARID1A Ser1707 and Asn1997. Considering the similar length and the similar H-bond propensity of asparagine and serine, this mutation is not supposed to cause large structural effects.
- p.Arg1833Cys (6LTH/6LTJ) (Case #16). This residue is located on a solvent-exposed loop. The side chain of Arg1833 is H-bonded to Gln70 and Leu71 backbone oxygen atoms of the SMARCC2 subunit and to ARID1A Glu1853. The observed mutation can damage these interactions considering the different lengths in the side chain of an arginine residue if compared with the length of a cysteine. Moreover, cysteine is not a good H-bond donor as an arginine.
- p.Arg1906Gln (00000056) (Case #17). The mutation is on a residue that is exposed to the solvent and that does not interact with other residues from ARID1A or from other SWI/SNF proteins. A polar residue is replaced by a similar equally polar residue. Apparently, this mutation is not supposed to cause any effect.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Esteve, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [Green Version]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jurgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef] [Green Version]
- De Leo, A.; de Biase, D.; Lenzi, J.; Barbero, G.; Turchetti, D.; Grillini, M.; Ravegnini, G.; Angelini, S.; Zamagni, C.; Coluccelli, S.; et al. ARID1A and CTNNB1/beta-Catenin Molecular Status Affects the Clinicopathologic Features and Prognosis of Endometrial Carcinoma: Implications for an Improved Surrogate Molecular Classification. Cancers 2021, 13, 950. [Google Scholar] [CrossRef]
- Ravegnini, G.; Gorini, F.; De Crescenzo, E.; De Leo, A.; De Biase, D.; Di Stanislao, M.; Hrelia, P.; Angelini, S.; De Iaco, P.; Perrone, A.M. Can miRNAs be useful biomarkers in improving prognostic stratification in endometrial cancer patients? An update review. Int. J. Cancer 2022, 150, 1077–1090. [Google Scholar] [CrossRef]
- Ravegnini, G.; De Leo, A.; Coada, C.; Gorini, F.; de Biase, D.; Ceccarelli, C.; Dondi, G.; Tesei, M.; De Crescenzo, E.; Santini, D.; et al. Identification of miR-499a-5p as a Potential Novel Biomarker for Risk Stratification in Endometrial Cancer. Front. Oncol. 2021, 11, 757678. [Google Scholar] [CrossRef]
- Perrone, A.M.; De Leo, A.; de Biase, D.; Ravegnini, G.; De Iaco, P. Endometrial carcinoma: Past, present, and future. Eur. J. Gynaecol. Oncol. 2021, 42, 610–612. [Google Scholar] [CrossRef]
- Dou, Y.; Kawaler, E.A.; Zhou, D.C.; Gritsenko, M.A.; Huang, C.; Blumenberg, L.; Karpova, A.; Petyuk, V.A.; Savage, S.R.; Satpathy, S.; et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell 2020, 180, 729–748.e26. [Google Scholar] [CrossRef]
