Abstract
Despite the efforts made in the management of PDAC, the 5-year relative survival rate of pancreatic ductal adenocarcinoma (PDAC) still remains very low (10%). To date, precision oncology is far from being ready to be applied in cases of PDAC, although studies exploring the molecular and genetic alterations have been conducted, and the genomic landscape of PDAC has been characterized. This study aimed to apply a next-generation sequencing (NGS) laboratory-developed multigene panel to PDAC samples to find molecular alterations that could be associated with histopathological features and clinical outcomes. A total of 68 PDACs were characterized by using a laboratory-developed multigene NGS panel. KRAS and TP53 mutations were the more frequent alterations in 75.0% and 44.6% of cases, respectively. In the majority (58.7%) of specimens, more than one mutation was detected, mainly in KRAS and TP53 genes. KRAS mutation was significantly associated with a shorter time in tumor recurrence compared with KRAS wild-type tumors. Intriguingly, KRAS wild-type cases had a better short-term prognosis despite the lymph node status. In conclusion, our work highlights that the combination of KRAS mutation with the age of the patient and the lymph node status may help in predicting the outcome in PDAC patients.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) still represents the major cause of death in Western countries. Despite all the efforts made regarding the comprehension of the cancerogenesis of PDAC and toward its genomic key alterations, the 5-year survival rate of these patients is still very low (below 10%) [1,2] and, unfortunately, only a minority are suitable for resection at the time of the diagnosis [3]. Several efforts have been performed in the management of the PDAC with advances in the surgical, as well as the oncological fields and its multimodal approach, but the prognosis remains poor and has not significantly changed in the last decades [3]. To date, personalized therapy is at the basis of the modern concept of oncological treatments and represents the precision medicine that is able to target different specific proteins that control cancer growth, apoptosis, and or spread in every single patient. These results have been obtained thanks to the advances in molecular biology coupled with improvements in laboratory technology devices that have led to the deepest insight into tumor biology. Currently, many types of tumors are treated with targeted approaches with several choice drugs (such as gastrointestinal stromal tumors, melanoma, breast tumors, ovarian cancers, colorectal cancers, and lung cancers [4,5,6]).
Several papers have demonstrated the association between clinicopathological features, such as lymph node spread and positive surgical margins, and a worse outcome [7,8,9,10]; however, these characteristics have been found also in 20–40% of patients with a good prognosis [7,8,9,10], suggesting that clinicopathological features are not the only determinant for predicting PDAC prognosis. Precision oncology is far from being ready to be applied in cases of PDAC, although studies exploring the molecular and genetic alterations have been conducted, and the genomic landscape of PDAC has been characterized [11,12,13]. Furthermore, relatively few clinically drugable mutations have been identified, and there are still not any targeted agents against the four major genes altered in PDAC that have been validated. In fact, to date, no targetable molecules are available for personalized treatment in the clinical practice of PDAC, as stated by the current ESMO (European Society for Medical Oncology) guidelines [14,15]. Different types of molecular alterations may be found in PDAC, from oncogenes mutations to inactivation of tumor suppressor genes/genes controlling the repair of DNA damage [16]. The most frequently involved and mutated genes in PDAC tumorigenesis are early events in cancerogenesis, such as KRAS mutations or CDKN2A/p16 loss of function, and alterations occurring later in the neoplastic progression, such as TP53 mutations and SMAD4/DPC4 loss [17,18,19,20,21]. Other alterations involved with less frequency have been identified, such as those in the mismatch repair deficiency (dMMR) genes (accounting for about 1–2% of PDAC), NTRK fusions (about 0.3%), or BRCA1/2 mutations (5–10%) [5,16,21,22,23].
Previous studies have demonstrated a worse prognosis in PDAC harboring mutations in KRAS, TP53, CDKN2A, or SMAD4 genes [5,22,23,24,25,26]. In particular, KRAS mutations have been associated with a decreased overall survival (OS), and TP53 alterations have been associated with a poor prognosis [12,24,25].
The aim of this study was to apply an NGS laboratory-developed multigene panel to treatment-naive PDAC samples in order to find any molecular alterations that could be associated with histopathological features and clinical outcomes helping in the choice of patient clinical management.
2. Materials and Methods
2.1. Case Selection
A total of 68 PDAC samples were retrieved from the archives of the Anatomic Pathology of Maggiore Hospital (Bologna, Italy) and the Molecular Pathology Laboratory of Bologna (Bologna, Italy).
