Aux-MVNet: Auxiliary Classifier-Based Multi-View Convolutional Neural Network for Maxillary Sinusitis Diagnosis on Paranasal Sinuses View
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Populations
2.2. Radiograph Acquisition
2.3. Labeling
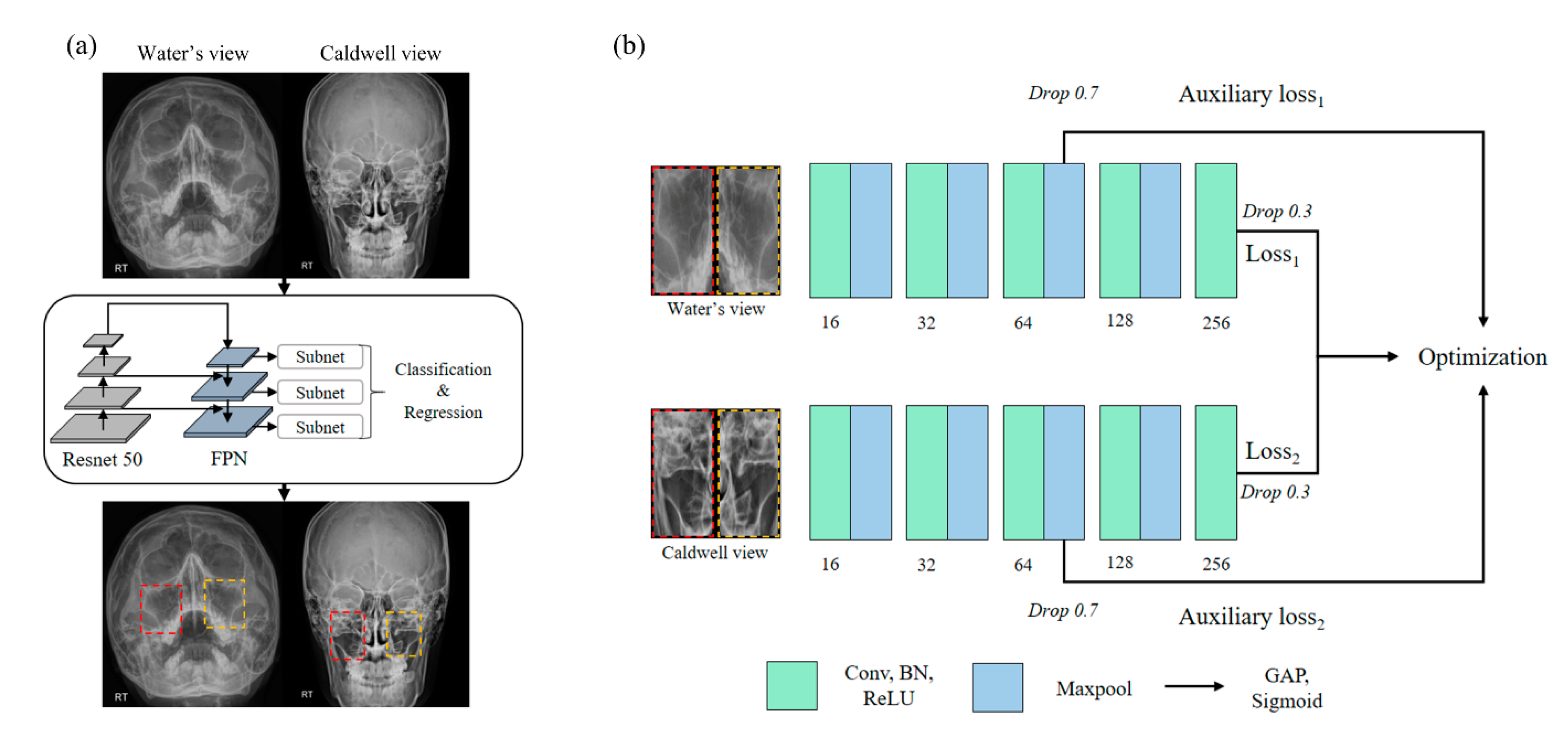
2.4. Experimental Design
2.5. Region of Sinus Detection
2.6. Sinusitis Classification
2.7. Implementation Details
3. Results
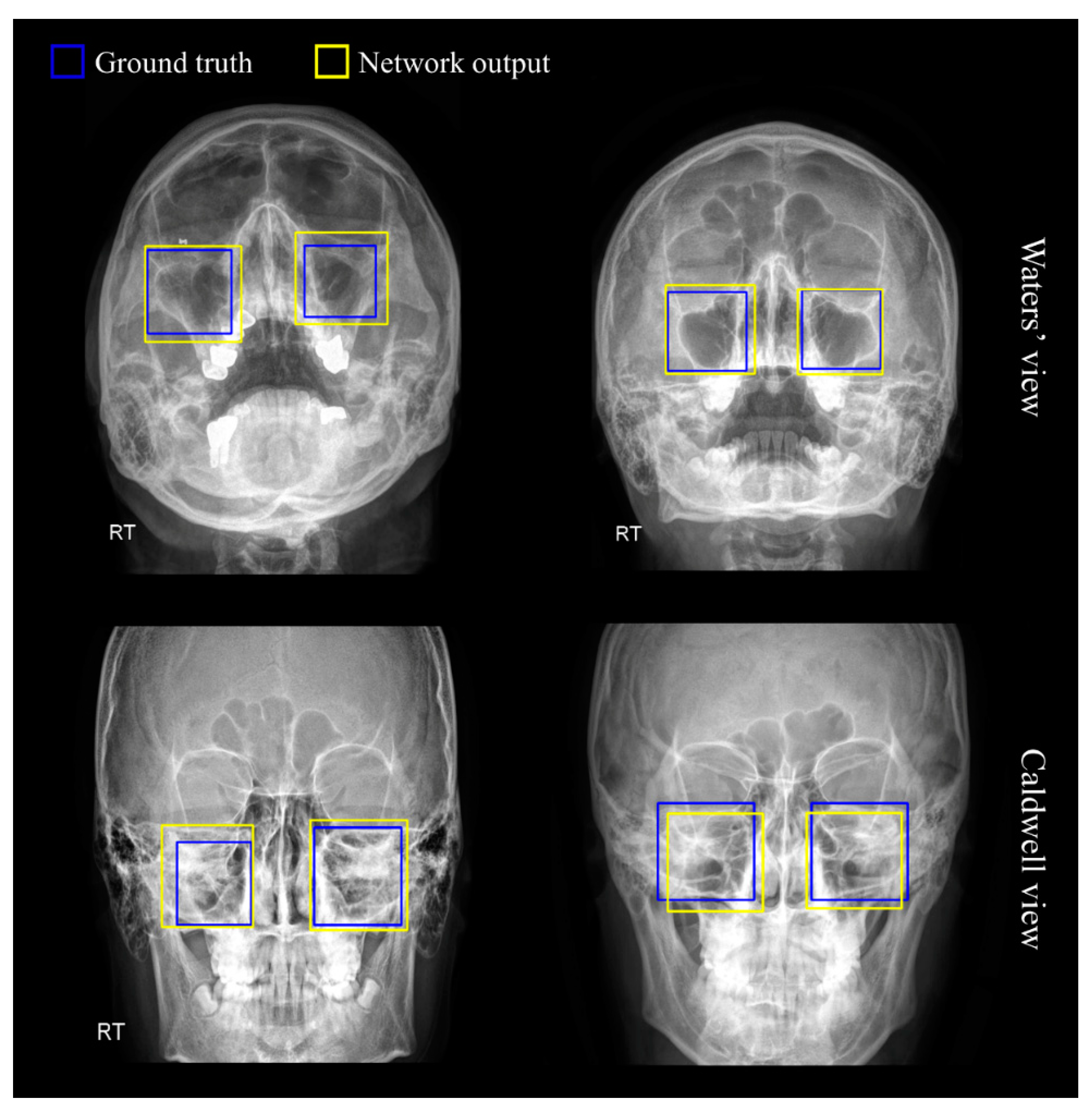
3.1. Region of Sinus Detection
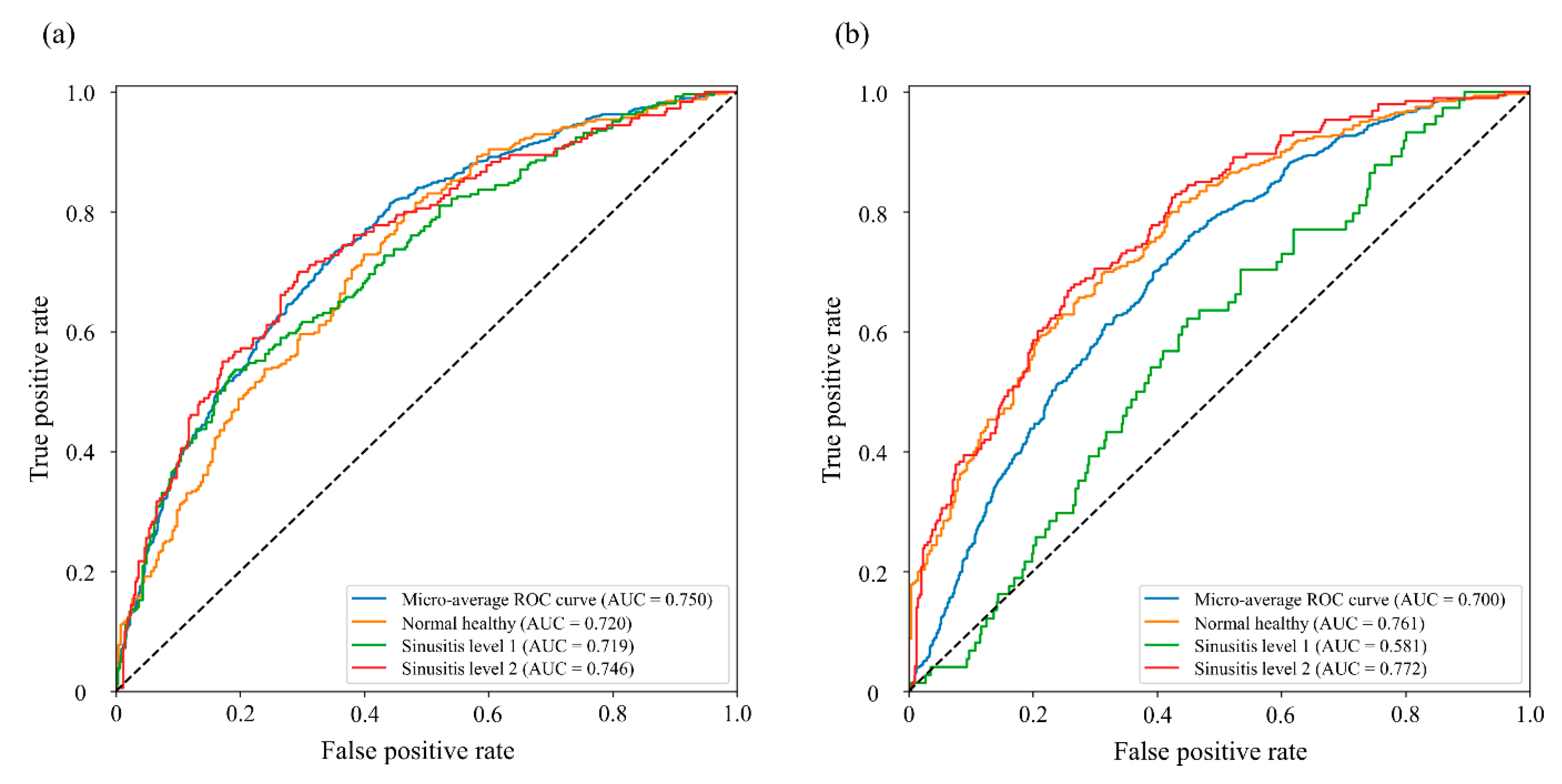
3.2. Sinusitis Classification
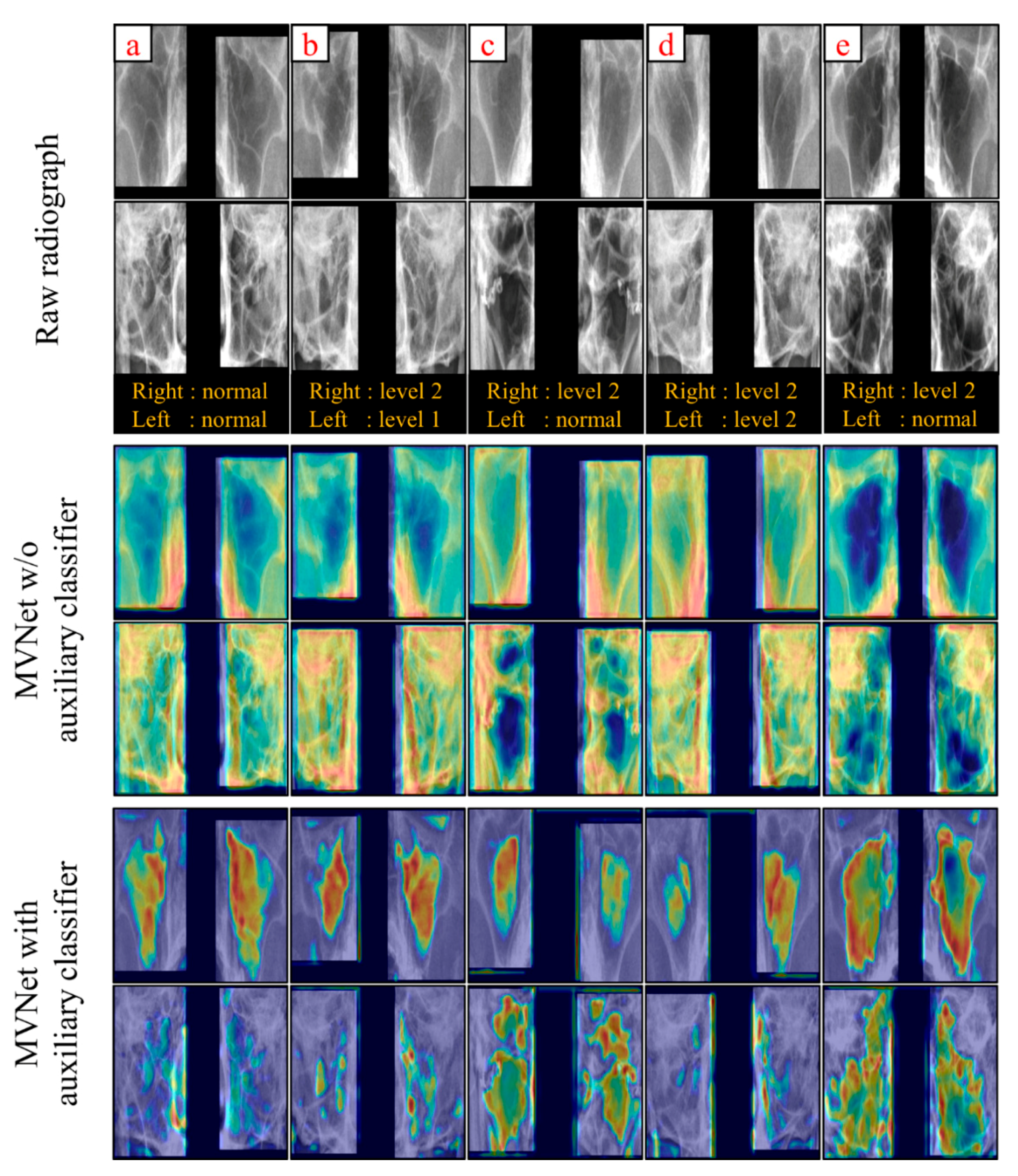
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Loos, D.D.; Lourijsen, E.S.; Wildeman, M.A.; Freling, N.J.M.; Wolvers, M.D.; Reitsma, S.; Fokkens, W.J. Prevalence of chronic rhinosinusitis in the general population based on sinus radiology and symptomatology. J. Allergy Clin. Immunol. 2019, 143, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Dass, K.; Peters, A.T. Diagnosis and Management of Rhinosinusitis: Highlights from the 2015 Practice Parameter. Curr. Allergy Asthma Rep. 2016, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, C.F.; Bykowski, J.; Aulino, J.M.; Berger, K.L.; Choudhri, A.F.; Conley, D.B.; Luttrull, M.D.; Nunez, D.; Shah, L.M.; Sharma, A.; et al. ACR Appropriateness Criteria® Sinonasal Disease. J. Am. Coll. Radiol. 