- Dondi, G.; Coluccelli, S.; De Leo, A.; Ferrari, S.; Gruppioni, E.; Bovicelli, A.; Godino, L.; Coada, C.A.; Morganti, A.G.; Giordano, A.; et al. An Analysis of Clinical, Surgical, Pathological and Molecular Characteristics of Endometrial Cancer According to Mismatch Repair Status. A Multidisciplinary Approach. Int. J. Mol. Sci. 2020, 21, 7188. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Mao, T.L.; Panuganti, P.K.; Kuhn, E.; Kurman, R.J.; Maeda, D.; Chen, E.; Jeng, Y.M.; Wang, T.L.; Shih Ie, M. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 2011, 35, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.; Nakayama, K.; Rahman, M.T.; Katagiri, H.; Katagiri, A.; Ishibashi, T.; Ishikawa, M.; Iida, K.; Miyazaki, K. Clinicopathologic analysis of loss of AT-rich interactive domain 1A expression in endometrial cancer. Hum. Pathol. 2013, 44, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, K.C.; Lee, A.F.; Al-Agha, O.M.; Chow, C.; Kalloger, S.E.; Scott, D.W.; Steidl, C.; Wiseman, S.M.; Gascoyne, R.D.; Gilks, B.; et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J. Pathol. 2011, 224, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Peng, Y.; Wei, L.; Zhang, W.; Yang, L.; Lan, L.; Kapoor, P.; Ju, Z.; Mo, Q.; Shih Ie, M.; et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov. 2015, 5, 752–767. [Google Scholar] [CrossRef] [Green Version]
- Mathur, R. ARID1A loss in cancer: Towards a mechanistic understanding. Pharmacol. Ther. 2018, 190, 15–23. [Google Scholar] [CrossRef]
- Santandrea, G.; Piana, S.; Valli, R.; Zanelli, M.; Gasparini, E.; De Leo, A.; Mandato, V.D.; Palicelli, A. Immunohistochemical Biomarkers as a Surrogate of Molecular Analysis in Ovarian Carcinomas: A Review of the Literature. Diagnostics 2021, 11, 199. [Google Scholar] [CrossRef]
- de Biase, D.; Malapelle, U.; De Leo, A.; Maloberti, T.; Visani, M.; Pisapia, P.; Acquaviva, G.; Pepe, F.; Russo, G.; Iaccarino, A.; et al. Multi-gene custom panels for the characterisation of metastatic colorectal carcinoma in clinical practice: Express the role of PIK3CA mutations. J. Clin. Pathol. 2021. [Google Scholar] [CrossRef]
- De Leo, A.; Santini, D.; Ceccarelli, C.; Santandrea, G.; Palicelli, A.; Acquaviva, G.; Chiarucci, F.; Rosini, F.; Ravegnini, G.; Pession, A.; et al. What Is New on Ovarian Carcinoma: Integrated Morphologic and Molecular Analysis Following the New 2020 World Health Organization Classification of Female Genital Tumors. Diagnostics 2021, 11, 697. [Google Scholar] [CrossRef]
- Visani, M.; Acquaviva, G.; De Leo, A.; Sanza, V.; Merlo, L.; Maloberti, T.; Brandes, A.A.; Franceschi, E.; Di Battista, M.; Masetti, M.; et al. Molecular alterations in pancreatic tumors. World J. Gastroenterol. 2021, 27, 2710–2726. [Google Scholar] [CrossRef]
- de Biase, D.; Acquaviva, G.; Visani, M.; Sanza, V.; Argento, C.M.; De Leo, A.; Maloberti, T.; Pession, A.; Tallini, G. Molecular Diagnostic of Solid Tumor Using a Next Generation Sequencing Custom-Designed Multi-Gene Panel. Diagnostics 2020, 10, 250. [Google Scholar] [CrossRef] [PubMed]
- Bosse, T.; ter Haar, N.T.; Seeber, L.M.; v Diest, P.J.; Hes, F.J.; Vasen, H.F.; Nout, R.A.; Creutzberg, C.L.; Morreau, H.; Smit, V.T. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod. Pathol. 2013, 26, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Zhang, Z.; Upchurch, S.; Isern, N.; Chen, Y. Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition. J. Biol. Chem. 2004, 279, 16670–16676. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Wu, Z.; Tian, Y.; Yu, Z.; Yu, J.; Wang, X.; Li, J.; Liu, B.; Xu, Y. Structure of nucleosome-bound human BAF complex. Science 2020, 367, 875–881. [Google Scholar] [CrossRef]
