Nucleic Acid Biomarkers in Waldenström Macroglobulinemia and IgM-MGUS: Current Insights and Clinical Relevance
Abstract
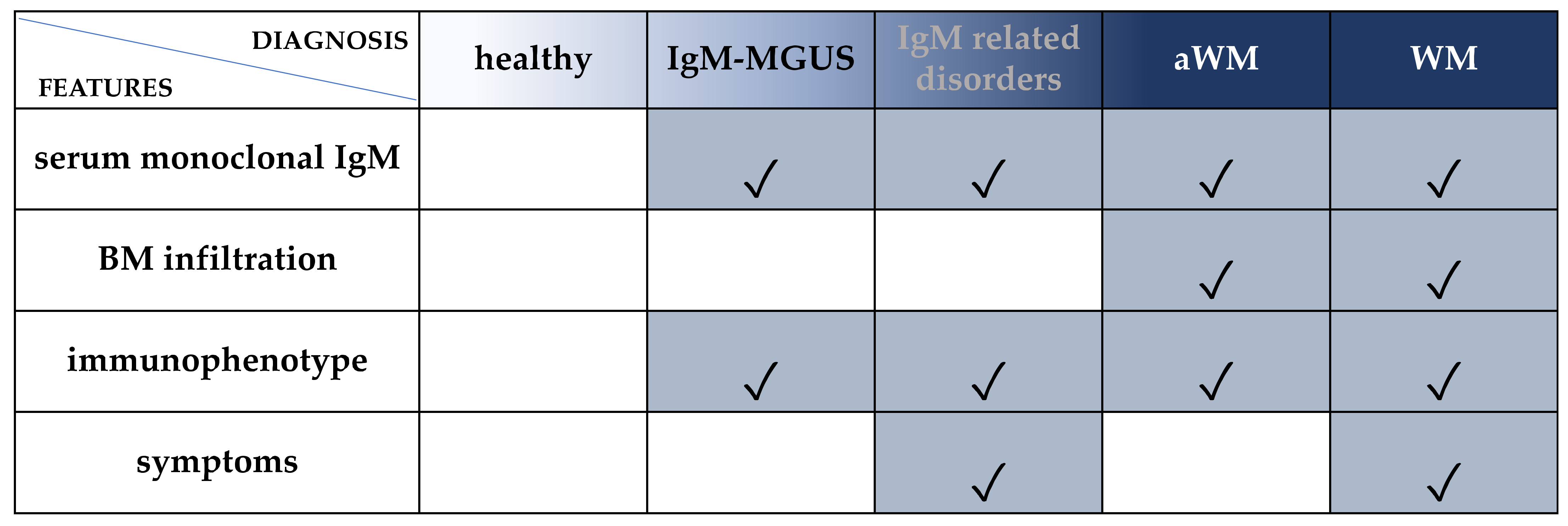
1. Introduction
2. DNA Biomarkers
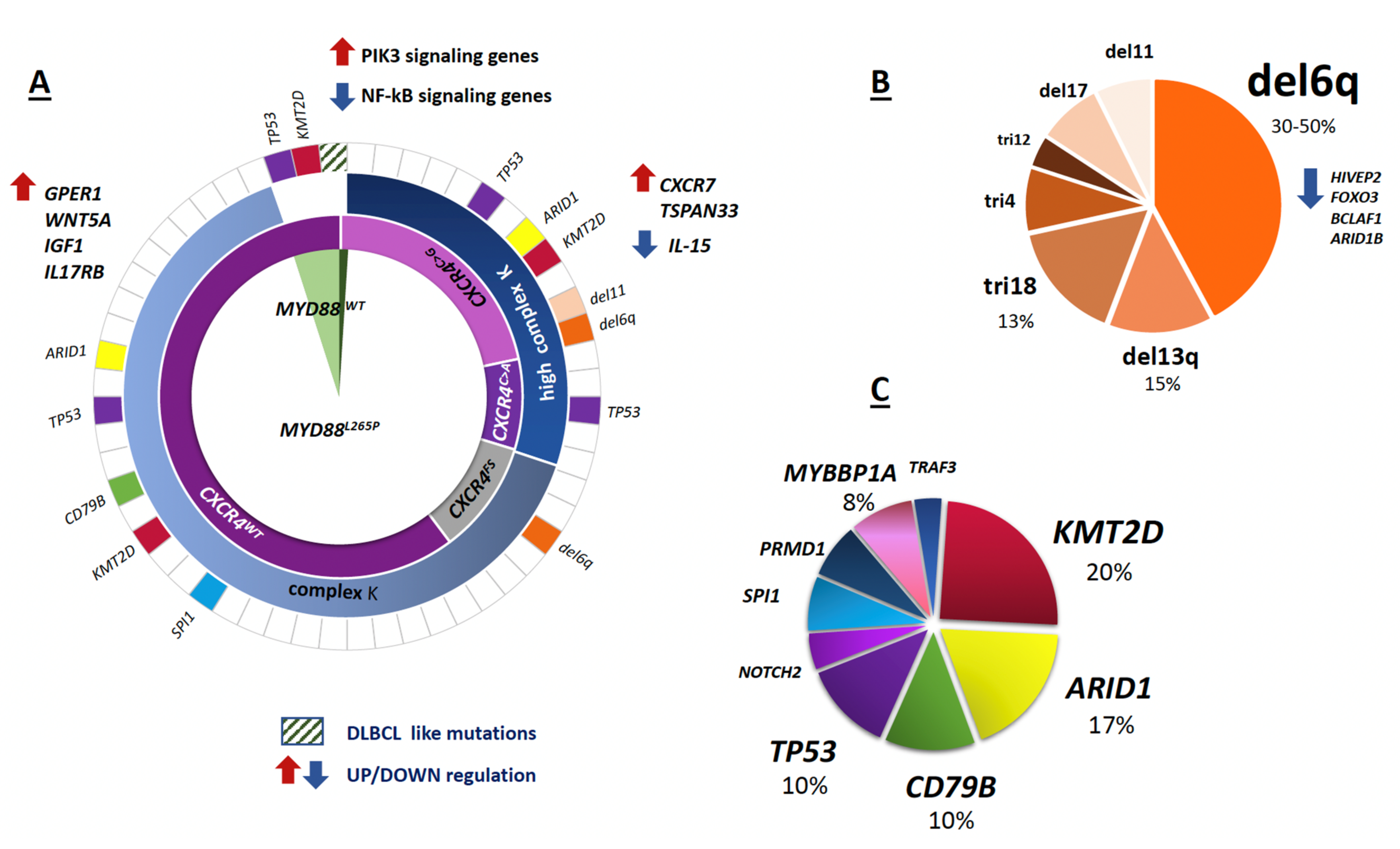
2.1. The Hallmark Genomic Alterations
2.1.1. MYD88L265P and CXCR4MUT
2.1.2. 6q21 Deletion
2.2. Infrequent DNA Mutations
2.3. Impact of Somatic Mutations on Outcome and Therapy Response
3. RNA Biomarkers
3.1. Coding mRNA
3.2. Non-Coding RNA
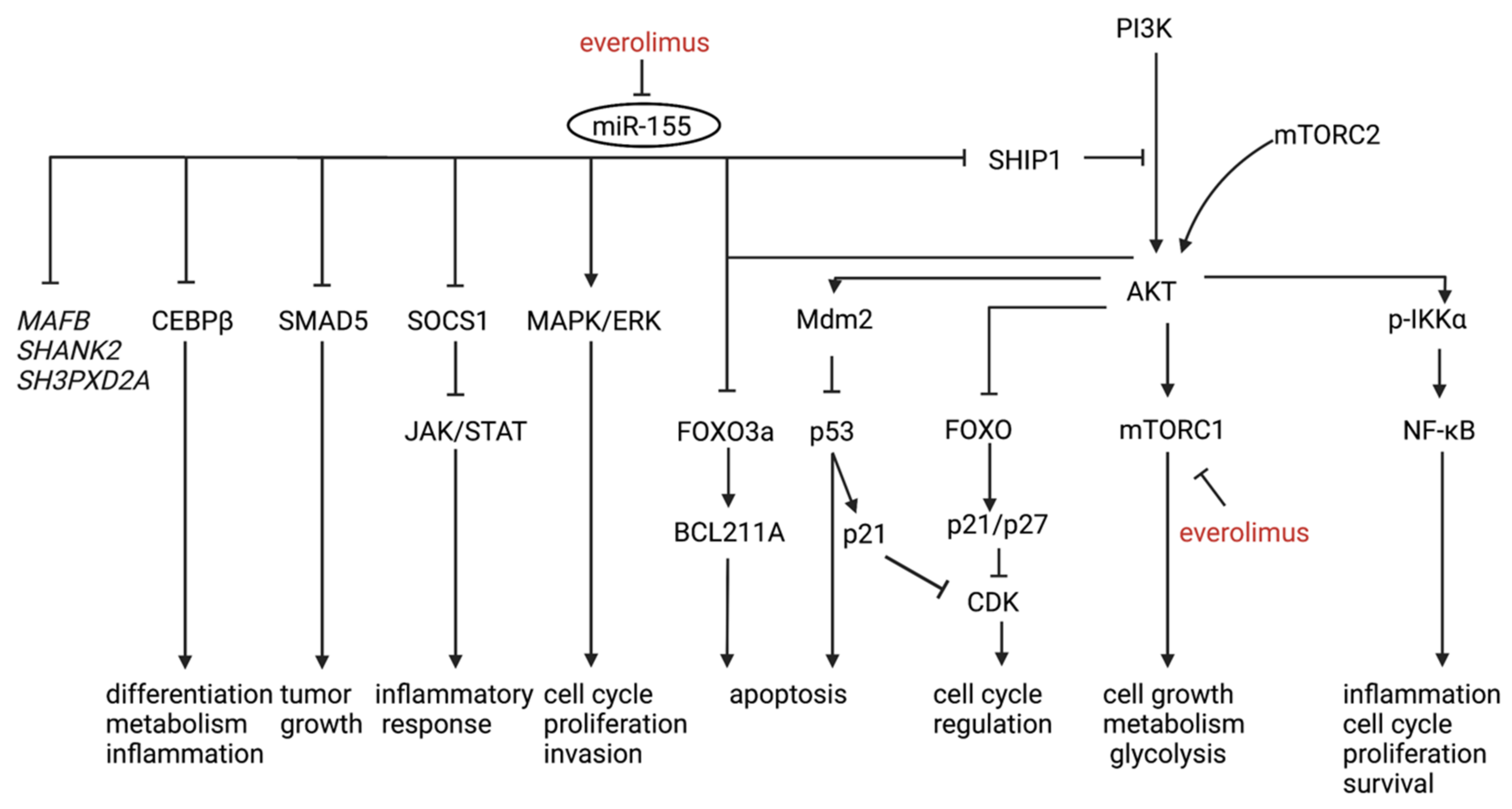
3.2.1. miRNA
3.2.2. Diagnostic Markers
3.2.3. Therapy Response and Prognostic Marker
3.2.4. miRNAs and Epigenetic Regulation
3.2.5. LncRNAs
3.2.6. CircRNAs
4. IgM-MGUS to WM Progression
5. Liquid Biopsy
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ziegler, A.; Koch, A.; Krockenberger, K.; Grosshennig, A. Personalized medicine using DNA biomarkers: A review. Hum. Genet. 2012, 131, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, T.; Garden, P.M.; Cohen, L. Single-molecule analysis of nucleic acid biomarkers—A review. Anal. Chim. Acta 2020, 1115, 61–85. [Google Scholar] [CrossRef] [PubMed]
- Braggio, E.; Philipsborn, C.; Novak, A.; Hodge, L.; Ansell, S.; Fonseca, R. Molecular pathogenesis of Waldenstrom’s macroglobulinemia. Haematologica 2012, 97, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Gertz, M. Waldenström macroglobulinemia: My way. Leuk. Lymphoma 2013, 54, 464–471. [Google Scholar] [CrossRef]
- Remstein, E.D.; Hanson, C.A.; Kyle, R.A.; Hodnefield, J.M.; Kurtin, P.J. Despite apparent morphologic and immunophenotypic heterogeneity, Waldenstrom’s macroglobulinemia is consistently composed of cells along a morphologic continuum of small lymphocytes, plasmacytoid lymphocytes, and plasma cells. Semin. Oncol. 2003, 30, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Kriangkum, J.; Taylor, B.J.; Treon, S.P.; Mant, M.J.; Belch, A.R.; Pilarski, L.M. Clonotypic IgM V/D/J sequence analysis in Waldenstrom macroglobulinemia suggests an unusual B-cell origin and an expansion of polyclonal B cells in peripheral blood. Blood 2004, 104, 2134–2142. [Google Scholar] [CrossRef]
- Chen, L.Y.; Keddie, S.; Lunn, M.P.; Bomsztyk, J.; Vitsaras, E.; Gupta, R.; D’Sa, S. IgM paraprotein-associated peripheral neuropathy: Small CD20-positive B-cell clones may predict a monoclonal gammopathy of neurological significance and rituximab responsiveness. Br. J. Haematol. 2020, 188, 511–515. [Google Scholar] [CrossRef]
- Leung, N.; Bridoux, F.; Nasr, S.H. Monoclonal Gammopathy of Renal Significance. N. Engl. J. Med. 2021, 384, 1931–1941. [Google Scholar] [CrossRef]
- Owen, R.G.; Treon, S.P.; Al-Katib, A.; Fonseca, R.; Greipp, P.R.; McMaster, M.L.; Morra, E.; Pangalis, G.A.; San Miguel, J.F.; Branagan, A.R.; et al. Clinicopathological definition of Waldenstrom’s macroglobulinemia: Consensus Panel Recommendations from the Second International Workshop on Waldenstrom’s Macroglobulinemia. Semin. Oncol. 2003, 30, 110–115. [Google Scholar] [CrossRef]
- Varettoni, M.; Zibellini, S.; Defrancesco, I.; Ferretti, V.V.; Rizzo, E.; Malcovati, L.; Gallì, A.; Della Porta, M.G.; Boveri, E.; Arcaini, L.; et al. Pattern of somatic mutations in patients with Waldenström macroglobulinemia or IgM monoclonal gammopathy of undetermined significance. Haematologica 2017, 102, 2077–2085. [Google Scholar] [CrossRef]
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood 2011, 117, 5019–5032. [Google Scholar] [CrossRef]
- Gertz, M.A. Waldenström macroglobulinemia: 2021 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2021, 96, 258–269. [Google Scholar] [CrossRef]
- Swerdlow, S.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.; Stein, H. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; IARC Publications: Lyon, France, 2008; Volume 2. [Google Scholar]
- Ansell, S.M.; Kyle, R.A.; Reeder, C.B.; Fonseca, R.; Mikhael, J.R.; Morice, W.G.; Bergsagel, P.L.; Buadi, F.K.; Colgan, J.P.; Dingli, D.; et al. Diagnosis and Management of Waldenström Macroglobulinemia: Mayo Stratification of Macroglobulinemia and Risk-Adapted Therapy (mSMART) Guidelines. Mayo Clin. Proc. 2010, 85, 824–833. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.-V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Kapoor, P.; Ansell, S.M.; Fonseca, R.; Chanan-Khan, A.; Kyle, R.A.; Kumar, S.K.; Mikhael, J.R.; Witzig, T.E.; Mauermann, M.; Dispenzieri, A.; et al. Diagnosis and Management of Waldenström Macroglobulinemia. JAMA Oncol. 2017, 3, 1257. [Google Scholar] [CrossRef]
- Maqbool, M.G.; Tam, C.S.; Morison, I.M.; Simpson, D.; Mollee, P.; Schneider, H.; Chan, H.; Juneja, S.; Harvey, Y.; Nath, L.; et al. A practical guide to laboratory investigations at diagnosis and follow up in Waldenström macroglobulinaemia: Recommendations from the Medical and Scientific Advisory Group, Myeloma Australia, the Pathology Sub-committee of the Lymphoma and Related Disease. Pathology 2020, 52, 167–178. [Google Scholar] [CrossRef]
- Pratt, G.; El-Sharkawi, D.; Kothari, J.; D’Sa, S.; Auer, R.; McCarthy, H.; Krishna, R.; Miles, O.; Kyriakou, C.; Owen, R. Guidelines on the diagnosis and management of Waldenström macroglobulinaemia—A British Society for Haematology guideline. Br. J. Haematol. 2022. [Google Scholar] [CrossRef]
- Swerdlow, S.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; IARC Publications: Lyon, France, 2017; ISBN 9789283244943. [Google Scholar]
- Morice, W.G.; Chen, D.; Kurtin, P.J.; Hanson, C.A.; McPhail, E.D. Novel immunophenotypic features of marrow lymphoplasmacytic lymphoma and correlation with Waldenström’s macroglobulinemia. Mod. Pathol. 2009, 22, 807–816. [Google Scholar] [CrossRef]
- Askari, E.; Rodriguez, S.; Garcia-Sanz, R. Waldenström’s Macroglobulinemia: An Exploration into the Pathology and Diagnosis of a Complex B-Cell Malignancy. J. Blood Med. 2021, 12, 795–807. [Google Scholar] [CrossRef]
- Kyle, R.A.; Larson, D.R.; Therneau, T.M.; Dispenzieri, A.; Kumar, S.; Cerhan, J.R.; Rajkumar, S.V. Long-Term Follow-up of Monoclonal Gammopathy of Undetermined Significance. N. Engl. J. Med. 2018, 378, 241–249. [Google Scholar] [CrossRef]
- Hobbs, M.; Fonder, A.; Hwa, Y.L. Waldenström Macroglobulinemia: Clinical Presentation, Diagnosis, and Management. J. Adv. Pract. Oncol. 2020, 11, 381–389. [Google Scholar] [CrossRef]
- Waldenström, J. Incipient myelomatosis or «essential» hyperglobulinemia with fibrinogenopenia-A new syndrome? Acta Med. Scand. 2009, 117, 216–247. [Google Scholar] [CrossRef]
- McMaster, M.L.; Goldin, L.R.; Bai, Y.; Ter-Minassian, M.; Boehringer, S.; Giambarresi, T.R.; Vasquez, L.G.; Tucker, M.A. Genomewide Linkage Screen for Waldenström Macroglobulinemia Susceptibility Loci in High-Risk Families. Am. J. Hum. Genet. 2006, 79, 695–701. [Google Scholar] [CrossRef]
- Nguyen-Khac, F.; Lambert, J.; Chapiro, E.; Grelier, A.; Mould, S.; Barin, C.; Daudignon, A.; Gachard, N.; Struski, S.; Henry, C.; et al. Chromosomal aberrations and their prognostic value in a series of 174 untreated patients with Waldenström’s macroglobulinemia. Haematologica 2013, 98, 649–654. [Google Scholar] [CrossRef]
- Treon, S.P.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Sheehy, P.; Manning, R.J.; Patterson, C.J.; Tripsas, C.; et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N. Engl. J. Med. 2012, 367, 826–833. [Google Scholar] [CrossRef]
- Hunter, Z.R.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Manning, R.J.; Tripsas, C.; Patterson, C.J.; Sheehy, P.; et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 2014, 123, 1637–1646. [Google Scholar] [CrossRef]
- Kastritis, E.; Morel, P.; Duhamel, A.; Gavriatopoulou, M.; Kyrtsonis, M.C.; Durot, E.; Symeonidis, A.; Laribi, K.; Hatjiharissi, E.; Ysebaert, L.; et al. A revised international prognostic score system for Waldenström’s macroglobulinemia. Leukemia 2019, 33, 2654–2661. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.J.; Garrido-Navas, M.C.; Diaz Mochon, J.J.; Cristofanilli, M.; Gil-Bazo, I.; Pauwels, P.; Malapelle, U.; Russo, A.; Lorente, J.A.; Ruiz-Rodriguez, A.J.; et al. Precision Prevention and Cancer Interception: The New Challenges of Liquid Biopsy. Cancer Discov. 2020, 10, 1635–1644. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K. Liquid Biopsy: Blood-Based Analyses of ctDNA and CTCs. Clin. Chem. 2021, 67, 1437–1439. [Google Scholar] [CrossRef]