We retrospectively analyzed a total of 68 cases of treatment-naive PDAC that underwent surgical resection upfront. The cases had been collected from the pathological archives of the Molecular Pathology Laboratory (Bologna, Italy): 35 patients underwent duodenocephalopancreasectomy, 20 patients had a total pancreatectomy, and in 13 patients a distal pancreatic resection was performed. All patients underwent computed tomography (CT) scan of the whole body, with contrast material, to evaluate the possible presence of metastases and the resectability of the tumor. Moreover, CT allowed for the evaluation of the presence of neoplastic dissemination in mesenteric and splenic vessels. In all cases, a histological diagnosis of ductal pancreatic adenocarcinoma had been performed.
All cases had been revised by a GI-dedicated pathologist (DM and AF), blind to each other, to collect the main histological variable (i.e., grading, lymph vascular invasion, perineural invasion, surgical margins, and lymph node metastases) and to determine the pathological stage according to the 8th Edition of AJCC (American Joint Committee on Cancer) [27]. In controversial cases, a definitive diagnosis was made after a collegial discussion and joint agreement.
In order to identify the main representative neoplastic area for NGS analysis, the tumor area was marked on the control slide, and the proportion of neoplastic cells vs. non-neoplastic cells (i.e., endothelial, stromal, and inflammatory cells) in the area marked on the slide and used for DNA extraction was estimated after microscopic evaluation, providing the tumor cell enrichment (i.e., neoplastic cells/total number of cells).
The mean follow-up was 44.2 + 54.3 months. At the end of the available follow-up, 32 of 64 (50.0%) died. Twenty-three of 32 (71.9%) died within the first 24 months after the date of surgery. Nineteen patients (29.7%) were still alive 60 months after the date of surgery.
2.2. DNA Extraction and Next-Generation Sequencing Analysis
For each sample, a pathologist (AF and DM) evaluated the most representative neoplastic area, and DNA was extracted starting from two or three 10 µm–thick sections, using the “QuickExtract™ FFPE DNA Extraction Solution” kit (Lucigen Corporation, Middleton, WI, USA), and quantified by using Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). An “extended version” of a previously published NGS lab-developed multigene panel [28] was used. The “extended version” of the panel includes the following genomic regions (human reference sequence hg19/GRCh37, total of 343 amplicons, 21.77 kb): BRAF (exons 11, 15), c-Kit (exons 8, 9, 11, 13, 14, 17), CTNNB1 (exons 3, 7, 8), DICER1 (exons 10, 21, 26, 27, 29), DPYD (exons 11, 13, 22, chr1 intronic regions: g.98187018- 98187098, g.98045419- 98045499, g.97915570-97915789), EGFR (exons 18, 19, 20, 21), EIF1AX (exons 1, 2, and chrX intronic region g.20148634–20148745), GNA11 (exons 4, 5), GNAQ (exons 4, 5), GNAS (exons 8, 9), H3F3A (exon 1), HRAS (exons 2–4), IDH1 (exon 4), IDH2 (exon 4), KRAS (exons 2–4), MED12 (exons 1, 2), MET (exons 2, 14), NRAS (exons 2–4), PDGFRα (exons 12, 14, 18), PIK3CA (exons 8, 10, 21), PTEN (exon 5), RET (exons 5, 8, 10, 11, 13, 15, 16), RNF43 (exons 2–10), SMAD4 (exons 2–12), TERT (promoter region, Chr5 g.1295141-1295471), and TP53 (exons 2–11), TSHR (exons 1–10), VHL (exons 1–3). Briefly, about 50 ng of input DNA was used for NGS libraries preparation with the AmpliSeq Plus Library Kit 2.0 (Thermo Fisher Scientific). Templates were then sequenced by using an Ion 530 chip, and the results were analyzed with the IonReporter tools (version 5.16, Thermo Fisher Scientific) and IGV software (Integrative Genome Viewer version 2.9.2—https://software.broadinstitute.org/software/igv/, accessed on 22 December 2021). According to the previously reported validation [28], only mutations present in at least 5% of the total number of reads analyzed and observed in both strands were considered for mutational calls. The significance of alterations was checked by using the Varsome database (https://varsome.com/, accessed on 22 December 2021) and “IARC TP53 Database” (http://p53.iarc.fr/, accessed on 22 December 2021).
2.3. Statistical Analysis
In this study, we chose to evaluate as our final end-point the occurrence of the death of the patients as the major event. The p-values less than 0.05 were considered statistically significant. Variables that were found to be significant on univariate analysis at p < 0.1 were included in multivariate analysis in a backward stepwise fashion. Cox proportional hazards models were generated for multivariate analysis. Statistical analysis was performed by using SPSS Statistics 20.