2017, 14, S550–S559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frerichs, N.; Brateanu, A. Rhinosinusitis and the role of imaging. Clevel. Clin. J. Med. 2020, 87, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Ohba, T.; Ogawa, Y.; Shinohara, Y.; Hiromatsu, T.; Uchida, A.; Toyoda, Y. Limitations of panoramic radiography in the detection of bone defects in the posterior wall of the maxillary sinus: An experimental study. Dentomaxillofac. Radiol. 1994, 23, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Konen, E.; Faibel, M.; Kleinbaum, Y.; Wolf, M.; Lusky, A.; Hoffman, C.; Eyal, A.; Tadmor, R. The value of the occipitomental (Waters’) view in diagnosis of sinusitis: A comparative study with computed tomography. Clin. Radiol. 2000, 55, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.-H.; Yoon, J.; Kim, Y.J.; Kang, C.-K.; Cho, S.-E.; Kim, K.G.; Kang, S.-G. Reproducibility of automated habenula segmentation via deep learning in major depressive disorder and normal controls with 7 Tesla MRI. Sci. Rep. 2021, 11, 13445. [Google Scholar] [CrossRef]
- Baltruschat, I.M.; Nickisch, H.; Grass, M.; Knopp, T.; Saalbach, A. Comparison of Deep Learning Approaches for Multi-Label Chest X-ray Classification. Sci. Rep. 2019, 9, 6381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Kim, Y.J.; Kim, K.G. Bone age estimation using deep learning and hand X-ray images. Biomed. Eng. Lett. 2020, 10, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, K.G. Applying Deep Learning in Medical Images: The Case of Bone Age Estimation. Healthc. Inform. Res. 2018, 24, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, K.J.; Sunwoo, L.; Choi, D.; Nam, C.-M.; Cho, J.; Kim, J.; Bae, Y.J.; Yoo, R.-E.; Choi, B.S.; et al. Deep Learning in Diagnosis of Maxillary Sinusitis Using Conventional Radiography. Investig. Radiol. 2019, 54, 7–15. [Google Scholar] [CrossRef]
- Jeon, Y.; Lee, K.; Sunwoo, L.; Choi, D.; Oh, D.; Lee, K.; Kim, Y.; Kim, J.-W.; Cho, S.; Baik, S.; et al. Deep Learning for Diagnosis of Paranasal Sinusitis Using Multi-View Radiographs. Diagnostics 2021, 11, 250. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Ren, S.; He, K.; Girshick, R. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Processing Syst. 2015, 28, 91–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornblith, S.; Shlens, J.; Le, Q.V. Do better imagenet models transfer better? In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 2656–2666. [Google Scholar]
- Meyes, R.; Lu, M.; de Puiseau, C.W.; Meisen, T. Ablation studies in artificial neural networks. arXiv preprint 2019, arXiv:1901.08644. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Su, H.; Maji, S.; Kalogerakis, E.; Learned-Miller, E. Multi-view convolutional neural networks for 3D shape recognition. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 945–953. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yu, M.; Chen, T.; Shi, Z.; Bao, C.; Ma, K. Auxiliary training: Towards accurate and robust models. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2020; pp. 369–378. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv preprint 2014, arXiv:1412.6980v9. [Google Scholar]
- Boyd, K.; Eng, K.H.; Page, C.D. Area under the precision-recall curve: Point estimates and confidence intervals. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Prague, Czech Republic, 23–27 September 2013; pp. 451–466. [Google Scholar]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2019, Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.D.; Davis, H.T. Receiver operating characteristics curves and related decision measures: A tutorial. Chemom. Intell. Lab. Syst. 2006, 80, 24–38. [Google Scholar] [CrossRef]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Mishra, D.; Chaudhury, S.; Sarkar, M.; Soin, A.S. Ultrasound Image Segmentation: A Deeply Supervised Network with Attention to Boundaries. IEEE Trans. Biomed. Eng. 2018, 66, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yao, A.; Zhou, A.; Zhao, H. Deeply-supervised knowledge synergy. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 6990–6999. [Google Scholar]
- Ge, Z.; Mahapatra, D.; Chang, X.; Chen, Z.; Chi, L.; Lu, H. Improving multi-label chest X-ray disease diagnosis by exploiting disease and health labels dependencies. Multimed. Tools Appl. 2020, 79, 14889–14902. [Google Scholar] [CrossRef]

| Basic MVNet | MVNet with Dense Module | MVNet with Inception-v1 Module |
|---|---|---|
| Input shape: (N, 512, 512, 1) | ||
| Conv5, 16 | ||
| Maxpool2, stride2 | ||
| Conv5, 32 | 6 dense block Transition layer | Inception-v1 block |
| Maxpool2, stride2 | ||
| Conv3, 64 | 12 dense block Transition layer | Inception-v1 block |
| Drop 0.7, * | ||
| Maxpool2, stride2 | ||
| Conv3, 128 | 24 dense block Transition layer | Inception-v1 block |
| Maxpool2, stride2 | ||
| Conv3, 256 | 16 dense block Transition layer | Inception-v1 block |
| Drop 0.3 * | ||
| Fully connected layer with sigmoid | ||
| Output shape: (N, 6) | ||
| Area Under the Curve | p Value * | |||
|---|---|---|---|---|
| Left | Right | Total | ||
| MVNet | 0.552 | 0.637 | 0.602 | <0.001 |
| MVNet with auxiliary classifier | 0.750 | 0.700 | 0.722 | - |
| Dense MVNet | 0.695 | 0.723 | 0.706 | =0.001 |
| Dense MVNet with auxiliary classifier | 0.703 | 0.722 | 0.709 | =0.005 |
| Inception MVNet | 0.583 | 0.671 | 0.621 | <0.001 |
| Inception MVNet with auxiliary classifier | 0.678 | 0.753 | 0.710 | =0.002 |
| Normal Sinus | Sinusitis Level 1 | Sinusitis Level 2 | |
|---|---|---|---|
| Left Sinus | |||
| Accuracy | 0.676 | 0.687 | 0.704 |