- Qi, F.; Cao, Z.; Chen, B.; Chai, Y.; Lin, J.; Ye, J.; Wei, Y.; Liu, H.; Han-Zhang, H.; Mao, X.; et al. Liquid biopsy in extranodal NK/T-cell lymphoma: A prospective analysis of cell-free DNA genotyping and monitoring. Blood Adv. 2021, 5, 2505–2514. [Google Scholar] [CrossRef]
- Landgren, O.; Staudt, L. MYD88 L265P somatic mutation in IgM MGUS. N. Engl. J. Med. 2012, 367, 2255–2256. [Google Scholar] [CrossRef]
- Gachard, N.; Parrens, M.; Soubeyran, I.; Petit, B.; Marfak, A.; Rizzo, D.; Devesa, M.; Delage-Corre, M.; Coste, V.; Laforêt, M.P.; et al. IGHV gene features and MYD88 L265P mutation separate the three marginal zone lymphoma entities and Waldenström macroglobulinemia/lymphoplasmacytic lymphomas. Leukemia 2013, 27, 183–189. [Google Scholar] [CrossRef]
- Xu, L.; Hunter, Z.R.; Yang, G.; Zhou, Y.; Cao, Y.; Liu, X.; Morra, E.; Trojani, A.; Greco, A.; Arcaini, L.; et al. MYD88 L265P in Waldenström macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood 2013, 121, 2051–2058. [Google Scholar] [CrossRef]
- Ondrejka, S.L.; Lin, J.J.; Warden, D.W.; Durkin, L.; Cook, J.R.; Hsi, E.D. MYD88 L265P somatic mutation: Its usefulness in the differential diagnosis of bone marrow involvement by B-cell lymphoproliferative disorders. Am. J. Clin. Pathol. 2013, 140, 387–394. [Google Scholar] [CrossRef]
- Jiménez, C.; Sebastián, E.; Chillón, M.C.; Giraldo, P.; Mariano Hernández, J.; Escalante, F.; González-López, T.J.; Aguilera, C.; de Coca, A.G.; Murillo, I.; et al. MYD88 L265P is a marker highly characteristic of, but not restricted to, Waldenström’s macroglobulinemia. Leukemia 2013, 27, 1722–1728. [Google Scholar] [CrossRef]
- Poulain, S.; Roumier, C.; Decambron, A.; Renneville, A.; Herbaux, C.; Bertrand, E.; Tricot, S.; Daudignon, A.; Galiègue-Zouitina, S.; Soenen, V.; et al. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood 2013, 121, 4504–4511. [Google Scholar] [CrossRef]
- Willenbacher, W.; Willenbacher, E.; Brunner, A.; Manzl, C. Improved accuracy of discrimination between IgM Multiple Myeloma and Waldenström Macroglobulinaemia by testing for MYD88 L265P mutations. Br. J. Haematol. 2013, 161, 902–904. [Google Scholar] [CrossRef]
- Mori, N.; Ohwashi, M.; Yoshinaga, K.; Mitsuhashi, K.; Tanaka, N.; Teramura, M.; Okada, M.; Shiseki, M.; Tanaka, J.; Motoji, T. L265P Mutation of the MYD88 Gene Is Frequent in Waldenström’s Macroglobulinemia and Its Absence in Myeloma. PLoS ONE 2013, 8, e80088. [Google Scholar] [CrossRef]
- Varettoni, M.; Arcaini, L.; Zibellini, S.; Boveri, E.; Rattotti, S.; Riboni, R.; Corso, A.; Orlandi, E.; Bonfichi, M.; Gotti, M.; et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom’s macroglobulinemia and related lymphoid neoplasms. Blood 2013, 121, 2522–2528. [Google Scholar] [CrossRef]
- Argentou, N.; Vassilopoulos, G.; Ioannou, M.; Germenis, A.E.; Speletas, M. Rapid detection of MYD88-L265P mutation by PCR-RFLP in B-cell lymphoproliferative disorders. Leukemia 2014, 28, 447–449. [Google Scholar] [CrossRef]
- Capaldi, I.B.; May, A.M.; Schmitt-Graeff, A.; Follo, M.; Aumann, K.; Kayser, G.; Perazzo, J.C.; Werner, M.; Fisch, P. Detection of MYD88 L265P mutations in formalin-fixed and decalcified BM biopsies from patients with lymphoplasmacytic lymphoma. Exp. Mol. Pathol. 2014, 97, 57–65. [Google Scholar] [CrossRef]
- Petrikkos, L.; Kyrtsonis, M.-C.; Roumelioti, M.; Georgiou, G.; Efthymiou, A.; Tzenou, T.; Panayiotidis, P. Clonotypic analysis of immunoglobulin heavy chain sequences in patients with Waldenström’s macroglobulinemia: Correlation with MYD88 L265P somatic mutation status, clinical features, and outcome. Biomed Res. Int. 2014, 2014, 809103. [Google Scholar] [CrossRef]
- Ansell, S.M.; Hodge, L.S.; Secreto, F.J.; Manske, M.; Braggio, E.; Price-Troska, T.; Ziesmer, S.; Li, Y.; Johnson, S.H.; Hart, S.N.; et al. Activation of TAK1 by MYD88 L265P drives malignant B-cell Growth in non-Hodgkin lymphoma. Blood Cancer J. 2014, 4, e183. [Google Scholar] [CrossRef]
- Xu, L.; Hunter, Z.R.; Yang, G.; Cao, Y.; Liu, X.; Manning, R.; Tripsas, C.; Chen, J.; Patterson, C.J.; Kluk, M.; et al. Detection of MYD88 L265P in peripheral blood of patients with Waldenström’s Macroglobulinemia and IgM monoclonal gammopathy of undetermined significance. Leukemia 2014, 28, 1698–1704. [Google Scholar] [CrossRef]
- Treon, S.P.; Cao, Y.; Xu, L.; Yang, G.; Liu, X.; Hunter, Z.R. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenström macroglobulinemia. Blood 2014, 123, 2791–2796. [Google Scholar] [CrossRef]
- Patkar, N.; Subramanian, P.G.; Deshpande, P.; Ghodke, K.; Tembhare, P.; Mascarenhas, R.; Muranjan, A.; Chaudhary, S.; Bagal, B.; Gujral, S.; et al. MYD88 mutant lymphoplasmacytic lymphoma/Waldenström macroglobulinemia has distinct clinical and pathological features as compared to its mutation negative counterpart. Leuk. Lymphoma 2015, 56, 420–425. [Google Scholar] [CrossRef]
- Schmidt, J.; Federmann, B.; Schindler, N.; Steinhilber, J.; Bonzheim, I.; Fend, F.; Quintanilla-Martinez, L. MYD88 L265P and CXCR4 mutations in lymphoplasmacytic lymphoma identify cases with high disease activity. Br. J. Haematol. 2015, 169, 795–803. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Lee, S.-T.; Kim, H.-Y.; Park, C.-H.; Kim, H.-J.; Kim, J.-W.; Kim, S.J.; Kim, W.S.; Kim, S.-H. Detection of MYD88 L265P in patients with lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia and other B-cell non-Hodgkin lymphomas. Blood Res. 2016, 51, 181–186. [Google Scholar] [CrossRef]
- Burnworth, B.; Wang, Z.; Singleton, T.P.; Bennington, A.; Fritschle, W.; Bennington, R.; Brodersen, L.E.; Wells, D.A.; Loken, M.R.; Zehentner, B.K. Clone-specific MYD88 L265P and CXCR4 mutation status can provide clinical utility in suspected Waldenström macroglobulinemia/lymphoplasmacytic lymphoma. Leuk. Res. 2016, 51, 41–48. [Google Scholar] [CrossRef]
- Correa, J.G.; Cibeira, M.T.; Tovar, N.; Isola, I.; Pedrosa, F.; Díaz, T.; Lozano, E.; Magnano, L.; Rosiñol, L.; Bladé, J.; et al. Prevalence and prognosis implication of MYD88 L265P mutation in IgM monoclonal gammopathy of undetermined significance and smouldering Waldenström macroglobulinaemia. Br. J. Haematol. 2017, 179, 849–851. [Google Scholar] [CrossRef]
- Baer, C.; Dicker, F.; Kern, W.; Haferlach, T.; Haferlach, C. Genetic characterization of MYD88-mutated lymphoplasmacytic lymphoma in comparison with MYD88-mutated chronic lymphocytic leukemia. Leukemia 2017, 31, 1355–1362. [Google Scholar] [CrossRef]
- Paludo, J.; Abeykoon, J.P.; Kumar, S.; Shreders, A.; Ailawadhi, S.; Gertz, M.A.; Kourelis, T.; King, R.L.; Reeder, C.B.; Leung, N.; et al. Dexamethasone, rituximab and cyclophosphamide for relapsed and/or refractory and treatment-naïve patients with Waldenstrom macroglobulinemia. Br. J. Haematol. 2017, 179, 98–105. [Google Scholar] [CrossRef]
- Cao, X.-X.; Meng, Q.; Cai, H.; He, T.-H.; Zhang, C.-L.; Su, W.; Sun, J.; Li, Y.; Xu, W.; Zhou, D.-B.; et al. Detection of MYD88 L265P and WHIM-like CXCR4 mutation in patients with IgM monoclonal gammopathy related disease. Ann. Hematol. 2017, 96, 971–976. [Google Scholar] [CrossRef]
- Abeykoon, J.P.; Paludo, J.; King, R.L.; Ansell, S.M.; Gertz, M.A.; LaPlant, B.R.; Halvorson, A.E.; Gonsalves, W.I.; Dingli, D.; Fang, H.; et al. MYD88 mutation status does not impact overall survival in Waldenström macroglobulinemia. Am. J. Hematol. 2018, 93, 187–194. [Google Scholar] [CrossRef]
- Drandi, D.; Genuardi, E.; Dogliotti, I.; Ferrante, M.; Jiménez, C.; Guerrini, F.; Lo Schirico, M.; Mantoan, B.; Muccio, V.; Lia, G.; et al. Highly sensitive MYD88L265P mutation detection by droplet digital PCR in Waldenström Macroglobulinemia. Haematologica 2018, 103, 1029–1037. [Google Scholar] [CrossRef]
