Tubulin Alpha 1b Is Associated with the Immune Cell Infiltration and the Response of HCC Patients to Immunotherapy
Abstract
:1. Introduction
2. Methods
2.1. Data Collection and Differential Expression Analysis
2.2. The Relationship of Survival and Clinicopathology with TUBA1B in HCC
2.3. Analysis of Immune Infiltrates, Immune Checkpoint, and Immune-Related Genes
2.4. Conduction of Protein Protein Interaction (PPI) Network
2.5. TUBA1B Enrichment Analyses in HCC
2.6. Statistical Analysis
3. Results
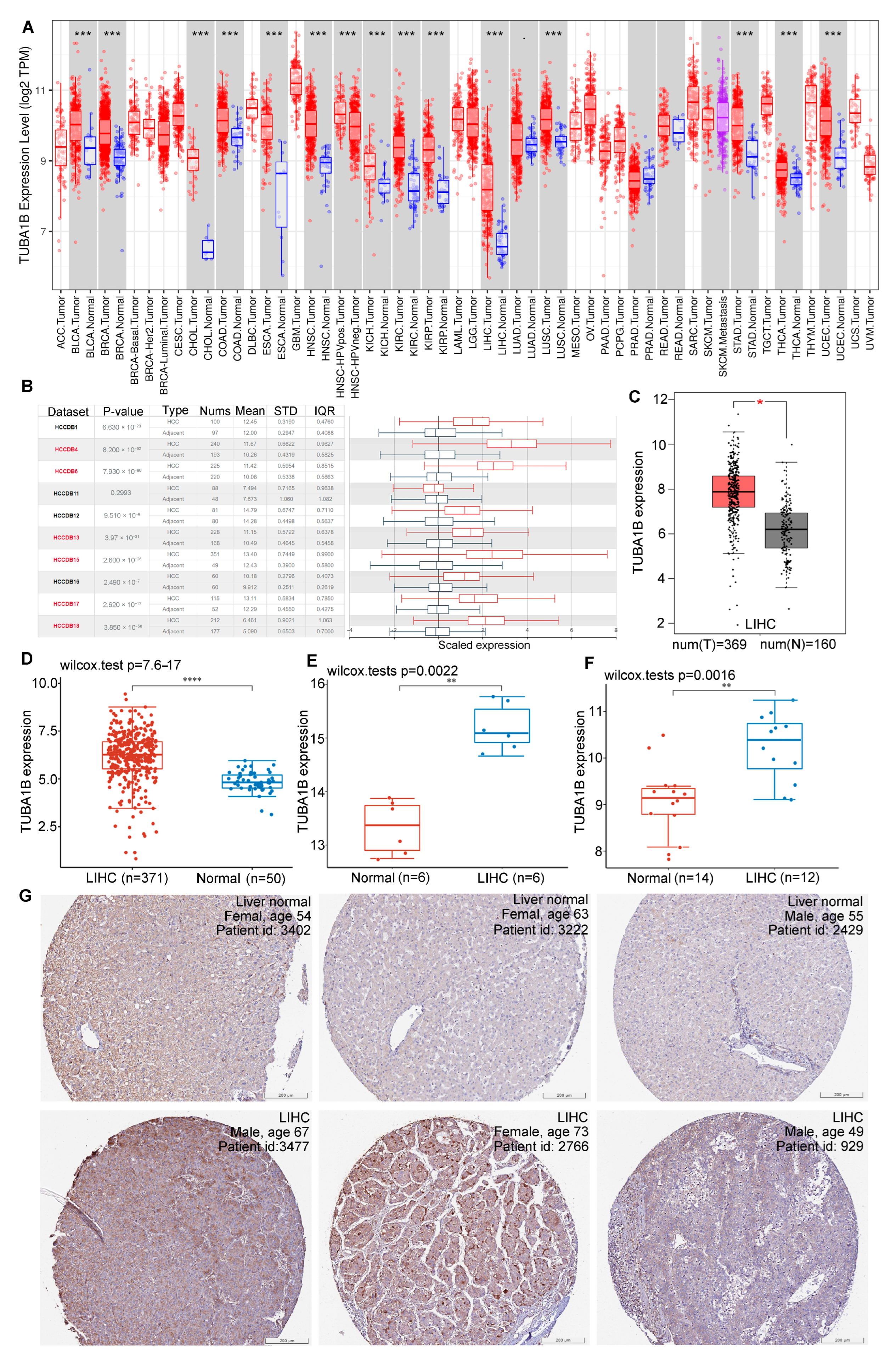
3.1. Differential Expression of TUBA1B in Normal and HCC Tissues
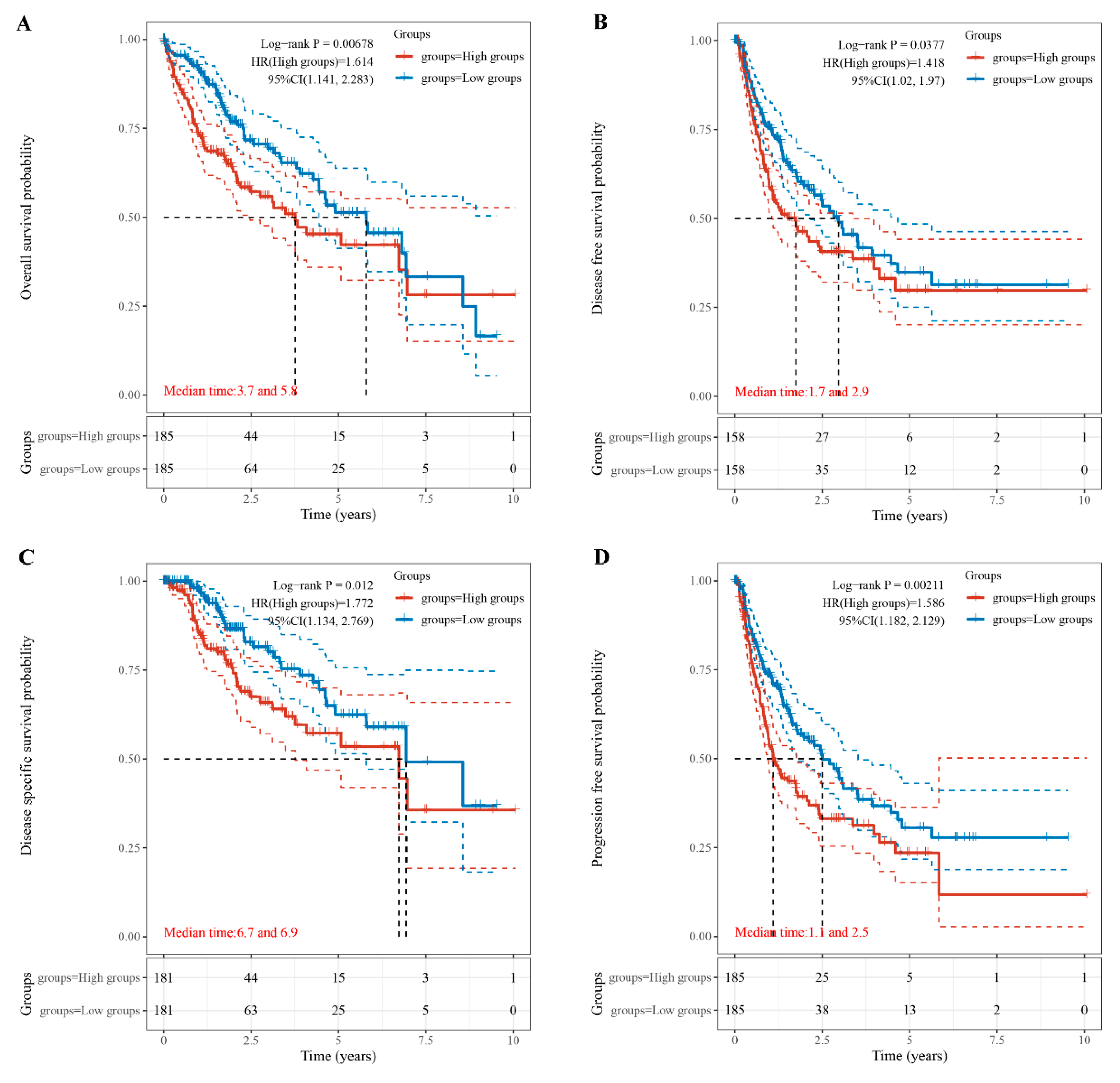
3.2. Relationship of TUBA1B Expression with OS, DFS, PFS, and DSS
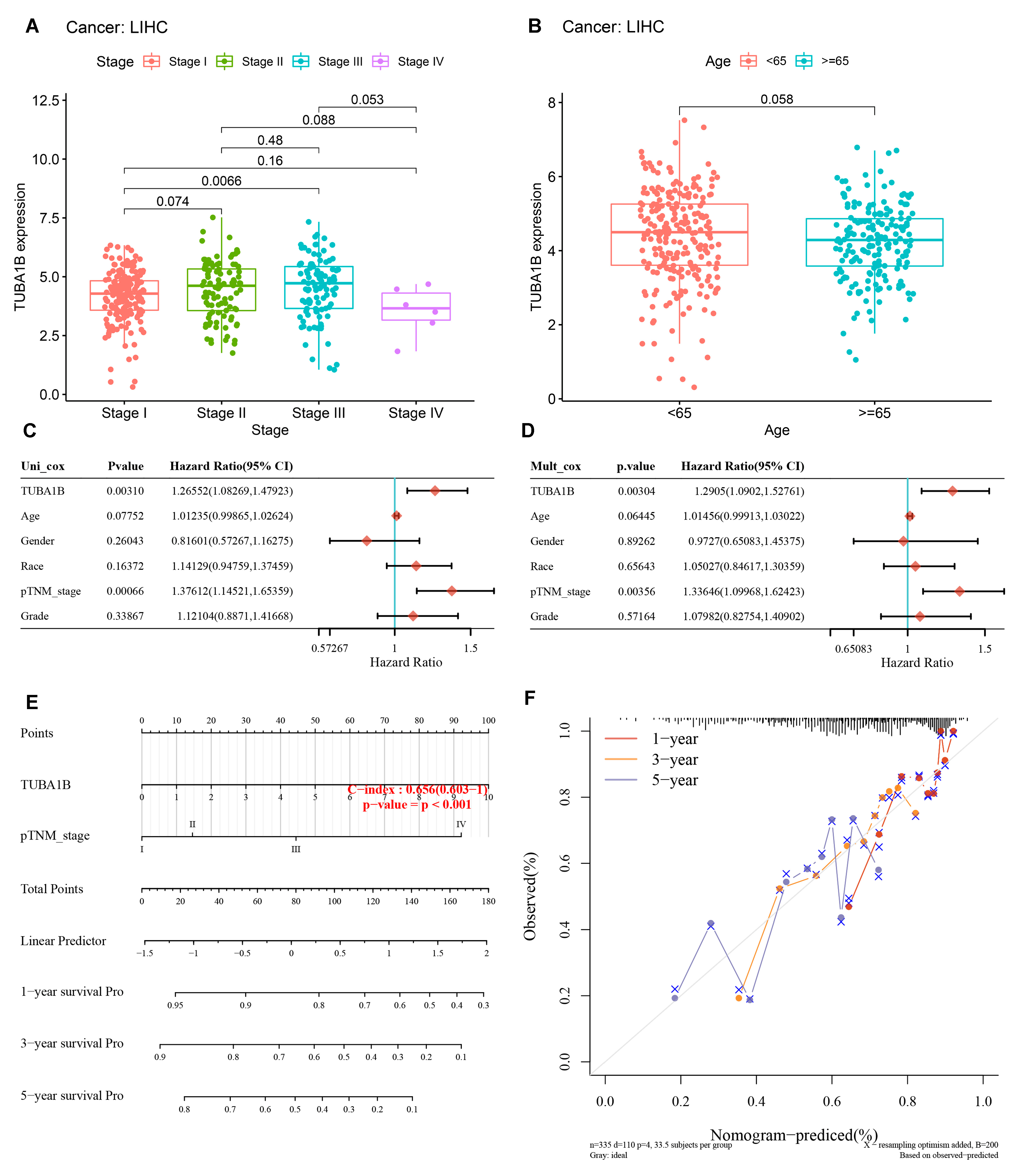
3.3. TUBA1B Correlated with Clinicopathology and Its Prognostic Potential
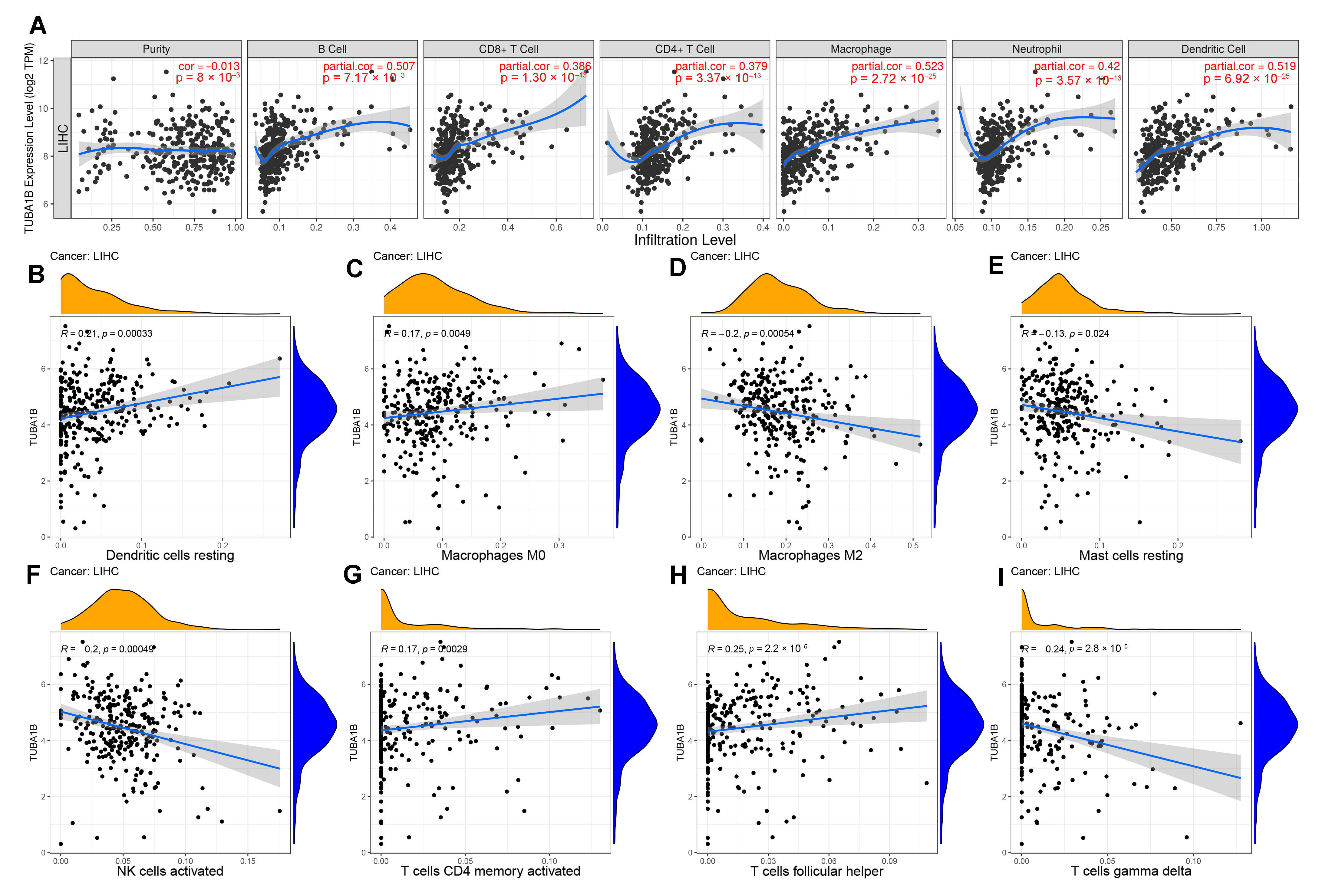
3.4. TUBA1B Expression Was Related to Immune Cell Infiltration in LIHC
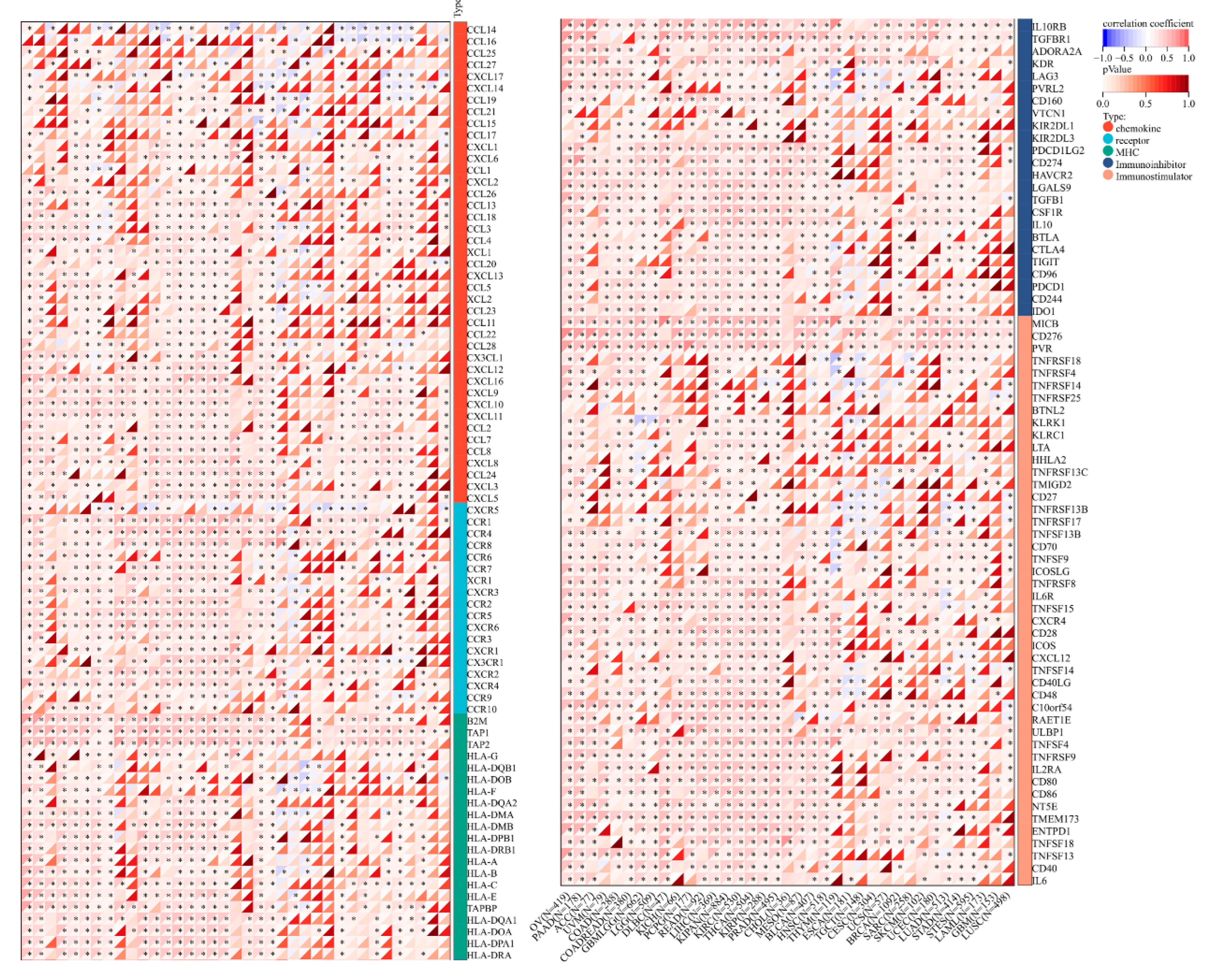
3.5. Correlation of TUBA1B Expression with Immune Checkpoints, Immune-Related Genes, and Patient Response to Immunotherapy