- Vallat, B.; Webb, B.; Westbrook, J.D.; Sali, A.; Berman, H.M. Development of a Prototype System for Archiving Integrative/Hybrid Structure Models of Biological Macromolecules. Structure 2018, 26, 894–904.e2. [Google Scholar] [CrossRef] [Green Version]
- Mashtalir, N.; Suzuki, H.; Farrell, D.P.; Sankar, A.; Luo, J.; Filipovski, M.; D’Avino, A.R.; St Pierre, R.; Valencia, A.M.; Onikubo, T.; et al. A Structural Model of the Endogenous Human BAF Complex Informs Disease Mechanisms. Cell 2020, 183, 802–817.e24. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berns, K.; Caumanns, J.J.; Hijmans, E.M.; Gennissen, A.M.C.; Severson, T.M.; Evers, B.; Wisman, G.B.A.; Meersma, G.J.; Lieftink, C.; Beijersbergen, R.L.; et al. ARID1A mutation sensitizes most ovarian clear cell carcinomas to BET inhibitors. Oncogene 2018, 37, 4611–4625. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Takenaka, M.; Okamoto, A.; Bowtell, D.D.L.; Kohno, T. Treatment Strategies for ARID1A-Deficient Ovarian Clear Cell Carcinoma. Cancers 2021, 13, 1769. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Veronese, N.; Solmi, M.; Cho, H.; Kim, J.H.; Chou, A.; Gill, A.J.; Faraj, S.F.; Chaux, A.; Netto, G.J.; et al. Prognostic role and implications of mutation status of tumor suppressor gene ARID1A in cancer: A systematic review and meta-analysis. Oncotarget 2015, 6, 39088–39097. [Google Scholar] [CrossRef] [PubMed]
- Pavlidou, E.N.; Balis, V. Diagnostic significance and prognostic role of the ARID1A gene in cancer outcomes (Review). World Acad. Sci. J. 2020, 2, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, M.; Kawashima, A.; Yasuhara, R.; Hayakawa, Y.; Miyamoto, S.; Iizuka, C.; Sekizawa, A. Massively parallel sequencing of cell-free DNA in plasma for detecting gynaecological tumour-associated copy number alteration. Sci. Rep. 2018, 8, 11205. [Google Scholar] [CrossRef]
- Baldi, S.; Khamgan, H.; Qian, Y.; Wu, H.; Zhang, Z.; Zhang, M.; Gao, Y.; Safi, M.; Al-Radhi, M.; Zuo, Y.F. Downregulated ARID1A by miR-185 Is Associated With Poor Prognosis and Adverse Outcomes in Colon Adenocarcinoma. Front. Oncol. 2021, 11, 679334. [Google Scholar] [CrossRef]
- Xu, S.; Tang, C. The Role of ARID1A in Tumors: Tumor Initiation or Tumor Suppression? Front. Oncol. 2021, 11, 745187. [Google Scholar] [CrossRef]
- Kwan, S.Y.; Cheng, X.; Tsang, Y.T.; Choi, J.S.; Kwan, S.Y.; Izaguirre, D.I.; Kwan, H.S.; Gershenson, D.M.; Wong, K.K. Loss of ARID1A expression leads to sensitivity to ROS-inducing agent elesclomol in gynecologic cancer cells. Oncotarget 2016, 7, 56933–56943. [Google Scholar] [CrossRef] [Green Version]
- Takeda, T.; Banno, K.; Okawa, R.; Yanokura, M.; Iijima, M.; Irie-Kunitomi, H.; Nakamura, K.; Iida, M.; Adachi, M.; Umene, K.; et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol. Rep. 2016, 35, 607–613. [Google Scholar] [CrossRef] [Green Version]