- Vinarkar, S.; Arora, N.; Chowdhury, S.S.; Saha, K.; Pal, B.; Parihar, M.; Radhakrishnan, V.S.; Chakrapani, A.; Bhartia, S.; Bhave, S.; et al. MYD88 and CXCR4 Mutation Profiling in Lymphoplasmacytic Lymphoma/Waldenstrom’s Macroglobulinaemia. Indian J. Hematol. Blood Transfus. 2019, 35, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Ohwada, C.; Takeuchi, M.; Takeda, Y.; Tsukamoto, S.; Mimura, N.; Nagisa, O.-H.; Sugita, Y.; Tanaka, H.; Wakita, H.; et al. Detection of MYD88 L265P mutation by next-generation deep sequencing in peripheral blood mononuclear cells of Waldenström’s macroglobulinemia and IgM monoclonal gammopathy of undetermined significance. PLoS ONE 2019, 14, e0221941. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Y.; Jia, M.-N.; Cai, H.; Qiu, Y.; Zhou, D.-B.; Li, J.; Cao, X.-X. Detection of the MYD88L265P and CXCR4S338X mutations by cell-free DNA in Waldenström macroglobulinemia. Ann. Hematol. 2020, 99, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gali, V.L.; Xu-Monette, Z.Y.; Sano, D.; Thomas, S.K.; Weber, D.M.; Zhu, F.; Fang, X.; Deng, M.; Zhang, M.; et al. Molecular and genetic biomarkers implemented from next-generation sequencing provide treatment insights in clinical practice for Waldenström macroglobulinemia. Neoplasia 2021, 23, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Kofides, A.; Hunter, Z.R.; Xu, L.; Tsakmaklis, N.; Demos, M.G.; Munshi, M.; Liu, X.; Guerrera, M.L.; Leventoff, C.R.; White, T.P.; et al. Diagnostic Next-generation Sequencing Frequently Fails to Detect MYD88L265P in Waldenström Macroglobulinemia. HemaSphere 2021, 5, e624. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef]
- Tan, Y.; Kagan, J.C. Innate Immune Signaling Organelles Display Natural and Programmable Signaling Flexibility. Cell 2019, 177, 384–398.e11. [Google Scholar] [CrossRef]
- Kagan, J.C.; Magupalli, V.G.; Wu, H. SMOCs: Supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 2014, 14, 821–826. [Google Scholar] [CrossRef]
- Balka, K.R.; Nardo, D. Understanding early TLR signaling through the Myddosome. J. Leukoc. Biol. 2019, 105, 339–351. [Google Scholar] [CrossRef]
- Deliz-Aguirre, R.; Cao, F.; Gerpott, F.H.U.; Auevechanichkul, N.; Chupanova, M.; Mun, Y.; Ziska, E.; Taylor, M.J. MyD88 oligomer size functions as a physical threshold to trigger IL1R Myddosome signaling. J. Cell Biol. 2021, 220, e202012071. [Google Scholar] [CrossRef]
- Wang, J.Q.; Jeelall, Y.S.; Beutler, B.; Horikawa, K.; Goodnow, C.C. Consequences of the recurrent MYD88L265P somatic mutation for B cell tolerance. J. Exp. Med. 2014, 211, 413–426. [Google Scholar] [CrossRef]
- O’Carroll, A.; Chauvin, B.; Brown, J.W.P.; Meagher, A.; Coyle, J.; Schill, J.; Bhumkhar, A.; Hunter, D.J.B.; Ve, T.; Kobe, B.; et al. Pathological mutations differentially affect the self-assembly and polymerisation of the innate immune system signalling adaptor molecule MyD88. BMC Biol. 2018, 16, 149. [Google Scholar] [CrossRef]
- Yang, G.; Buhrlage, S.J.; Tan, L.; Liu, X.; Chen, J.; Xu, L.; Tsakmaklis, N.; Chen, J.G.; Patterson, C.J.; Brown, J.R.; et al. HCK is a survival determinant transactivated by mutated MYD88, and a direct target of ibrutinib. Blood 2016, 127, 3237–3252. [Google Scholar] [CrossRef]
- Manček-Keber, M.; Lainšček, D.; Benčina, M.; Chen, J.G.; Romih, R.; Hunter, Z.R.; Treon, S.P.; Jerala, R. Extracellular vesicle–mediated transfer of constitutively active MyD88L265P engages MyD88wt and activates signaling. Blood 2018, 131, 1720–1729. [Google Scholar] [CrossRef]
- Varettoni, M.; Zibellini, S.; Arcaini, L.; Boveri, E.; Rattotti, S.; Pascutto, C.; Mangiacavalli, S.; Gotti, M.; Pochintesta, L.; Paulli, M.; et al. MYD88 (L265P) mutation is an independent risk factor for progression in patients with IgM monoclonal gammopathy of undetermined significance. Blood 2013, 122, 2284–2285. [Google Scholar] [CrossRef]
- Sewastianik, T.; Guerrera, M.L.; Adler, K.; Dennis, P.S.; Wright, K.; Shanmugam, V.; Huang, Y.; Tanton, H.; Jiang, M.; Kofides, A.; et al. Human MYD88L265P is insufficient by itself to drive neoplastic transformation in mature mouse B cells. Blood Adv. 2019, 3, 3360–3374. [Google Scholar] [CrossRef]
- Schmidt, K.; Sack, U.; Graf, R.; Winkler, W.; Popp, O.; Mertins, P.; Sommermann, T.; Kocks, C.; Rajewsky, K. B-Cell-Specific Myd88 L252P Expression Causes a Premalignant Gammopathy Resembling IgM MGUS. Front. Immunol. 2020, 11, 602868. [Google Scholar] [CrossRef]
- Rodriguez, S.; Celay, J.; Goicoechea, I.; Jimenez, C.; Botta, C.; Garcia-Barchino, M.-J.; Garces, J.-J.; Larrayoz, M.; Santos, S.; Alignani, D.; et al. Preneoplastic somatic mutations including MYD88 L265P in lymphoplasmacytic lymphoma. Sci. Adv. 2022, 8, eabl4644. [Google Scholar] [CrossRef]
- Treon, S.P.; Gustine, J.; Xu, L.; Manning, R.J.; Tsakmaklis, N.; Demos, M.; Meid, K.; Guerrera, M.L.; Munshi, M.; Chan, G.; et al. MYD88 wild-type Waldenstrom Macroglobulinaemia: Differential diagnosis, risk of histological transformation, and overall survival. Br. J. Haematol. 2018, 180, 374–380. [Google Scholar] [CrossRef]
- Hunter, Z.R.; Xu, L.; Tsakmaklis, N.; Demos, M.G.; Kofides, A.; Jimenez, C.; Chan, G.G.; Chen, J.; Liu, X.; Munshi, M.; et al. Insights into the genomic landscape of MYD88 wild-Type Waldenström macroglobulinemia. Blood Adv. 2018, 2, 2937–2946. [Google Scholar] [CrossRef]
- Guerrera, M.L.; Tsakmaklis, N.; Xu, L.; Yang, G.; Demos, M.; Kofides, A.; Chan, G.G.; Manning, R.J.; Liu, X.; Chen, J.G.; et al. MYD88 mutated and wild-type Waldenström’s Macroglobulinemia: Characterization of chromosome 6q gene losses and their mutual exclusivity with mutations in CXCR4. Haematologica 2018, 103, e408–e411. [Google Scholar] [CrossRef]
- Poulain, S.; Roumier, C.; Venet-Caillault, A.; Figeac, M.; Herbaux, C.; Marot, G.; Doye, E.; Bertrand, E.; Geffroy, S.; Lepretre, F.; et al. Genomic Landscape of CXCR4 Mutations in Waldenstro m Macroglobulinemia. Clin. Cancer Res. 2016, 22, 1480–1488. [Google Scholar] [CrossRef]
- Castillo, J.J.; Xu, L.; Gustine, J.N.; Keezer, A.; Meid, K.; Dubeau, T.E.; Liu, X.; Demos, M.G.; Kofides, A.; Tsakmaklis, N.; et al. CXCR4 mutation subtypes impact response and survival outcomes in patients with Waldenström macroglobulinaemia treated with ibrutinib. Br. J. Haematol. 2019, 187, 356–363. [Google Scholar] [CrossRef]
- Pozzobon, T.; Goldoni, G.; Viola, A.; Molon, B. CXCR4 signaling in health and disease. Immunol. Lett. 2016, 177, 6–15. [Google Scholar] [CrossRef]
- Xu, L.; Hunter, Z.R.; Tsakmaklis, N.; Cao, Y.; Yang, G.; Chen, J.; Liu, X.; Kanan, S.; Castillo, J.J.; Tai, Y.-T.; et al. Clonal architecture of CXCR4 WHIM-like mutations in Waldenström Macroglobulinaemia. Br. J. Haematol. 2016, 172, 735–744. [Google Scholar] [CrossRef]
- Milanesi, S.; Locati, M.; Borroni, E.M. Aberrant CXCR4 Signaling at Crossroad of WHIM Syndrome and Waldenstrom’s Macroglobulinemia. Int. J. Mol. Sci. 2020, 21, 5696. [Google Scholar] [CrossRef]
- Kaiser, L.M.; Hunter, Z.R.; Treon, S.P.; Buske, C. CXCR4 in Waldenström’s Macroglobulinema: Chances and challenges. Leukemia 2021, 35, 333–345. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Jimenez, C.; Maiso, P.; Moschetta, M.; Mishima, Y.; Aljawai, Y.; Sahin, I.; Kuhne, M.; Cardarelli, P.; et al. C1013G/CXCR4 acts as a driver mutation of tumor progression and modulator of drug resistance in lymphoplasmacytic lymphoma. Blood 2014, 123, 4120–4131. [Google Scholar] [CrossRef]
- Cao, Y.; Hunter, Z.R.; Liu, X.; Xu, L.; Yang, G.; Chen, J.; Patterson, C.J.; Tsakmaklis, N.; Kanan, S.; Rodig, S.; et al. The WHIM-like CXCR4(S338X) somatic mutation activates AKT and ERK, and promotes resistance to ibrutinib and other agents used in the treatment of Waldenstrom’s Macroglobulinemia. Leukemia 2015, 29, 169–176. [Google Scholar] [CrossRef]
- Gustine, J.N.; Xu, L.; Yang, G.; Liu, X.; Kofides, A.; Tsakmaklis, N.; Munshi, M.; Demos, M.; Guerrera, M.L.; Meid, K.; et al. Bone marrow involvement and subclonal diversity impairs detection of mutated CXCR4 by diagnostic next-generation sequencing in Waldenström macroglobulinaemia. Br. J. Haematol. 2021, 194, 730–733. [Google Scholar] [CrossRef]
- Jiménez, C.; Prieto-Conde, M.I.; García-Álvarez, M.; Alcoceba, M.; Escalante, F.; Del Carmen Chillón, M.; García de Coca, A.; Balanzategui, A.; Cantalapiedra, A.; Aguilar, C.; et al. Unraveling the heterogeneity of IgM monoclonal gammopathies: A gene mutational and gene expression study. Ann. Hematol. 2018, 97, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Ballester, L.Y.; Loghavi, S.; Kanagal-Shamanna, R.; Barkoh, B.A.; Lin, P.; Medeiros, L.J.; Luthra, R.; Patel, K.P. Clinical Validation of a CXCR4 Mutation Screening Assay for Waldenstrom Macroglobulinemia. Clin. Lymphoma. Myeloma Leuk. 2016, 16, 395–403.e1. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, C.; Xu, L.; Tsakmaklis, N.; Demos, M.G.; Kofides, A.; Chan, G.G.; Guerrera, M.L.; Chen, J.G.; Liu, X.; Munshi, M.; et al. Comparative genomics of CXCR4MUT and CXCR4WT single cells in Waldenström’s macroglobulinemia. Blood Adv. 2020, 4, 4550–4553. [Google Scholar] [CrossRef] [PubMed]