3.6. Construction of PPI Networks for TUBA1B, Analysis of GO Function Annotation, and KEGG Pathway Enrichment of TUBA1B
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bruix, J.; Han, K.H.; Gores, G.; Llovet, J.M.; Mazzaferro, V. Liver cancer: Approaching a personalized care. J. Hepatol. 2015, 62, S144–S156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Jin, T.; Zhu, Y.; Dai, C. Immune checkpoint therapy in liver cancer. J. Exp. Clin. Cancer Res. 2018, 37, 110. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhu, H.; Shen, Y.; Zhang, X.; He, X.; Xu, X. The Role of Non-Coding RNAs in the Sorafenib Resistance of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 696705. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Y.; Wang, D.S.; Lin, H.C.; Chen, X.X.; Yang, H.; Zheng, Y.; Li, Y.H. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int. J. Biol. Sci. 2020, 16, 2430–2441. [Google Scholar] [CrossRef]
- Akhmanova, A.; Maiato, H. Closing the tubulin detyrosination cycle. Science 2017, 358, 1381–1382. [Google Scholar] [CrossRef]
- Snelleksz, M.; Dean, B. Lower levels of tubulin alpha 1b in the frontal pole in schizophrenia supports a role for changed cytoskeletal dynamics in the aetiology of the disorder. Psychiatry Res. 2021, 303, 114096. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, J.; He, S.; Wan, C.; Shan, A.; Wang, Y.; Yu, L.; Liu, G.; Chen, K.; Shi, J.; et al. Increased α-tubulin1b expression indicates poor prognosis and resistance to chemotherapy in hepatocellular carcinoma. Dig. Dis. Sci. 2013, 58, 2713–2720. [Google Scholar] [CrossRef]
- Tian, Y.; Arai, E.; Makiuchi, S.; Tsuda, N.; Kuramoto, J.; Ohara, K.; Takahashi, Y.; Ito, N.; Ojima, H.; Hiraoka, N.; et al. Aberrant DNA methylation results in altered gene expression in non-alcoholic steatohepatitis-related hepatocellular carcinomas. J. Cancer Res. Clin. Oncol. 2020, 146, 2461–2477. [Google Scholar] [CrossRef]
- Xu, Q.Q.; Qin, L.T.; Liang, S.W.; Chen, P.; Gu, J.H.; Huang, Z.G.; Yang, X.; Gao, L.; Wang, S.S.; Luo, Y.G.; et al. The Expression and Potential Role of Tubulin Alpha 1b in Wilms’ Tumor. BioMed Res. Int. 2020, 2020, 9809347. [Google Scholar] [CrossRef]