- Takano, M.; Kikuchi, Y.; Kudoh, K.; Goto, T.; Furuya, K.; Kikuchi, R.; Kita, T.; Fujiwara, K.; Shiozawa, T.; Aoki, D. Weekly administration of temsirolimus for heavily pretreated patients with clear cell carcinoma of the ovary: A report of six cases. Int. J. Clin. Oncol. 2011, 16, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Ikeda, Y.; Kudoh, K.; Kita, T.; Sasaki, N.; Kikuchi, Y. Complete remission of recurrent ovarian clear cell carcinoma by chemotherapy with bevacizumab, trabectedin and oxaliplatin. J. Obstet. Gynaecol. Res. 2013, 39, 872–875. [Google Scholar] [CrossRef] [PubMed]



| Clinicopathologic Characteristics | n = 50 (%) |
|---|---|
| Age, years | 63 ± 11 |
| (34–80) | |
| Body mass index, kg/m2 | 27.5 ± 6.6 |
| (22.8–30.1) | |
| Tumor type | |
| Endometrioid | 38 (76.0) |
| Dedifferentiated/Undifferentiated | 4 (8.0) |
| Serous | 7 (14.0) |
| Clear cell | 1 (2.0) |
| Grade | |
| 1 | 13 (26.0) |
| 2 | 15 (30.0) |
| 3 | 22 (44.0) |
| Depth of invasion | |
| <50% | 43 (86.0) |
| ≥50% | 7 (14.0) |
| Lymphovascular space invasion (LVSI) | |
| Absent/Focal | 40 (80.0) |
| Diffuse | 10 (20.0) |
| Lymph node status | |
| Negative | 44 (88.0) |
| Positive | 6 (12.0) |
| FIGO stage | |
| IA | 33 (66.0) |
| IB | 4 (8.0) |
| II | 1 (2.0) |
| III | 12 (24.0) |
| ARID1A Alteration | 20 (40.0) |
| Endometrioid | 17 (34.0) |
| Dedifferentiated/Undifferentiated | 2 (4.0) |
| Serous | 1 (2.0) |
| Clear cell | 0 (0.0) |
| Case | ARID1A Protein Mutation | Exon | PolyPhen2 Score | Varsome Verdict |
|---|---|---|---|---|
| 1 | p.Asn209Ser | 1 | 0.049 | Likely Benign |
| 2 | p.Ala226Asp | 1 | 0.037 | Likely Benign |
| 3 | p.Gly455Glu | 3 | 0.998 | VUS |
| 4 | p.Ser530fs | 3 | 1.000 | Pathogenic |
| 5 | p.Arg596His | 3 | 0.998 | Likely Benign |
| p.Leu2195Arg | 20 | 1.000 | VUS | |
| 6 | p.Arg693Gln | 5 | 0.999 | VUS |
| p.Ala1272Val | 15 | 0.913 | Likely Benign | |
| 7 | p.Arg693Ter | 5 | 1.000 | Pathogenic |
| 8 | p.Pro728fs | 6 | 1.000 | Pathogenic |
| 9 | p.Gly768Asp | 7 | 0.181 | VUS |
| 10 | p.Ala900Thr | 8 | 0.984 | Benign |
| 11 | p.Lys996fs | 10 | 1.000 | Pathogenic |
| 12 | p.Leu1100Phe | 12 | 1.000 | VUS |
| p.Arg1446Gln | 18 | 0.999 | VUS | |
| p.Arg1989Ter | 20 | 1.000 | Pathogenic | |
| 13 | p.Gln1519fs | 18 | 1.000 | Pathogenic |
| 14 | p.Asn1705Ser | 19 | 0.137 | Benign |
| 15 | p.Arg1722Ter | 20 | 1.000 | Pathogenic |
| 16 | p.Arg1833Cys | 20 | 0.999 | VUS |
| 17 | p.Arg1906Gln | 20 | 0.996 | Benign |
| 18 | p.Arg1989Ter | 20 | 1.000 | Pathogenic |
| 19 | p.Arg1989Ter | 20 | 1.000 | Pathogenic |
| 20 | p.Ser2262fs | 20 | 1.000 | Pathogenic |
| Case | Aminoacidic Change | PolyPhen2 Score | Varsome Verdict | IHC |
|---|---|---|---|---|
| 1 | p.Asn209Ser | 0.049 | Likely Benign | Positive |
| 3 | p.Gly455Glu | 0.998 | VUS | Positive |
| 6 | p.Arg693Gln p.Ala1272Val | 0.999 0.913 | VUS Likely Benign | Positive |
| 14 | p.Asn1705Ser | 0.137 | Benign | Positive |
| Case | Aminoacidic Change | PolyPhen2 Score | Varsome Verdict | IHC |