- Alegría-Landa, V.; Prieto-Torres, L.; Santonja, C.; Córdoba, R.; Manso, R.; Requena, L.; Rodríguez-Pinilla, S.M. MYD88 L265P mutation in cutaneous involvement by Waldenström macroglobulinemia. J. Cutan. Pathol. 2017, 44, 625–631. [Google Scholar] [CrossRef]
- Gustine, J.; Meid, K.; Xu, L.; Hunter, Z.R.; Castillo, J.J.; Treon, S.P. To select or not to select? The role of B-cell selection in determining the MYD88 mutation status in Waldenström Macroglobulinaemia. Br. J. Haematol. 2017, 176, 822–824. [Google Scholar] [CrossRef]
- Poulain, S.; Boyle, E.M.; Roumier, C.; Demarquette, H.; Wemeau, M.; Geffroy, S.; Herbaux, C.; Bertrand, E.; Hivert, B.; Terriou, L.; et al. MYD88 L265P mutation contributes to the diagnosis of Bing Neel syndrome. Br. J. Haematol. 2014, 167, 506–513. [Google Scholar] [CrossRef]
- Ferrante, M.; Furlan, D.; Zibellini, S.; Borriero, M.; Candido, C.; Sahnane, N.; Uccella, S.; Genuardi, E.; Alessandria, B.; Bianchi, B.; et al. MYD88L265P Detection in IgM Monoclonal Gammopathies: Methodological Considerations for Routine Implementation. Diagnostics 2021, 11, 779. [Google Scholar] [CrossRef]
- Paiva, B.; Corchete, L.A.; Vidriales, M.-B.; Garcia-Sanz, R.; Perez, J.J.; Aires-Mejia, I.; Sanchez, M.-L.; Barcena, P.; Alignani, D.; Jimenez, C.; et al. The cellular origin and malignant transformation of Waldenstrom macroglobulinemia. Blood 2015, 125, 2370–2380. [Google Scholar] [CrossRef]
- Ocio, E.M.; Schop, R.F.J.; Gonzalez, B.; Van Wier, S.A.; Hernandez-Rivas, J.M.; Gutierrez, N.C.; Garcia-Sanz, R.; Moro, M.J.; Aguilera, C.; Hernandez, J.; et al. 6q deletion in Waldenström macroglobulinemia is associated with features of adverse prognosis. Br. J. Haematol. 2007, 136, 80–86. [Google Scholar] [CrossRef]
- Chang, H.; Qi, C.; Trieu, Y.; Jiang, A.; Young, K.H.; Chesney, A.; Jani, P.; Wang, C.; Reece, D.; Chen, C. Prognostic Relevance of 6q Deletion in Waldenström’s Macroglobulinemia: A Multicenter Study. Clin. Lymphoma Myeloma 2009, 9, 36–38. [Google Scholar] [CrossRef]
- García-Sanz, R.; Dogliotti, I.; Zaccaria, G.M.; Ocio, E.M.; Rubio, A.; Murillo, I.; Escalante, F.; Aguilera, C.; García-Mateo, A.; García de Coca, A.; et al. 6q deletion in Waldenström macroglobulinaemia negatively affects time to transformation and survival. Br. J. Haematol. 2021, 192, 843–852. [Google Scholar] [CrossRef]
- Hunter, Z.R.; Xu, L.; Yang, G.; Tsakmaklis, N.; Vos, J.M.; Liu, X.; Chen, J.J.G.; Manning, R.J.; Chen, J.J.G.; Brodsky, P.; et al. Transcriptome sequencing reveals a profile that corresponds to genomic variants in Waldenström macroglobulinemia. Blood 2016, 128, 827–838. [Google Scholar] [CrossRef]
- Schop, R.F.J.; Van Wier, S.A.; Xu, R.; Ghobrial, I.; Ahmann, G.J.; Greipp, P.R.; Kyle, R.A.; Dispenzieri, A.; Lacy, M.Q.; Rajkumar, S.V.; et al. 6q deletion discriminates Waldenström macroglobulinemia from IgM monoclonal gammopathy of undetermined significance. Cancer Genet. Cytogenet. 2006, 169, 150–153. [Google Scholar] [CrossRef]
- Terré, C.; Nguyen-Khac, F.; Barin, C.; Mozziconacci, M.J.; Eclache, V.; Léonard, C.; Chapiro, E.; Farhat, H.; Bouyon, A.; Rousselot, P.; et al. Trisomy 4, a new chromosomal abnormality in Waldenström’s macroglobulinemia: A study of 39 cases. Leukemia 2006, 20, 1634–1636. [Google Scholar] [CrossRef][Green Version]
- Braggio, E.; Keats, J.J.; Leleu, X.; Van Wier, S.; Jimenez-Zepeda, V.H.; Valdez, R.; Schop, R.F.J.J.; Price-Troska, T.; Henderson, K.; Sacco, A.; et al. Identification of Copy Number Abnormalities and Inactivating Mutations in Two Negative Regulators of Nuclear Factor- B Signaling Pathways in Waldenstrom’s Macroglobulinemia. Cancer Res. 2009, 69, 3579–3588. [Google Scholar] [CrossRef]
- Poulain, S.; Braggio, E.; Roumier, C.; Aijjou, R.; Broucqsault, N.; Galiègue-Zouitina, S.; Manier, S.; Soenen, V.; Nibourel, O.; Duthilleul, P.; et al. High-Throughput Genomic Analysis in Waldenström’s Macroglobulinemia. Clin. Lymphoma Myeloma Leuk. 2011, 11, 106–108. [Google Scholar] [CrossRef]
- Sekiguchi, N.; Nomoto, J.; Nagata, A.; Kiyota, M.; Fukuda, I.; Yamada, K.; Takezako, N.; Kobayashi, Y. Gene Expression Profile Signature of Aggressive Waldenström Macroglobulinemia with Chromosome 6q Deletion. Biomed Res. Int. 2018, 2018, 6728128. [Google Scholar] [CrossRef]
- Krzisch, D.; Guedes, N.; Boccon-Gibod, C.; Baron, M.; Bravetti, C.; Davi, F.; Armand, M.; Smagghe, L.; Caron, J.; Bernard, O.A.; et al. Cytogenetic and molecular abnormalities in Waldenström’s macroglobulinemia patients: Correlations and prognostic impact. Am. J. Hematol. 2021, 96, 1569–1579. [Google Scholar] [CrossRef]
- Poulain, S.; Roumier, C.; Bertrand, E.; Renneville, A.; Caillault-Venet, A.; Doye, E.; Geffroy, S.; Sebda, S.; Nibourel, O.; Nudel, M.; et al. TP53 Mutation and Its Prognostic Significance in Waldenstrom’s Macroglobulinemia. Clin. Cancer Res. 2017, 23, 6325–6335. [Google Scholar] [CrossRef]
- Roos-Weil, D.; Decaudin, C.; Armand, M.; Della-Valle, V.; Diop, M.K.; Ghamlouch, H.; Ropars, V.; Hérate, C.; Lara, D.; Durot, E.; et al. A Recurrent Activating Missense Mutation in Waldenström Macroglobulinemia Affects the DNA Binding of the ETS Transcription Factor SPI1 and Enhances Proliferation. Cancer Discov. 2019, 9, 796–811. [Google Scholar] [CrossRef]
- Treon, S.P.; Meid, K.; Gustine, J.; Yang, G.; Xu, L.; Liu, X.; Patterson, C.J.; Hunter, Z.R.; Branagan, A.R.; Laubach, J.P.; et al. Long-Term Follow-Up of Ibrutinib Monotherapy in Symptomatic, Previously Treated Patients With Waldenström Macroglobulinemia. J. Clin. Oncol. 2021, 39, 565–575. [Google Scholar] [CrossRef]
- Treon, S.P.; Tripsas, C.K.; Meid, K.; Warren, D.; Varma, G.; Green, R.; Argyropoulos, K.V.; Yang, G.; Cao, Y.; Xu, L.; et al. Ibrutinib in Previously Treated Waldenström’s Macroglobulinemia. N. Engl. J. Med. 2015, 372, 1430–1440. [Google Scholar] [CrossRef]
- Treon, S.P.; Xu, L.; Guerrera, M.L.; Jimenez, C.; Hunter, Z.R.; Liu, X.; Demos, M.; Gustine, J.; Chan, G.; Munshi, M.; et al. Genomic landscape of Waldenström macroglobulinemia and its impact on treatment strategies. J. Clin. Oncol. 2020, 38, 1198–1208. [Google Scholar] [CrossRef]
- Castillo, J.J.; Moreno, D.F.; Arbelaez, M.I.; Hunter, Z.R.; Treon, S.P. CXCR4 mutations affect presentation and outcomes in patients with Waldenström macroglobulinemia: A systematic review. Expert Rev. Hematol. 2019, 12, 873–881. [Google Scholar] [CrossRef]
- Owen, R.G.; McCarthy, H.; Rule, S.; D’Sa, S.; Thomas, S.K.; Tournilhac, O.; Forconi, F.; Kersten, M.J.; Zinzani, P.L.; Iyengar, S.; et al. Acalabrutinib monotherapy in patients with Waldenström macroglobulinemia: A single-arm, multicentre, phase 2 study. Lancet Haematol. 2020, 7, e112–e121. [Google Scholar] [CrossRef]
- Tam, C.S.; Opat, S.; D’Sa, S.; Jurczak, W.; Lee, H.-P.; Cull, G.; Owen, R.G.; Marlton, P.; Wahlin, B.E.; Sanz, R.G.; et al. A randomized phase 3 trial of zanubrutinib vs. ibrutinib in symptomatic Waldenström macroglobulinemia: The ASPEN study. Blood 2020, 136, 2038–2050. [Google Scholar] [CrossRef]
- Dimopoulos, M.; Sanz, R.G.; Lee, H.-P.P.; Trneny, M.; Varettoni, M.; Opat, S.; D’Sa, S.; Owen, R.G.; Cull, G.; Mulligan, S.; et al. Zanubrutinib for the treatment of MYD88 wild-type Waldenström macroglobulinemia: A substudy of the phase 3 ASPEN trial. Blood Adv. 2020, 4, 6009–6018. [Google Scholar] [CrossRef]
- Jiménez, C.; Alonso-Álvarez, S.; Alcoceba, M.; Ordóñez, G.R.; García-Álvarez, M.; Prieto-Conde, M.I.; Chillón, M.C.; Balanzategui, A.; Corral, R.; Marín, L.A.; et al. From Waldenström’s macroglobulinemia to aggressive diffuse large B-cell lymphoma: A whole-exome analysis of abnormalities leading to transformation. Blood Cancer J. 2017, 7, e591. [Google Scholar] [CrossRef]