- Hu, J.; Han, C.; Zhong, J.; Liu, H.; Liu, R.; Luo, W.; Chen, P.; Ling, F. Dynamic Network Biomarker of Pre-Exhausted CD8(+) T Cells Contributed to T Cell Exhaustion in Colorectal Cancer. Front. Immunol. 2021, 12, 691142. [Google Scholar] [CrossRef]
- Dou, Y.; Zhu, K.; Sun, Z.; Geng, X.; Fang, Q. Screening of disorders associated with osteosarcoma by integrated network analysis. Biosci. Rep. 2019, 39, BSR20190235. [Google Scholar] [CrossRef] [Green Version]
- Qin, D.; Zhao, Y.; Guo, Q.; Zhu, S.; Zhang, S.; Min, L. Detection of Pancreatic Ductal Adenocarcinoma by A qPCR-based Normalizer-free Circulating Extracellular Vesicles RNA Signature. J. Cancer 2021, 12, 1445–1454. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, H.; Zhang, X.; He, X.; Xu, X. Comprehensive analysis of pan-cancer reveals potential of ASF1B as a prognostic and immunological biomarker. Cancer Med. 2021, 10, 6897–6916. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, X.; Ye, Y.; Jian, Z.; Zhong, Y.; Gu, L.; Xiong, X. Pan-Cancer Analysis of PIMREG as a Biomarker for the Prognostic and Immunological Role. Front. Genet. 2021, 12, 687778. [Google Scholar] [CrossRef]