3. Results
Clinicopathological and molecular characteristics of the cohort analyzed are reported in Table 1.

Table 1.
Clinicopathological and molecular characteristics of the cohort analyzed by NSG.
3.1. Molecular Alteration in PDAC
DNA was amplifiable in 64 of 68 samples (94.1%), while, in four cases, the quality/quantity of DNA was not enough for us to obtain reliable results. In these cases, the coverage obtained after the NGS analysis was too low (<100 reads per amplicon), and then the samples were excluded from the following analyses. This inadequacy of the specimens may be due to an over-degradation of the samples as a result of pre-analytic conditions (e.g., prolonged formalin fixation). At least one gene alteration was detected in 52 of 64 cases (81.2%), while, in 12 cases (18.8%), no alterations were detected the in genes analyzed.
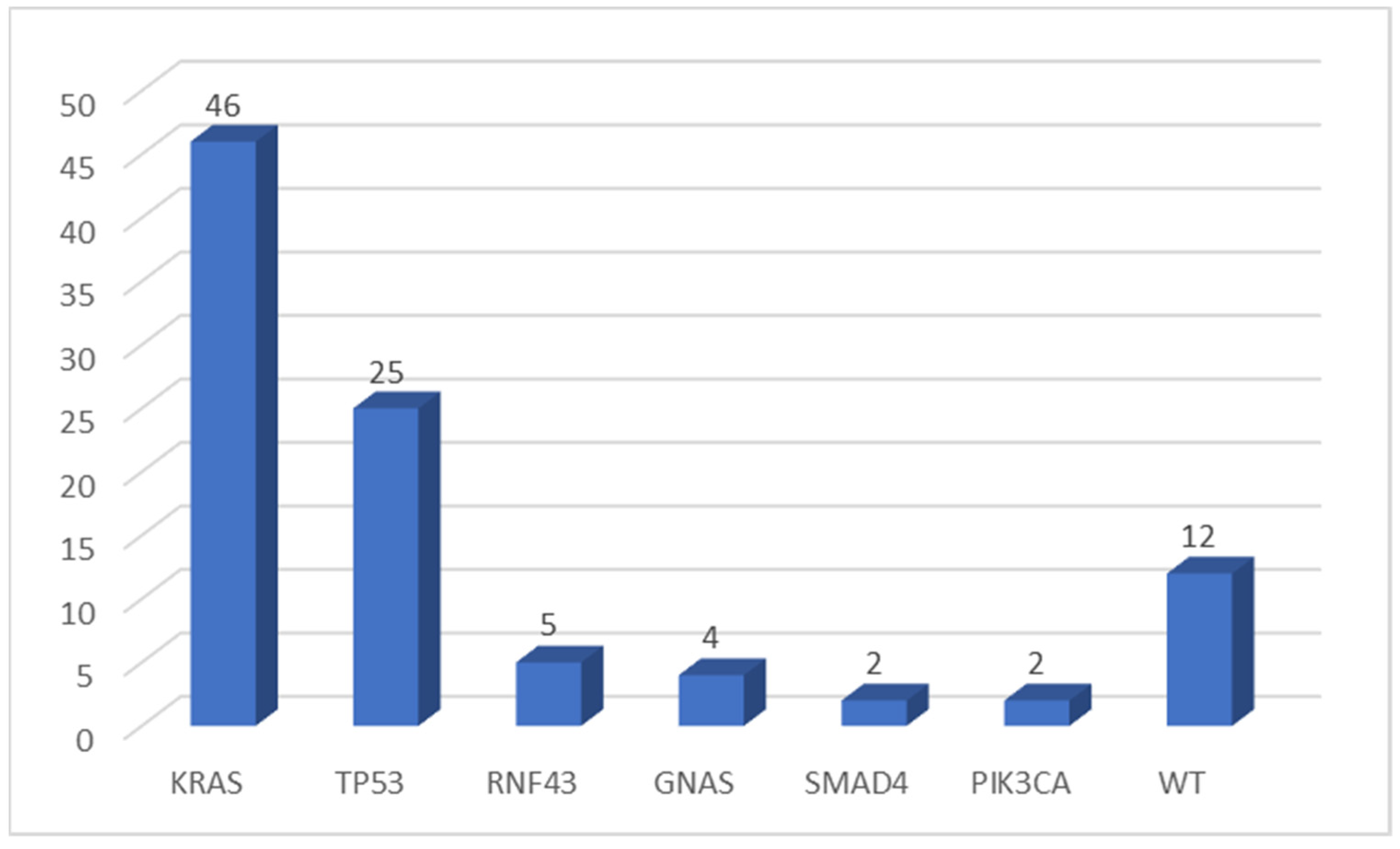
KRAS was the most frequently mutated gene (46 of 64 evaluable samples, 71.9%), while TP53 was altered in 25 cases (39.0%). The more frequent KRAS mutation was the p.Gly12Asp substitution (34.8% of KRAS mutated samples). No p.Gly12Cys mutation was detected in our cohort. Other genes were found to be mutated in a few numbers of cases: RNF43 in 5 of 64 (7.8%), GNAS in 4 (6.3%), SMAD4 in 2 samples (3.1%), and PIK3CA altered in two tumors (3.1%) (Figure 1). All variants found in this cohort of pancreatic tumors were pathogenic mutations (according to the Varsome database), except for one TP53 mutation, two RNF43 variants, and two SMAD4 alterations (classified as VUS—variant of uncertain significance).