- Gustine, J.N.; Tsakmaklis, N.; Demos, M.G.; Kofides, A.; Chen, J.G.; Liu, X.; Munshi, M.; Guerrera, M.L.; Chan, G.G.; Patterson, C.J.; et al. TP 53 mutations are associated with mutated MYD 88 and CXCR 4, and confer an adverse outcome in Waldenström macroglobulinaemia. Br. J. Haematol. 2019, 184, 242–245. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Klein, U.; Tu, Y.; Stolovitzky, G.A.; Mattioli, M.; Cattoretti, G.; Husson, H.; Freedman, A.; Inghirami, G.; Cro, L.; Baldini, L.; et al. Gene Expression Profiling of B Cell Chronic Lymphocytic Leukemia Reveals a Homogeneous Phenotype Related to Memory B Cells. J. Exp. Med. 2001, 194, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Chng, W.J.; Schop, R.F.; Price-Troska, T.; Ghobrial, I.; Kay, N.; Jelinek, D.F.; Gertz, M.A.; Dispenzieri, A.; Lacy, M.; Kyle, R.A.; et al. Gene-expression profiling of Waldenstrom macroglobulinemia reveals a phenotype more similar to chronic lymphocytic leukemia than multiple myeloma. Blood 2006, 108, 2755–2763. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jackson, D.A.; Smith, T.D.; Amarsaikhan, N.; Han, W.; Neil, M.S.; Boi, S.K.; Vrabel, A.M.; Tolosa, E.J.; Almada, L.L.; Fernandez-Zapico, M.E.; et al. Modulation of the IL-6 Receptor α Underlies GLI2-Mediated Regulation of Ig Secretion in Waldenström Macroglobulinemia Cells. J. Immunol. 2015, 195, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Elsawa, S.F.; Novak, A.J.; Ziesmer, S.C.; Almada, L.L.; Hodge, L.S.; Grote, D.M.; Witzig, T.E.; Fernandez-Zapico, M.E.; Ansell, S.M. Comprehensive analysis of tumor microenvironment cytokines in Waldenstrom macroglobulinemia identifies CCL5 as a novel modulator of IL-6 activity. Blood 2011, 118, 5540–5549. [Google Scholar] [CrossRef]
- Hatzimichael, E.C.; Christou, L.; Bai, M.; Kolios, G.; Kefala, L.; Bourantas, K.L. Serum levels of IL-6 and its soluble receptor (sIL-6R) in Waldenström’s macroglobulinemia. Eur. J. Haematol. 2001, 66, 1–6. [Google Scholar] [CrossRef]
- Gutiérrez, N.C.; Ocio, E.M.; de las Rivas, J.; Maiso, P.; Delgado, M.; Fermiñán, E.; Arcos, M.J.; Sánchez, M.L.; Hernández, J.M.; San Miguel, J.F. Gene expression profiling of B lymphocytes and plasma cells from Waldenström’s macroglobulinemia: Comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma and normal individuals. Leukemia 2007, 21, 541–549. [Google Scholar] [CrossRef]
- Han, W.; Matissek, S.J.; Jackson, D.A.; Sklavanitis, B.; Elsawa, S.F. Targeting IL-6 receptor reduces IgM levels and tumor growth in Waldenström macroglobulinemia. Oncotarget 2019, 10, 3400–3407. [Google Scholar] [CrossRef][Green Version]
- Gaudette, B.T.; Dwivedi, B.; Chitta, K.S.; Poulain, S.; Powell, D.; Vertino, P.; Leleu, X.; Lonial, S.; Chanan-Khan, A.A.; Kowalski, J.; et al. Low expression of pro-apoptotic Bcl-2 family proteins sets the apoptotic threshold in Waldenström macroglobulinemia. Oncogene 2016, 35, 479–490. [Google Scholar] [CrossRef]
- Leleu, X.; Eeckhoute, J.; Jia, X.; Roccaro, A.M.; Moreau, A.-S.; Farag, M.; Sacco, A.; Ngo, H.T.; Runnels, J.; Melhem, M.R.; et al. Targeting NF-kappaB in Waldenstrom macroglobulinemia. Blood 2008, 111, 5068–5077. [Google Scholar] [CrossRef]
- Hunter, Z.; Cao, Y.; Lewicki, M.; Sun, J.; Tseng, H.; Hanzis, C.; Brodsky, P.; Manning, R.; Xu, L.; Yang, G.; et al. Aberrant Expression of Regulatory miRNAs and Transcripts for IRS-PI3K Growth and Survival Signaling In Waldenstrom’s Macroglobulinemia. Blood 2010, 116, 1912. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Jia, X.; Azab, A.K.; Maiso, P.; Ngo, H.T.; Azab, F.; Runnels, J.; Quang, P.; Ghobrial, I.M. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood 2010, 116, 1506–1514. [Google Scholar] [CrossRef]
- Sun, J.Y.; Xu, L.; Tseng, H.; Ciccarelli, B.; Fulciniti, M.; Hunter, Z.R.; Maghsoudi, K.; Hatjiharissi, E.; Zhou, Y.; Yang, G.; et al. Histone deacetylase inhibitors demonstrate significant preclinical activity as single agents, and in combination with bortezomib in Waldenström’s macroglobulinemia. Clin. Lymphoma Myeloma Leuk. 2011, 11, 152–156. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Chen, C.; Runnels, J.; Leleu, X.; Azab, F.; Azab, A.K.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNA expression in the biology, prognosis, and therapy of Waldenström macroglobulinemia. Blood 2009, 113, 4391–4402. [Google Scholar] [CrossRef]
- Fulciniti, M.; Amodio, N.; Bandi, R.L.; Cagnetta, A.; Samur, M.K.; Acharya, C.; Prabhala, R.; D’Aquila, P.; Bellizzi, D.; Passarino, G.; et al. MiR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J. 2016, 6, e380. [Google Scholar] [CrossRef]
- Caivano, A.; La Rocca, F.; Simeon, V.; Girasole, M.; Dinarelli, S.; Laurenzana, I.; De Stradis, A.; De Luca, L.; Trino, S.; Traficante, A.; et al. MicroRNA-155 in serum-derived extracellular vesicles as a potential biomarker for hematologic malignancies—A short report. Cell. Oncol. 2017, 40, 97–103. [Google Scholar] [CrossRef]
- Bouyssou, J.M.; Liu, C.-J.; Bustoros, M.; Sklavenitis-Pistofidis, R.; Aljawai, Y.; Manier, S.; Yosef, A.; Sacco, A.; Kokubun, K.; Tsukamoto, S.; et al. Profiling of circulating exosomal miRNAs in patients with Waldenström Macroglobulinemia. PLoS ONE 2018, 13, e0204589. [Google Scholar] [CrossRef]
- Hodge, L.S.; Elsawa, S.F.; Grote, D.M.; Price-Troska, T.L.; Asmann, Y.W.; Fonseca, R.; Gertz, M.A.; Witzig, T.E.; Novak, A.J.; Ansell, S.M. MicroRNA expression in tumor cells from Waldenstrom’s macroglobulinemia reflects both their normal and malignant cell counterparts. Blood Cancer J. 2011, 1, e24. [Google Scholar] [CrossRef][Green Version]
- Kubiczkova Besse, L.; Sedlarikova, L.; Kryukov, F.; Nekvindova, J.; Radova, L.; Almasi, M.; Pelcova, J.; Minarik, J.; Pika, T.; Pikalova, Z.; et al. Combination of serum microRNA-320a and microRNA-320b as a marker for Waldenström macroglobulinemia. Am. J. Hematol. 2015, 90, E51–E52. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Jia, X.; Banwait, R.; Maiso, P.; Azab, F.; Flores, L.; Manier, S.; Azab, A.K.; Ghobrial, I.M. Mechanisms of Activity of the TORC1 Inhibitor Everolimus in Waldenstrom Macroglobulinemia. Clin. Cancer Res. 2012, 18, 6609. [Google Scholar] [CrossRef]
- Barh, D.; Malhotra, R.; Ravi, B.; Sindhurani, P. Microrna Let-7: An Emerging Next-Generation Cancer Therapeutic. Curr. Oncol. 2010, 17, 70–80. [Google Scholar] [CrossRef]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.-J.; van den Berg, A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Roccaro, A.M.; Rombaoa, C.; Flores, L.; Obad, S.; Fernandes, S.M.; Sacco, A.; Liu, Y.; Ngo, H.; Quang, P.; et al. LNA-mediated anti–miR-155 silencing in low-grade B-cell lymphomas. Blood 2012, 120, 1678–1686. [Google Scholar] [CrossRef] [PubMed]
- Morel, P.; Duhamel, A.; Gobbi, P.; Dimopoulos, M.A.; Dhodapkar, M.V.; McCoy, J.; Crowley, J.; Ocio, E.M.; Garcia-Sanz, R.; Treon, S.P.; et al. International prognostic scoring system for Waldenström macroglobulinemia. Blood 2009, 113, 4163–4170. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Otero, D.; Kao, E.; Miletic, A.V.; Hother, C.; Ralfkiaer, E.; Rickert, R.C.; Gronbaek, K.; David, M. Onco-miR-155 targets SHIP1 to promote TNFα-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009, 1, 288–295. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in Cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Q.; Chang, C. Epigenetics in Health and Disease. Adv. Exp. Med. Biol. 2020, 1253, 3–55. [Google Scholar] [CrossRef]
- Amodio, N.; Rossi, M.; Raimondi, L.; Pitari, M.R.; Botta, C.; Tagliaferri, P.; Tassone, P. miR-29s: A family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 2015, 6, 12837–12861. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br. J. Cancer 2006, 94, 179–183. [Google Scholar] [CrossRef]
- Sampath, D.; Liu, C.; Vasan, K.; Sulda, M.; Puduvalli, V.K.; Wierda, W.G.; Keating, M.J. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 2012, 119, 1162–1172. [Google Scholar] [CrossRef]
- Braga, T.V.; Evangelista, F.C.G.; Gomes, L.C.; da Silva Araújo, S.S.; das Graças Carvalho, M.; de Paula, A. Evaluation of MiR-15a and MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia. Biomed. Pharmacother. 2017, 92, 864–869. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Thompson, B.; Leleu, X.; Azab, A.K.; Azab, F.; Runnels, J.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009, 113, 6669–6680. [Google Scholar] [CrossRef]
- Ghobrial, I.M.; Campigotto, F.; Murphy, T.J.; Boswell, E.N.; Banwait, R.; Azab, F.; Chuma, S.; Kunsman, J.; Donovan, A.; Masood, F.; et al. Results of a phase 2 trial of the single-agent histone deacetylase inhibitor panobinostat in patients with relapsed/refractory Waldenström macroglobulinemia. Blood 2013, 121, 1296–1303. [Google Scholar] [CrossRef]