- Long, J.; Lin, J.; Wang, A.; Wu, L.; Zheng, Y.; Yang, X.; Wan, X.; Xu, H.; Chen, S.; Zhao, H. PD-1/PD-L blockade in gastrointestinal cancers: Lessons learned and the road toward precision immunotherapy. J. Hematol. Oncol. 2017, 10, 146. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Lian, Q.; Wang, S.; Zhang, G.; Wang, D.; Luo, G.; Tang, J.; Chen, L.; Gu, J. HCCDB: A Database of Hepatocellular Carcinoma Expression Atlas. Genom. Proteom. Bioinform. 2018, 16, 269–275. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Foerster, F.; Gairing, S.J.; Müller, L.; Galle, P.R. NAFLD-driven HCC: Safety and efficacy of current and emerging treatment options. J. Hepatol. 2022, 76, 446–457. [Google Scholar] [CrossRef]
- Ruf, B.; Heinrich, B.; Greten, T.F. Immunobiology and immunotherapy of HCC: Spotlight on innate and innate-like immune cells. Cell. Mol. Immunol. 2021, 18, 112–127. [Google Scholar] [CrossRef]
- Ringelhan, M.; Pfister, D.; O’Connor, T.; Pikarsky, E.; Heikenwalder, M. The immunology of hepatocellular carcinoma. Nat. Immunol. 2018, 19, 222–232. [Google Scholar] [CrossRef]