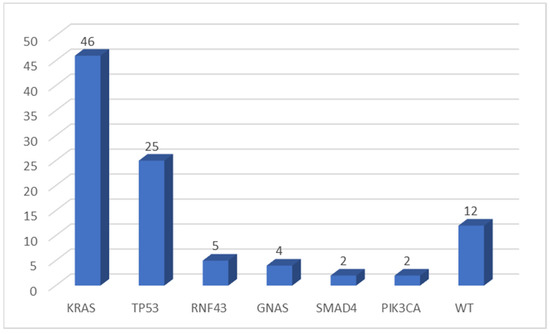
Figure 1.
Number of mutations detected by multigene NGS panel. WT, wild type.
Concomitant Mutations in PDAC
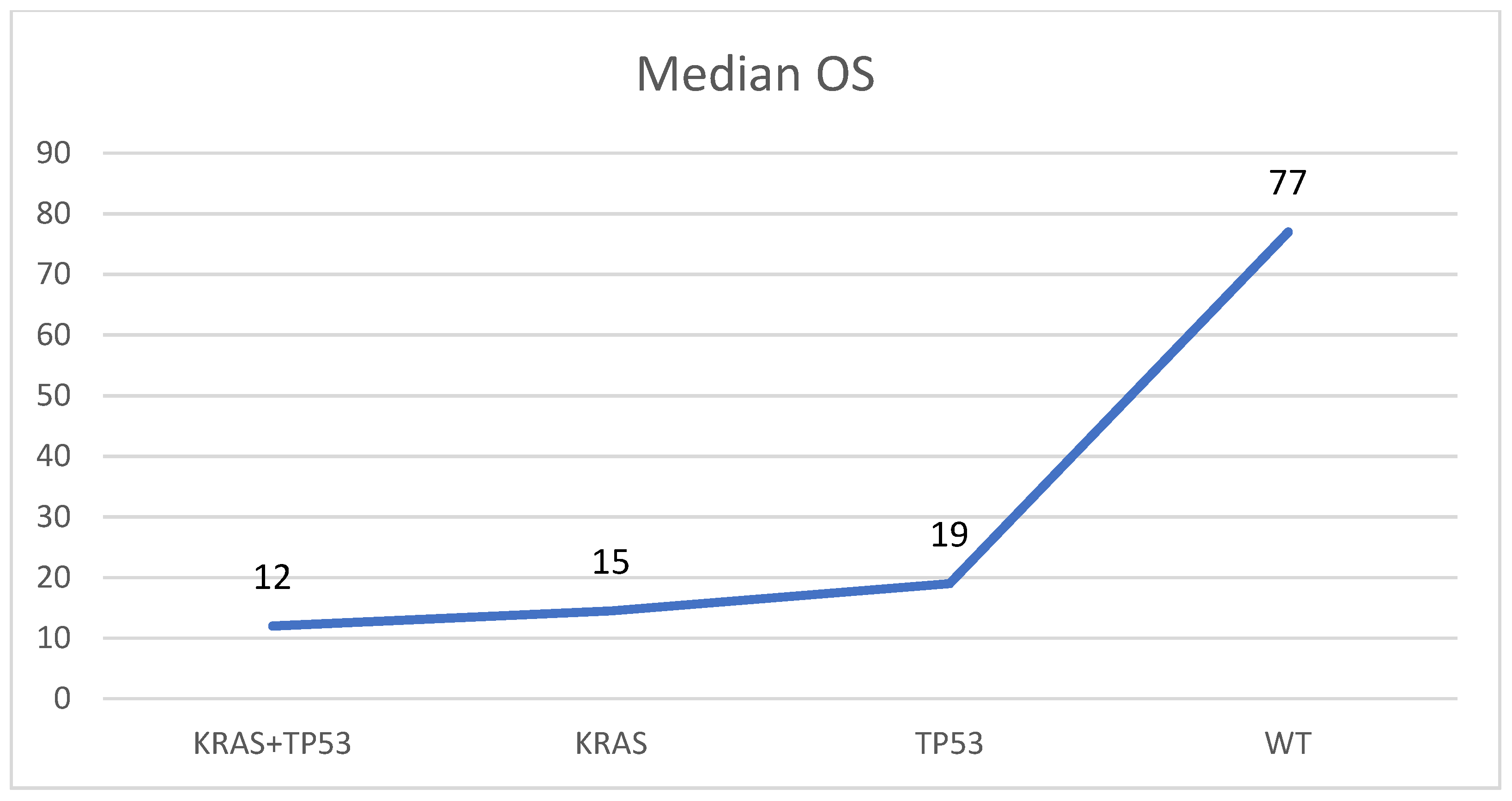
In 28 of 52 mutated samples (53.8%), concomitant alterations were detected. The most frequent combination was between KRAS and TP53, found altered together in 20 of 52 mutated specimens (28.5%). In four samples (8.7%) another two alterations were found (one KRAS + GNAS, two KRAS + RNF43, and one KRAS + SMAD4). In three samples (4.3%) three concomitant mutations were observed (one KRAS + GNAS + RNF43, one KRAS + TP53 + RNF43, and one KRAS + RNF43 + PIK3CA), and in one specimen (2.2%), four variants were observed (KRAS + GNAS + SMAD4 + PIK3CA). In the remnant 24 samples, single alterations were observed: 18 samples (75.0%) harbored a single KRAS mutation, 4 (16.7%) harbored only TP53 variants, and 2 cases (8.3%) contained one GNAS alteration. The median overall survival (OS) was higher in patients with PDAC harboring no KRAS/TP53 mutations (77 months) if compared to those with TP53 (OS: 19 ms), KRAS (OS: 15 ms), or KRAS and TP53 (OS: 12 ms) mutations (Figure 2).
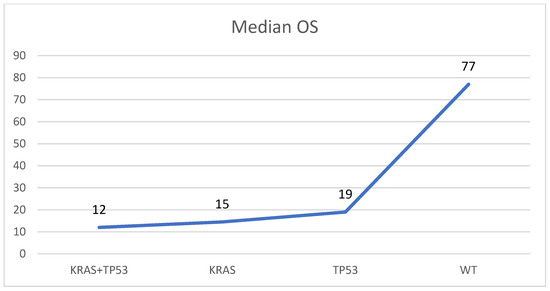
Figure 2.
Median overall survival (OS) in patients with PDAC without KRAS/TP53 mutations (WT), with a mutation in KRAS or TP53, and with mutations in KRAS and TP53 (KRAS + TP53). Months are reported on the Y-axis.
3.2. Correlation between Molecular/Clinicopathological Features and Outcome
As far as the histopathological variable is concerned, the tumor stage (represented by tumor size and lymph node status), as well as perineural invasion, vascular invasion, and margin resection status, is a well-known predictive marker of recurrence and seems to be related to survival in PDCA patients [29,30,31,32,33].