| Sensitivity | 0.796 | 0.536 | 0.700 |
| Specificity | 0.536 | 0.809 | 0.705 |
| Right Sinus | |||
| Accuracy | 0.692 | 0.559 | 0.712 |
| Sensitivity | 0.697 | 0.622 | 0.679 |
| Specificity | 0.689 | 0.552 | 0.734 |
| Total | |||
| Accuracy | 0.681 | 0.571 | 0.724 |
| Sensitivity | 0.708 | 0.656 | 0.625 |
| Specificity | 0.647 | 0.536 | 0.770 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, S.-H.; Kim, J.H.; Kim, Y.J.; Cho, M.Y.; Jung, J.U.; Ha, R.; Jung, J.H.; Kim, S.T.; Kim, K.G. Aux-MVNet: Auxiliary Classifier-Based Multi-View Convolutional Neural Network for Maxillary Sinusitis Diagnosis on Paranasal Sinuses View. Diagnostics 2022, 12, 736. https://doi.org/10.3390/diagnostics12030736
Lim S-H, Kim JH, Kim YJ, Cho MY, Jung JU, Ha R, Jung JH, Kim ST, Kim KG. Aux-MVNet: Auxiliary Classifier-Based Multi-View Convolutional Neural Network for Maxillary Sinusitis Diagnosis on Paranasal Sinuses View. Diagnostics. 2022; 12(3):736. https://doi.org/10.3390/diagnostics12030736
Chicago/Turabian StyleLim, Sang-Heon, Jong Hoon Kim, Young Jae Kim, Min Young Cho, Jin Uk Jung, Ryun Ha, Joo Hyun Jung, Seon Tae Kim, and Kwang Gi Kim. 2022. "Aux-MVNet: Auxiliary Classifier-Based Multi-View Convolutional Neural Network for Maxillary Sinusitis Diagnosis on Paranasal Sinuses View" Diagnostics 12, no. 3: 736. https://doi.org/10.3390/diagnostics12030736