- Matissek, S.J.; Han, W.; Karbalivand, M.; Sayed, M.; Reilly, B.M.; Mallat, S.; Ghazal, S.M.; Munshi, M.; Yang, G.; Treon, S.P.; et al. Epigenetic targeting of Waldenström macroglobulinemia cells with BET inhibitors synergizes with BCL2 or histone deacetylase inhibition. Epigenomics 2021, 13, 129–144. [Google Scholar] [CrossRef]
- Peng, W.-X.; Koirala, P.; Mo, Y.-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Garding, A.; Bhattacharya, N.; Claus, R.; Ruppel, M.; Tschuch, C.; Filarsky, K.; Idler, I.; Zucknick, M.; Caudron-Herger, M.; Oakes, C.; et al. Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the In Cis Downregulation of a Gene Cluster That Targets NF-kB. PLoS Genet. 2013, 9, e1003373. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wong, K.Y.; Li, Z.H.; Chim, C.S. Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget 2016, 7, 82400–82410. [Google Scholar] [CrossRef]
- Blume, C.J.; Hotz-Wagenblatt, A.; Hüllein, J.; Sellner, L.; Jethwa, A.; Stolz, T.; Slabicki, M.; Lee, K.; Sharathchandra, A.; Benner, A.; et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia 2015, 29, 2015–2023. [Google Scholar] [CrossRef]
- Isin, M.; Ozgur, E.; Cetin, G.; Erten, N.; Aktan, M.; Gezer, U.; Dalay, N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin. Chim. Acta 2014, 431, 255–259. [Google Scholar] [CrossRef]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef]
- Sattari, A.; Siddiqui, H.; Moshiri, F.; Ngankeu, A.; Nakamura, T.; Kipps, T.J.; Croce, C.M. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget 2016, 7, 54174–54182. [Google Scholar] [CrossRef]
- Ronchetti, D.; Manzoni, M.; Agnelli, L.; Vinci, C.; Fabris, S.; Cutrona, G.; Matis, S.; Colombo, M.; Galletti, S.; Taiana, E.; et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016, 6, e468. [Google Scholar] [CrossRef]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 2016, 7, 14814–14830. [Google Scholar] [CrossRef]
- Zhuang, W.; Ge, X.; Yang, S.; Huang, M.; Zhuang, W.; Chen, P.; Zhang, X.; Fu, J.; Qu, J.; Li, B. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells 2015, 33, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Chen, H.; Shen, X.; Wang, X.; Ju, S.; Lu, M.; Cong, H. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin. Chim. Acta 2018, 480, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, Y.; Liu, W.; Huang, Y.; Shen, X.; Jing, R.; Pu, J.; Wang, X.; Ju, S.; Cong, H.; et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sun, H.; Liu, W.; Zhu, H.; Fu, J.; Yang, C.; Fan, L.; Wang, L.; Liu, Y.; Xu, W.; et al. Circ-RPL15: A plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia 2020, 34, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Z.; Xia, Y.; Qin, S.; Li, Y.; Wu, J.; Liang, J.; Wang, L.; Zhu, H.; Fan, L.; et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging 2019, 11, 3561–3573. [Google Scholar] [CrossRef]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Investig. 2018, 98, 1657–1669. [Google Scholar] [CrossRef]
- Liu, H.; Wu, Y.; Wang, S.; Jiang, J.; Zhang, C.; Jiang, Y.; Wang, X.; Hong, L.; Huang, H. Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer 2019, 19, 937. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Wu, J.; Khadka, B.; Fang, Z.; Gu, J.; Tang, B.; Xiao, R.; Pan, G.; Liu, J. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 54. [Google Scholar] [CrossRef]
- Kyle, R.A.; Benson, J.T.; Larson, D.R.; Therneau, T.M.; Dispenzieri, A.; Kumar, S.; Melton, L.J.; Rajkumar, S.V. Progression in smoldering Waldenstrom macroglobulinemia: Long-term results. Blood 2012, 119, 4462–4466. [Google Scholar] [CrossRef]
- Morra, E.; Cesana, C.; Klersy, C.; Barbarano, L.; Miqueleiz, S.; Varettoni, M.; Lucchesini, C.; Ricci, F.; Lazzarino, M. Prognostic Factors for Transformation in Asymptomatic Immunoglobulin M Monoclonal Gammopathies. Clin. Lymphoma 2005, 5, 265–269. [Google Scholar] [CrossRef]
- Baldini, L.; Goldaniga, M.; Guffanti, A.; Broglia, C.; Cortelazzo, S.; Rossi, A.; Morra, E.; Colombi, M.; Callea, V.; Pogliani, E.; et al. Immunoglobulin M monoclonal gammopathies of undetermined significance and indolent Waldenstrom’s macroglobulinemia recognize the same determinants of evolution into symptomatic lymphoid disorders: Proposal for a common prognostic scoring system. J. Clin. Oncol. 2005, 23, 4662–4668. [Google Scholar] [CrossRef]
- Greco, A.; Tedeschi, A.; Varettoni, M.; Nichelatti, M.; Paris, L.; Ricci, F.; Vismara, E.; Morra, E. Factors predicting transformation of asymptomatic IgM monoclonal gammopathy. Clin. Lymphoma Myeloma Leuk. 2011, 11, 77–79. [Google Scholar] [CrossRef]
- Trojani, A.; Greco, A.; Tedeschi, A.; Lodola, M.; Di Camillo, B.; Ricci, F.; Turrini, M.; Varettoni, M.; Rattotti, S.; Morra, E. Microarray Demonstrates Different Gene Expression Profiling Signatures Between Waldenström Macroglobulinemia and IgM Monoclonal Gammopathy of Undetermined Significance. Clin. Lymphoma Myeloma Leuk. 2013, 13, 208–210. [Google Scholar] [CrossRef]
- Trojani, A.; Di Camillo, B.; Bossi, L.E.; Leuzzi, L.; Greco, A.; Tedeschi, A.; Frustaci, A.M.; Deodato, M.; Zamprogna, G.; Beghini, A.; et al. Identification of a Candidate Gene Set Signature for the Risk of Progression in IgM MGUS to Smoldering/Symptomatic Waldenström Macroglobulinemia (WM) by a Comparative Transcriptome Analysis of B Cells and Plasma Cells. Cancers 2021, 13, 1837. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Decruyenaere, P.; Offner, F.; Vandesompele, J. Circulating RNA biomarkers in diffuse large B-cell lymphoma: A systematic review. Exp. Hematol. Oncol. 2021, 10, 13. [Google Scholar] [CrossRef]
- Rossi, D.; Diop, F.; Spaccarotella, E.; Monti, S.; Zanni, M.; Rasi, S.; Deambrogi, C.; Spina, V.; Bruscaggin, A.; Favini, C.; et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017, 129, 1947–1957. [Google Scholar] [CrossRef]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef]
- Yeh, P.; Hunter, T.; Sinha, D.; Ftouni, S.; Wallach, E.; Jiang, D.; Chan, Y.-C.; Wong, S.Q.; Silva, M.J.; Vedururu, R.; et al. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nat. Commun. 2017, 8, 14756. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Bagratuni, T.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Mavrianou-Koutsoukou, N.; Liacos, C.; Patseas, D.; Kanellias, N.; Migkou, M.; Ziogas, D.C.; Eleutherakis-Papaiakovou, E.; et al. Detection of MYD88 and CXCR4 mutations in cell-free DNA of patients with IgM monoclonal gammopathies. Leukemia 2018, 32, 2617–2625. [Google Scholar] [CrossRef]
- Demos, M.G.; Hunter, Z.R.; Xu, L.; Tsakmaklis, N.; Kofides, A.; Munshi, M.; Liu, X.; Guerrera, M.L.; Leventoff, C.R.; White, T.P.; et al. Cell-free DNA analysis for detection of MYD88L265P and CXCR4S338X mutations in Waldenström macroglobulinemia. Am. J. Hematol. 2021, 96, E250–E253. [Google Scholar] [CrossRef] [PubMed]
- Ntanasis-Stathopoulos, I.; Bagratuni, T.; Gavriatopoulou, M.; Patseas, D.; Liacos, C.; Kanellias, N.; Fotiou, D.; Tsiligkeridou, E.; Andreatou, A.; Mavrianou-Koutsoukou, N.; et al. Cell-free DNA analysis for the detection of MYD88 and CXCR4 mutations in IgM monoclonal gammopathies; an update with clinicopathological correlations. Am. J. Hematol. 2020, 95, E148–E150. [Google Scholar] [CrossRef] [PubMed]
- Bagratuni, T.; Markou, A.; Patseas, D.; Mavrianou-Koutsoukou, N.; Aktypi, F.; Liacos, C.I.; Sklirou, A.D.; Theodorakakou, F.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; et al. Determination of MYD88L265P mutation fraction in IgM monoclonal gammopathies. Blood Adv. 2022, 6, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019, 58, 100–108. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Caivano, A.; Laurenzana, I.; De Luca, L.; La Rocca, F.; Simeon, V.; Trino, S.; D’Auria, F.; Traficante, A.; Maietti, M.; Izzo, T.; et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumor Biol. 2015, 36, 9739–9752. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Mitsiades, C.S.; Mitsiades, N.S.; Munshi, N.C.; Richardson, P.G.; Anderson, K.C. The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: Interplay of growth factors, their receptors and stromal interactions. Eur. J. Cancer 2006, 42, 1564–1573. [Google Scholar] [CrossRef]