|---|---|---|---|---|
| 2 | p.Ala226Asp | 0.037 | Likely Benign | Loss |
| 5 | p.Arg596His p.Leu2195Arg | 0.998 1.000 | Likely Benign VUS | Loss |
| 9 | p.Gly768Asp | 0.181 | VUS | Loss |
| 10 | p.Ala900Thr | 0.984 | Benign | Loss |
| 16 | p.Arg1833Cys | 0.999 | VUS | Loss |
| 17 | p.Arg1906Gln | 0.996 | Benign | Loss |
| # | ARID1A Mutational Status | POLYPHEN2 Score | Varsome Verdict | IHC Staining | Consensus |
|---|---|---|---|---|---|
| 24 cases | WT | / | / | Positive | OK |
| 6 cases | WT | / | / | Loss | NO |
| 1 | p.Asn209Ser | 0.049 | Likely Benign | Positive | OK |
| 2 | p.Ala226Asp | 0.037 | Likely Benign | Loss | NO |
| 3 | p.Gly455Glu | 0.998 | VUS | Positive | ? |
| 4 | p.Ser530fs | 1.000 | Pathogenic | Loss | OK |
| 5 | p.Arg596His | 0.998 | Likely Benign | Positive | ? |
| p.Leu2195Arg | 1.000 | VUS | |||
| 6 | p.Arg693Gln | 0.999 | VUS | Positive | ? |
| p.Ala1272Val | 0.913 | Likely Benign | |||
| 7 | p.Arg693Ter | 1.000 | Pathogenic | Loss | OK |
| 8 | p.Pro728fs | 1.000 | Pathogenic | Loss | OK |
| 9 | p.Gly768Asp | 0.181 | VUS | Loss | ? |
| 10 | p.Ala900Thr | 0.984 | Benign | Loss | ? |
| 11 | p.Lys996fs | 1.000 | Pathogenic | Loss | OK |
| 12 | p.Leu1100Phe | 1.000 | VUS | Loss | OK |
| p.Arg1446Gln | 0.999 | VUS | |||
| p.Arg1989Ter | 1.000 | Pathogenic | |||
| 13 | p.Gln1519fs | 1.000 | Pathogenic | Loss | OK |
| 14 | p.Asn1705Ser | 0.137 | Benign | Positive | OK |
| 15 | p.Arg1722Ter | 1.000 | Pathogenic | Loss | OK |
| 16 | p.Arg1833Cys | 0.999 | VUS | Loss | ? |
| 17 | p.Arg1906Gln | 0.996 | Benign | Loss | ? |
| 18 | p.Arg1989Ter | 1.000 | Pathogenic | Loss | OK |
| 19 | p.Arg1989Ter | 1.000 | Pathogenic | Loss | OK |
| 20 | p.Ser2262fs | 1.000 | Pathogenic | Loss | OK |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Leo, A.; Ravegnini, G.; Musiani, F.; Maloberti, T.; Visani, M.; Sanza, V.; Angelini, S.; Perrone, A.M.; De Iaco, P.; Corradini, A.G.; et al. Relevance of ARID1A Mutations in Endometrial Carcinomas. Diagnostics 2022, 12, 592. https://doi.org/10.3390/diagnostics12030592
De Leo A, Ravegnini G, Musiani F, Maloberti T, Visani M, Sanza V, Angelini S, Perrone AM, De Iaco P, Corradini AG, et al. Relevance of ARID1A Mutations in Endometrial Carcinomas. Diagnostics. 2022; 12(3):592. https://doi.org/10.3390/diagnostics12030592
Chicago/Turabian StyleDe Leo, Antonio, Gloria Ravegnini, Francesco Musiani, Thais Maloberti, Michela Visani, Viviana Sanza, Sabrina Angelini, Anna Myriam Perrone, Pierandrea De Iaco, Angelo Gianluca Corradini, and et al. 2022. "Relevance of ARID1A Mutations in Endometrial Carcinomas" Diagnostics 12, no. 3: 592. https://doi.org/10.3390/diagnostics12030592
APA StyleDe Leo, A., Ravegnini, G., Musiani, F., Maloberti, T., Visani, M., Sanza, V., Angelini, S., Perrone, A. M., De Iaco, P., Corradini, A. G., Rosini, F., Grillini, M., Santini, D., Ceccarelli, C., Zamagni, C., Tallini, G., & de Biase, D. (2022). Relevance of ARID1A Mutations in Endometrial Carcinomas. Diagnostics, 12(3), 592. https://doi.org/10.3390/diagnostics12030592