- Baig, M.S.; Roy, A.; Rajpoot, S.; Liu, D.; Savai, R.; Banerjee, S.; Kawada, M.; Faisal, S.M.; Saluja, R.; Saqib, U.; et al. Tumor-derived exosomes in the regulation of macrophage polarization. Inflamm. Res. 2020, 69, 435–451. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, H.; Wang, Y.; Brown, Z.J.; Xia, Y.; Huang, Z.; Shen, C.; Hu, Z.; Beane, J.; Ansa-Addo, E.A.; et al. Regulatory T-cell and neutrophil extracellular trap interaction contributes to carcinogenesis in non-alcoholic steatohepatitis. J. Hepatol. 2021, 75, 1271–1283. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef]
- Wang, D.Y.; Kamuda, K.; Montoya, G.; Mesa, P. The TRiC/CCT Chaperonin and Its Role in Uncontrolled Proliferation. Adv. Exp. Med. Biol. 2020, 1243, 21–40. [Google Scholar] [CrossRef]
- Gadadhar, S.; Bodakuntla, S.; Natarajan, K.; Janke, C. The tubulin code at a glance. J. Cell Sci. 2017, 130, 1347–1353. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Zhu, H.; Chen, B.; He, X.; Shen, Y.; Zhang, X.; Chen, W.; Liu, X.; Xu, Y.; Xu, X. Tubulin Alpha 1b Is Associated with the Immune Cell Infiltration and the Response of HCC Patients to Immunotherapy. Diagnostics 2022, 12, 858. https://doi.org/10.3390/diagnostics12040858
Hu X, Zhu H, Chen B, He X, Shen Y, Zhang X, Chen W, Liu X, Xu Y, Xu X. Tubulin Alpha 1b Is Associated with the Immune Cell Infiltration and the Response of HCC Patients to Immunotherapy. Diagnostics. 2022; 12(4):858. https://doi.org/10.3390/diagnostics12040858
Chicago/Turabian StyleHu, Xinyao, Hua Zhu, Biao Chen, Xiaoqin He, Yang Shen, Xiaoyu Zhang, Wenliang Chen, Xin Liu, Yangtao Xu, and Ximing Xu. 2022. "Tubulin Alpha 1b Is Associated with the Immune Cell Infiltration and the Response of HCC Patients to Immunotherapy" Diagnostics 12, no. 4: 858. https://doi.org/10.3390/diagnostics12040858
APA StyleHu, X., Zhu, H., Chen, B., He, X., Shen, Y., Zhang, X., Chen, W., Liu, X., Xu, Y., & Xu, X. (2022). Tubulin Alpha 1b Is Associated with the Immune Cell Infiltration and the Response of HCC Patients to Immunotherapy. Diagnostics, 12(4), 858. https://doi.org/10.3390/diagnostics12040858