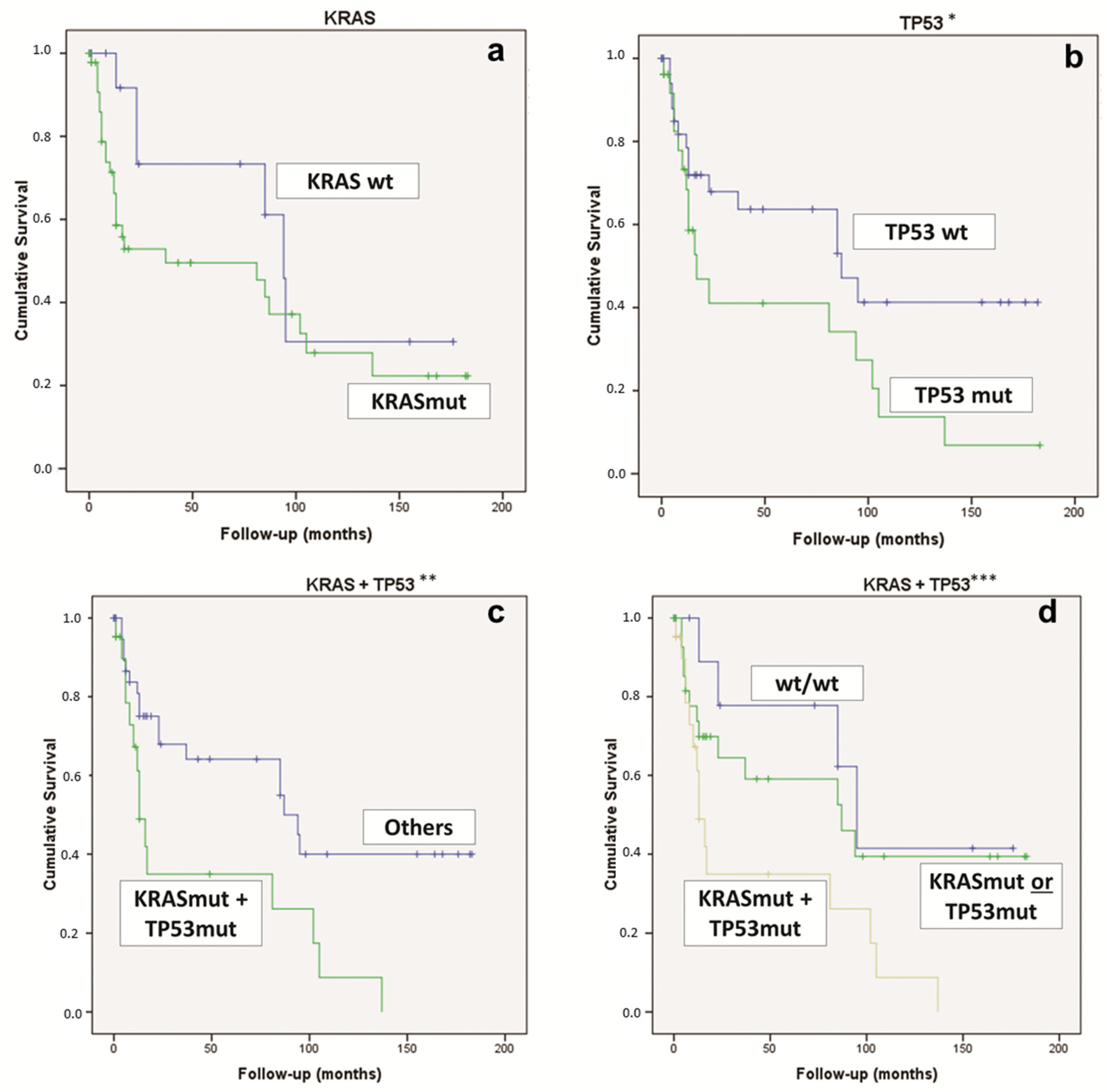
Considering the whole overall survival, for the univariate analysis, the statistically significant features were TP53 mutations (p = 0.037) and the concomitant KRAS & TP53 mutations (p = 0.023) (Figure 3). On the contrary, the KRAS mutation alone was not statistically significant (Figure 3).
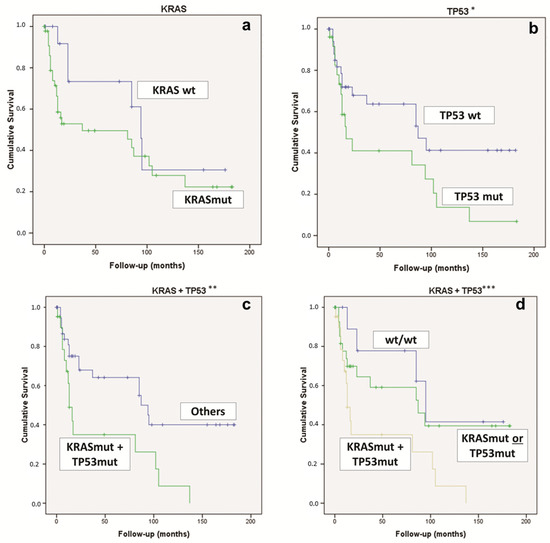
Figure 3.