- Jalali, S.; Ansell, S.M. Bone marrow microenvironment in Waldenstrom’s Macroglobulinemia. Best Pract. Res. Clin. Haematol. 2016, 29, 148–155. [Google Scholar] [CrossRef]

| Reference | Technique | Tissue | WM | IgM-MGUS | Diagnostic Criteria | ||
|---|---|---|---|---|---|---|---|
| pts | MYD88L265P | pts | MYD88L265P | ||||
| Treon et al., 2012 | WGS Sanger | BM CD19+ | 30 | 91% | 21 | 10% | 2° IWWM |
| [27] | |||||||
| Landgren et al., 2012 | Sanger | BM CD19+ | 9 | 56%7 | 2° IWWM | ||
| [38] | |||||||
| Gachard et al., 2013 | PCR | BM | 27 | 67% | WHO 2008 | ||
| [39] | |||||||
| Xu et al., 2013 | SYBR AS-qPCR | BM CD19+ | 104 | 93% | 24 | 54% | 2° IWWM |
| [40] | |||||||
| Ondrejka et al., 2013 | AS-PCR | BM biopsy FFPE | 13 | 100% | WHO 2008 | ||
| [41] | |||||||
| Jimenez et al., 2013 | AS-qPCR | BM/PB WBC | 117 | 86% | 31 | 87% | WHO 2011 |
| [42] | |||||||
| Poulain et al., 2013 | PCR | BM CD19+ | 67 | 79% | 2° IWWM | ||
| [43] | |||||||
| Willenbacher et al., 2013 | Sanger | BM biopsy FFPE | 7 | 86% | 2° IWWM | ||
| [44] | |||||||
| Mori et al., 2013 | AS-PCR Sanger | BM MNC | 25 | 76% | 2° IWWM | ||
| [45] | |||||||
| Varettoni et al., 2013 | AS-PCR | BM MNC | 58 | 100% | 77 | 47% | 2° IWWM |
| [46] | |||||||
| Argentou et al., 2014 | PCR-RFLP | BM-PB WBC, | 12 | 92% | 1 | 100% | WHO 2008 |
| [47] | BM CD19+ | ||||||
| Capaldi et al., 2014 | AS-PCR | BM biopsy FFPE | 32 | 97% | 21 | 43% | ND |
| [48] | |||||||
| Petrikkos et al., 2014 | AS-PCR | BM biopsy-MNC-slides | 29 | 66% | 2° IWWM | ||
| [49] | |||||||
| Ansell et al., 2014 | WES, Sanger AS-qPCR | LN-BM biopsy PC | 39 | 97% | ND | ||
| [50] | |||||||
| Hunter et al., 2014 | WGS | BM CD19+ | 30 | 90% | 2° IWWM | ||
| [28] | |||||||
| Xu et al., 2014 | AS-qPCR | BM-PB CD19+ | 118 | 97% | 12 | 42% | 2° IWWM |
| [51] | |||||||
| Treon et al., 2014 | AS-PCR | BM CD19+ | 175 | 90% | 2° IWWM | ||
| [52] | |||||||
| Patkar et al., 2015 | AS-PCR | BM slides | 32 | 84% | WHO 2008 | ||
| [53] | |||||||
| Schmidt et al., 2015 | LNA-clamped PCR | BM biopsy FFPE | 51 | 96% | 2° IWWM WHO 2008 | ||
| [54] | |||||||
| Shin et al., 2016 | MEMO-PCR | BM slides | 28 | 75% | ND | ||
| [55] | |||||||
| Burnworth et al., 2016 | PCR | BM C19+ | 21 | 100% | WHO 2008 | ||
| [56] | PC | ||||||
| Correa et al., 2017 | ARMS qPCR | BM biopsy FFPE | 42 | 82% | 55 | 27% | mSMART |
| [57] | |||||||
| Varettoni et al., 2017 | RT-qPCR | BM CD19+ | 130 | 86% | 130 | 60% | 2° IWWM |
| [10] | MPS | 62 | 85% | 57 | 47% | ||
| Baer et al., 2017 | AS-qPCR | BM/PB MNC | 78 | 86% | ND | ||
| [58] | MPS | 78 | 69% | ||||
| Paludo et al., 2017 | ARMS AS-PCR | BM | 29 | 86% | 2° IWWM | ||
| [59] | |||||||
| Cao et al., 2017 | AS-qPCR Sanger | BM CD19+ | 42 | 93% | 18 | 44% | 2° IWWM |
| [60] | |||||||
| Abeykoon et al., 2018 | AS-PCR | BM | 219 | 79% | mSMART | ||
| [61] | |||||||
| Drandi et al., 2018 | dPCR | BM/PB WBC | 133 | 96% | 4 | 100% | WHO 2011 |
| [62] | |||||||
| Vinarkar et al., 2019 | AS-PCR Sanger | BM/PB—BM slides | 33 | 85% | WHO 2008 | ||
| [63] | |||||||
| Nakamura et al., 2019 | MPS | PB MNC | 19 | 74% | 21 | 67% | WHO 2008 |
| [64] | |||||||
| Wu et al., 2020 | AS-qPCR | BM/PB MNC | 27 | 89% | 2° IWWM | ||
| [65] | |||||||
| Wang et al., 2021 | MPS | BM | 68 | 84% | 2° IWWM | ||
| [66] | |||||||
| Kofides et al., 2021 | AS-PCR | BM | 391 | 96% | 2° IWWM | ||
| [67] | MPS | 66% | |||||
| Reference | Technique | Tissue | WM | IgM-MGUS | Diagnostic Criteria | ||
|---|---|---|---|---|---|---|---|
| pts | CXCR4MUT | pts | CXCR4MUT | ||||
| Treon et al., 2014 [52] | Sanger | BM CD19+ | 175 | 29% | 2° IWWM | ||
| Roccaro et al., 2014 [90] | AS-qPCR | BM CD19+ | 131 | 28% | 40 | 20% | WHO 2011 |
| Hunter et al., 2014 [28] | WGS Sanger | BM CD19+ | 177 | 29% | 2° IWWM | ||
| Schmidt et al., 2015 [54] | Sanger | BM biopsy FFPE | 47 | 36% | 2° IWWM WHO 2008 | ||
| Xu et al., 2016 [87] | AS-PCR Sanger | BM CD19+ | 164 | 40% | 12 | 17% | 2° IWWM |
| Poulain et al., 2016 [84] | MPS Sanger | BM CD19+ | 98 | 25% | 2° IWWM | ||
| Burnworth et al., 2016 [56] | PCR | BM CD19+ PC | 27 | 47% | WHO 2008 | ||
| Cao et al., 2017 [60] | Sanger AS-qPCR | BM CD19+ | 42 | 24% | 18 | 6% | 2° IWWM |
| Varettoni et al., 2017 [10] | Sanger | BM CD19+ | 130 | 22% | 130 | 4% | 2° IWWM |
| MPS | 62 | 23% | 57 | 9% | |||
| Baer et al., 2017 [58] | MPS | BM/PB MNC | 69 | 25% | ND | ||
| Guerrera et al., 2018 [83] | AS-PCR Sanger | BM CD19+ | 33 | 66% | 2° IWWM | ||
| Vinarkar et al., 2019 [63] | Sanger | BM/PB or BM slides | 28 | 7% | WHO 2008 | ||
| Castillo et al., 2019 [85] | AS-PCR Sanger | BM CD19+ | 180 | 38% | 2° IWWM | ||
| Wu et al., 2020 [65] | AS-qPCR | BM/PB MNC | 27 | 4% | 2° IWWM | ||
| Wang et al., 2021 [66] | AS-qPCR | BM | 68 | 37% | 2° IWWM | ||
| Gustine et al., 2021 [92] | AS-PCR, Sanger | BM CD19+ | 107 | 40% | 2° IWWM | ||
| MPS | BM | 107 | 15% | ||||
| Gene | Technique | WM | IgM-MGUS | Reference | ||
|---|---|---|---|---|---|---|
| pts | MUTs | pts | MUTs | |||
| KMT2D | WGS | 18 | 22% | Hunter et al., 2018 [82] | ||
| MPS | 62 | 24% | 57 | 5% | Varettoni et al., 2017 [10] | |
| TP53 | WGS | 30 | 7% | Hunter et al., 2014 [28] | ||
| MPS | 125 | 7% | 10 | 0% | Poulain et al., 2017 [111] | |
| MPS | 62 | 10% | 57 | 5% | Varettoni et al., 2017 [10] Wang et al., 2021 [66] | |
| MPS | 68 | 12% | ||||
| ARID1A | WGS, Sanger | 30 | 17% | Treon et al., 2012 [27] | ||
| WGS | 30 | 17% | Hunter et al., 2014 [28] | |||
| MPS | 62 | 5% | 57 | 2% | Varettoni et al., 2017 [10] | |
| WGS, targeted MPS | 85 | 8% | Roos-Weil et al., 2019 [112] | |||
| CD79B | WGS | 30 | 7% | Hunter et al., 2014 [28] | ||
| MPS | 98 | 12% | Poulain et al., 2016 [84] | |||
| MPS | 62 | 3% | 57 | 2% | Varettoni et al., 2017 [10] | |
| MYBBP1A | WGS | 30 | 7% | Hunter et al., 2014 [28] | ||
| NOTCH2 | WGS | 30 | 3% | Hunter et al., 2014 [28] | ||
| MPS | 62 | 5% | 57 | 9% | Varettoni et al., 2017 [10] | |
| PRDM1 | MPS | 62 | 6% | 57 | 2% | Varettoni et al., 2017 [10] |
| TRAF3 | WGS | 30 | 3% | Hunter et al., 2014 [28] | ||
| MPS | 62 | 2% | Varettoni et al., 2017 [10] | |||
| SPI1 | WGS, targeted MPS | 85 | 6% | Roos-Weil et al., 2019 [112] | ||
| TWIST custom capture | 239 | 4% | Krzisch et al., 2021 [110] | |||
| Reference | Method | Sample | RNA | Level | Result |
|---|---|---|---|---|---|
| Chng et al. 2006 [124] | microarray | BM: 23 WM (CD19+/ CD138+); 101 MM (CD138+); 24 SMM (CD138+); 22 MGUS (1 IgM-MGUS: CD19+/CD138+); 15 NPC (CD138+) PB: 7 NBL (CD19+)/8 CLL (CD19+) | 48 mRNA (top 10: IL6, NRGN, P311, OSBPL3, CD1C, GPR30, HSU54999, GPR30, SLC2A3, TIP-1, WHSC1) | up | upregulated in WM compared to CLL/MM |
| 25 mRNA (top 10: DKFZP564A2416, KLF13, WBSCR14, PDE1C, CLDN1, DD96, CHRNA4, CST4, LY9, OPRK1) | down | downregulated in WM compared to CLL/MM | |||
| Gutiérrez et al, 2007 [128] | microarray | BM: 10 WM BL/PC (combination of CD10/CD19/CD38/CD34/CD45/K-L); 12 MM, 11 CLL (CD19+/CD5+); 5 NPC (CD38+) PB: 8 NBL (CD19+) | ABCB4, IL4R, ADAM28, ITPR1, SESN1, BACH2, ABCB1, ADARB1, APLP2, GABBR1 | down | downregulated in WM-BL compared to CLL/ NBL |
| IL6, NR4A2, HCK, DUSP1, EBI2, FAM46C, TNFRSF13B, FOSB, S100A8 | up | upregulated in WM-BL compared to CLL/NBL | |||
| IGLV2-14, DEK, HLA-DMA, HMGB1, CPA3, MS4A3, MYB, HLA-DPA1, RNASE2, CLC, EBI2, SYK, HLA-DRB1 | up | upregulated in WM-PC compared to MM-PC/NPC | |||
| LEF1, ATXN1 and FMOD (down), MARCKS (up) | this signature discriminated between clonal WM-BL and CLL | ||||
| Hunter et al, 2010 [132] | microarray RT-qPCR | BM: 40 WM/15 normal B cells (CD19+) | IRS2, PIK3R1 | down | downregulated in WM compared to NBL |
| Roccaro et al, 2010 [133] | microarray | BM:6 WM (CD19+) PB: 2 NBL (CD19+) | HDAC-2, -4, -5, -6, -8, and -9 | up | upregulated in primary WM-BL |
| HAT-1, -2, and -3 | down | downregulated in primary WM-BL | |||
| Sun et al, 2011 [134] | microarray RT-qPCR | BM: 30 treated WM-BL (CD19+) PB:C 10 HC (CD19+) BM:5 treated WM (CD19+) PB: 5 HC (CD19+) | HDAC4, HDAC9, Sirt5 | up | upregulated in WM compared to NBL |
| HDAC9 | up | upregulated in WM compared to NBL, no differential expression for HDAC4 and Sirt5 in RT-qPCR validation | |||
| Gaudette et al, 2016 [130] | microarray | 10 WM-BL/PC; 11 CLL; 12 MM; 8 NBL; 5 NPC | BAK1, BCL2L11, MCL1, BCL2L2 | down | downregulated in WM-PC compared to MM |