Kaplan–Meier curve comparing (a) PDAC KRAS-WT vs. KRAS-mut (p = 0.151); (b) PDAC TP53-WT vs. TP53-mut (* p = 0.037); (c) PDAC KRAS & TP53-mut vs. “other” PDAC (** p = 0.023); (d) PDAC KRAS & TP53-mut vs. KRAS-mut or TP53-mut vs. KRAS-WT and TP53-WT (*** p = 0.016). mut: mutated; WT, wild type.
We evaluated the clinicopathological and molecular features of the patients who died within the first 24 months after surgery (n = 23 patients—71.9%). Of these 23 PDAC patients, 21 (91.3%) harbored KRAS mutations (p = 0.021), 13 (56.5%) had a TP53 mutation (p = 0.047), and 12 (52.2%) had concomitant KRAS and TP53 alterations (p = 0.031) (Table 2). Intriguingly, 15 PDACs (65.2%) showed perineural invasion (p = 0.028).

Table 2.
Molecular characteristics of patients who died within 24 months compared to those of patients with OS higher than 24 months.
Taking into consideration the 19 patients with PDAC who were still alive after 60 months, in concordance with data reported in the literature, we found that these patients, compared with those with overall survival of fewer than 60 months, showed the following: (i) fewer incidences of vascular invasion (52.6% vs. 91.7%, respectively, p = 0.018) and (ii) a lower incidence of lymph-nodal involvement (36.8% vs. 81.8%, p = 0.001).
As confirmation, at logistic regression, a multivariate analysis considering only the histopathological variables of the lymph-nodal involvement was the most important parameter toward cancer-related death (Exp(B) = 1.349).
Including also molecular characterization of PDAC, the strongest feature was the presence of concomitant KRAS & TP53 mutations (Exp(B) = 1.600).
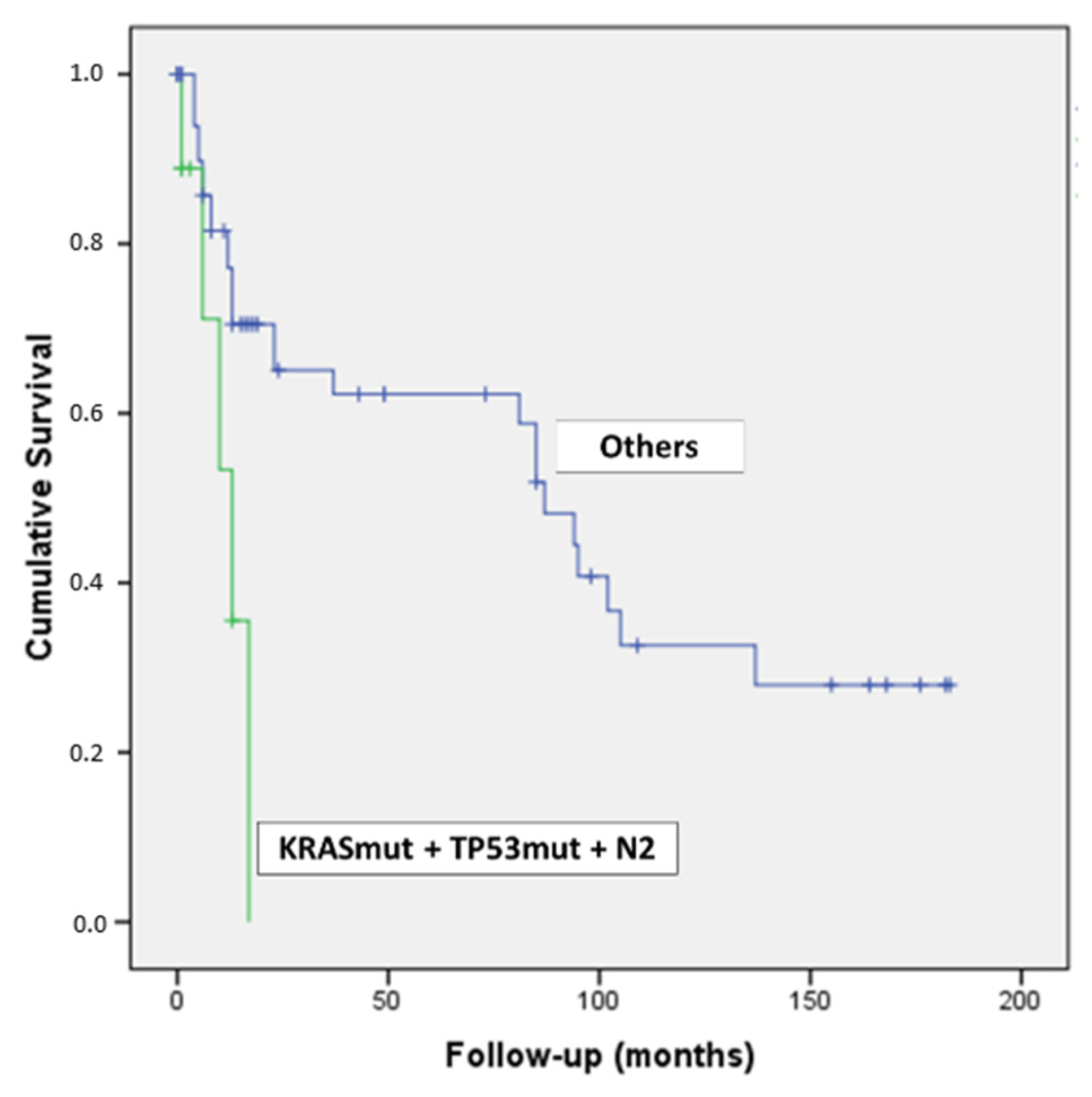
Intriguingly, by combining the concomitant presence of KRAS & TP53 mutations and N2 lymph-nodal status, we detected nine patients with a dramatic outcome (p = 0.007) (Figure 4).