| BID | up | upregulated in WM-PC compared to MM | |||
| BID, BCL2A1 | up | upregulated in WM-BL compared to CLL | |||
| BAK1 | down | downregulated in WM-BL compared to CLL | |||
| BAX, BCL2A1, BBC3, BCL2, NOXA | up | upregulated in WM-BL compared to NBL | |||
| Hunter et al, 2016 [104] | RNA-seq | BM: 57 WM-BL (CD19+) PB: normal nonmemory B-cells (CD19+/CD27-)/memory B-cells (CD19+/CD27+) | DNTT, RAG1, RAG2, IGF1, BMP3, CD5L, CXCL12, VCAM1, CXCR4, B2M, BCL2, BCL2L1 CXCR4, CD79A, CD79B (among 13 571 DE genes) | up | upregulated in WM-BL compared to NBL |
| BAX (among 13 571 DE genes) | down | downregulated in WM-BL compared to NBL | |||
| IL6, IRAK2, TNFAIP3, NFKBIZ, NFKB2, TIRAP, PIM1, PIM2, CD40 (among 1155 DE genes) | up | upregulated in MYD88L265P WM-BL compared to MYD88WT WM-BL | |||
| PTBP3, CD86, CXCR3, IGF1R, PIK3AP1, AKT2 among 1155 DE genes | down | downregulated in MYD88L265P WM-BL compared to MYD88WT WM-BL | |||
| TLR4, IL15, WNT5A, PRDM5, CXXC4, CKDN1C, WNK2, CABLES1, IL17RB, GPER1, IGF1, PMAIP1, RGS1, RGS2, RGS13, DUSP1, DUSP2, DUSP4, DUSP5, DUSP10, DUSP16, DUSP22, ERRFI1 (among others) | down | downregulated in MYD88L265P/CXCR4WHIM WM-BL versus MYD88L265P/CXCR4WT WM-BL | |||
| IRAK3, CXCR7, TLR7, TSPAN33, PIK3R5, PIK3CG (among others) | up | upregulated in MYD88L265P/CXCR4WHIM WM-BL versus MYD88L265P/CXCR4WT WM-BL | |||
| HIVEP2, BCLAF1, FOXO3, ARID1B (among 131 DE genes) | down | downregulated in WM-BL with 6q deletions |
| Reference | Method | EV Purification (QC) | Sample | RNA | Level | Result |
|---|---|---|---|---|---|---|
| Diagnosis | ||||||
| Roccaro et al, 2009 [135] | liquid phase Luminex microbead miRNA profiling RT-qPCR | NA | BM: 15 R/R WM (CD19+); 5 untreated WM (CD19+), 3 NBL (CD19+) PB: 3 NBL (CD19+) | miR-363-5p, miR-206, miR-494, miR-155, miR-184, miR-542-3p | up | upregulated in WM compared to NBL |
| miR-9-3p | down | downregulated in WM compared to NBL | ||||
| Hunter et al, 2010 [132] | microaray RT-qPCR | NA | BM: 11 WM (CD19+); 5 NBL (CD19+) | miR-21, miR-29c, miR-155 | up | upregulated in WM compared to NBL |
| miR-9-3p, miR-27b, miR-126-3p, miR-126-5p, miR-145, miR-223, miR-886-5p | down | downregulated in WM compared to NBL | ||||
| Roccaro et al, 2010 [133] | liquid-phase Luminex microbead miRNA profiling RT-qPCR | NA | BM:6 WM (CD19+) PB: 2 NBL (CD19+) BCWM.1 cell line | miR-206-3p | up | upregulated in WM-BL compared to NBL |
| miR-9-3p | down | downregulated in WM-BL compared to NBL | ||||
| Fulciniti et al, 2016 [136] | microarray | NA | BM: WM (CD19+) PB: NBL (CD19+) | miR-23b | down | downregulated in WM compared to NBL |
| Caivano et al, 2017 [137] | RT-qPCR | DC (AFM/TEM) | PB:14 WM;18 HC | miR-155 | up | upregulated in WM compared to HC |
| Gaudette et al, 2016 [130] | RT-qPCR | NA | BCWM.1, MWCL-1, RPCI-WM11 cell lines | miR-155-5p | up | upregulated in BCWM.1 and MWCL-1 cells but not RPCI-WM1 cells |
| Bouyssou et al, 2018 [138] | microarray | DC (TEM/particle size analysis) | BM: 6 WM (CD19+) PB: 30 smouldering WM; 44 symptomatic WM; 10 HC | miR-192-5p, miR-93-5p, miR-15a-5p, miR-16-5p, miR-20a-5p, miR-378a-3p | up | upregulated in smouldering WM compared to HC |
| miR-199a-5p, miR-145-5p, miR199a-3p, miR-221-3p, miR-335-5p, let-7d-5p | down | downregulated in smouldering WM compared to HC | ||||
| Hodge et al, 2011 [139] | microarray | NA | BM/PB: 8 WM (CD19+/CD138+); 6 WM-BL (CD19+); 3 WM-PC (CD138+), 5 MM (CD138+); 5 CLL (CD19+); 3 NBL (CD19+); 6 NPC (CD138+); 4 normal CD19+/CD138+ B-cells | miR-193b-3p, miR-126-3p, miR-181a-5p, miR-125b-5p, miR-451a | up | upregulated in combined WM (CD19+, CD 138+, CD19+/CD138+) vs CLL |
| miR-92a-3p, miR-223-3p, miR-92b-3p, miR-363-3p | up | upregulated in combined WM vs MM | ||||
| miR-9-3p, miR-193b-3p, miR-182-5p, miR-152-3p | down | downregulated in combined WM vs MM | ||||
| miR-21-5p, miR-142-3p | up | upregulated in combined WM (CD19+, CD 138+, CD19+/CD138+) vs NBL | ||||
| miR-182-5p, miR-152-3p, miR-373-5p, miR-575-3p | down | downregulated in combined WM (CD19+, CD 138+, CD19+/CD138+) vs NBL | ||||
| Kubiczkova et al, 2015 [140] | Microarray RT-qPCR | ExoQuick | PB: 21 WM (CD19+ and CD19-); 15 igM-MGUS; 10 IgM MM; 18 HC | miR-320a-3p, miR-320b-3p | down | downregulated in WM vs HC vs IgM-MGUS and vs IgM-MM |
| miR-151-5p, let-7a-5p | down | downregulated in WM vs. HC and vs. IgM-MGUS | ||||
| Therapy Response | ||||||
| Bouyssou et al, 2018 [138] | microarray | DC (TEM/particle size analysis) | PB: 30 smouldering WM; 44 symptomatic WM; 10 HC | miR-21-5p, miR-192-5p, miR-320b-3 | up | increased expression with disease progression |
| let-7d-5p | down | decreased expression with disease progression | ||||
| Roccaro et al, 2012 [141] | RT-qPCR | BM: 4 R/R WM (CD19+) PB: NBL (CD19+) BCWM.1, MEC.1, and RL cell lines | miR-155 | NA | everolimus exerts anti-WM activity by targeting miR-155 | |
| Prognosis | ||||||
| Roccaro et al, 2009 [135] | liquid phase Luminex microbead miRNA profiling RT-qPCR | NA | BM: 15 R/R WM (CD19+); 5 untreated WM (CD19+); 3 NBL (CD19+) PB: 3 NBL (CD19+) | miR-363-5p, miR-206, miR-494, miR-155, miR-184, miR-542-3p | up | upregulation is associated with worse IPSS score |
| References | Technique | Tissue | WM | IgM-MGUS | Diagnostic Criteria | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pts | MYD88L265P | Pts | CXCR4MUT | Pts | MYD88L265P | Pts | CXCR4MUT | ||||
| Bagratuni et al., 2018 | AS-PCR | plasma | 79 | 80% | 16 | 17% | 7 | 86% | 9 | 22% | ND |
| [189] | |||||||||||
| Drandi et al., 2018 | dPCR | plasma | 60 | 88% | WHO 2011 | ||||||
| [62] | |||||||||||
| Wu et al., 2020 | AS-qPCR | plasma | 27 | 85% | 27 | 4% | 2° IWWM | ||||
| [65] | |||||||||||
| Ntanasis-Stathopoulos et al., 2020 | AS-PCR Sanger | plasma | 188 | 89% | 131 | 36% | ND | ||||
| [191] | |||||||||||
| Ferrante et al., 2021 | dPCR | plasma | 32 | 78% | 4 | 75% | 2° IWWM | ||||
| [99] | |||||||||||
| Demos et al., 2021 | AS-qPCR | plasma | 28 | 68% | 23 | 17% | ND | ||||
| [190] | |||||||||||
| Bagratuni et al., 2022 | Cast-PCR | plasma | 92 | 88% | 51 | 80% | ND | ||||
| [192] | |||||||||||
| Reference | Method | EV Purification (QC) | Sample | RNA | Level | Result |
|---|---|---|---|---|---|---|
| Diagnosis | ||||||
| Caivano et al, 2017 [137] | RT-qPCR/ serum | DC (AFM/TEM) | PB:14 WM;18 HC | miR-155 | up | upregulated in WM compared to HC |
| Bouyssou et al, 2018 [138] | microarray/ plasma | DC (TEM/particle size analysis) | PB: 30 smouldering WD; 44 symptomatic WM; 10 HC | miR-192-5p, miR-93-5p, miR-15a-5p, miR-16-5p, miR-20a-5p, miR-378a-3p | up | upregulated in smouldering WM compared to HC |
| miR-199a-5p, miR-145-5p, miR199a-3p, miR-221-3p, miR-335-5p, let-7d-5p | down | downregulated in smouldering WM compared to HC | ||||
| Kubiczkova et al, 2015 [140] | microarray RT-qPCR/ serum | ExoQuick | PB: 21 WM (CD19+ and CD19-); 15 IgM-MGUS; 10 IgM-MM; 18 HC | miR-320a-3p, miR-320b-3p | down | downregulated in WM vs HC vs IgM-MGUS and vs IgM-MM |
| miR-151-5p, let-7a-5p | down | downregulated in WM vs HC and vs IgM-MGUS | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drandi, D.; Decruyenaere, P.; Ferrante, M.; Offner, F.; Vandesompele, J.; Ferrero, S. Nucleic Acid Biomarkers in Waldenström Macroglobulinemia and IgM-MGUS: Current Insights and Clinical Relevance. Diagnostics 2022, 12, 969. https://doi.org/10.3390/diagnostics12040969
Drandi D, Decruyenaere P, Ferrante M, Offner F, Vandesompele J, Ferrero S. Nucleic Acid Biomarkers in Waldenström Macroglobulinemia and IgM-MGUS: Current Insights and Clinical Relevance. Diagnostics. 2022; 12(4):969. https://doi.org/10.3390/diagnostics12040969
Chicago/Turabian StyleDrandi, Daniela, Philippe Decruyenaere, Martina Ferrante, Fritz Offner, Jo Vandesompele, and Simone Ferrero. 2022. "Nucleic Acid Biomarkers in Waldenström Macroglobulinemia and IgM-MGUS: Current Insights and Clinical Relevance" Diagnostics 12, no. 4: 969. https://doi.org/10.3390/diagnostics12040969
APA StyleDrandi, D., Decruyenaere, P., Ferrante, M., Offner, F., Vandesompele, J., & Ferrero, S. (2022). Nucleic Acid Biomarkers in Waldenström Macroglobulinemia and IgM-MGUS: Current Insights and Clinical Relevance. Diagnostics, 12(4), 969. https://doi.org/10.3390/diagnostics12040969