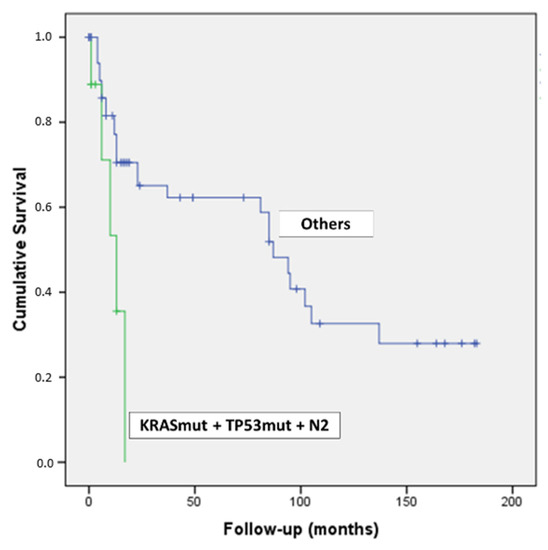
Figure 4.
Kaplan–Meier curve of patients with PDAC harboring KRAS & TP53 mutations and N2 lymph-nodal status.
4. Discussion
Pancreatic ductal adenocarcinoma (PDAC) is usually associated with a poor prognosis and a low survival rate, less than 10% at five years [12]. Despite all the efforts in these years to investigate and study the genetic and molecular alterations in pancreatic cancer, the majority of patients present with locally advanced or metastatic disease at the time of the diagnosis [12], and there are not still powerful druggable mutations that could be used in the clinical practice.
The characterization of multiple markers in solid tumors, using the NGS approach, has allowed for the translation of several molecular alterations into the clinical practice of metastatic tumors, as in lung adenocarcinomas, colorectal cancers, prostate cancer, ovarian carcinoma, and cholangiocarcinomas [34]. The clinical significance of molecular alterations found in pancreatic ductal adenocarcinoma is still controversial. PDACs are usually characterized by KRAS mutations, TP53 alterations, and/or loss of SMAD4 or CDKN2A/p16 proteins [11,12,13,17,18,19,20,21].
In this study, we tested a multigene laboratory-developed NGS panel used to characterize PDAC in daily clinical practice. Herein, we wanted to investigate the utility of using a laboratory-developed multigene panel for the molecular characterization of PDAC, aiming to identify prognostic biomarkers to improve the clinical management of PDAC patients.
Our data confirm that KRAS and TP53 are the most common genes altered in a series of PDACs, as previously described in the literature [23,25,28,35]. Other mutations (i.e., RNF43, GNAS, SMAD4, and PIK3CA) have been rarely found in our cohort. As previously reported [24], several PDAC samples showed concomitant alterations, mainly in KRAS and TP53 genes.
In our study, we demonstrated that PDCA patients’ survival is related to the presence of the TP53 or KRAS mutation: the presence of PDAC harboring TP53 mutations worsens the survival if compared to the TP53 wild-type tumors. Even if KRAS mutations are not significantly associated with overall survival, it should be considered that it seems to affect the outcome of the patients in the first part of the survival curve (Figure 3). Considering that patients died within the first 24 months from surgery, almost all (91.3%) harbored a KRAS mutation, more than half (56.5%) of them had a TP53 alteration, and concomitant KRAS and TP53 alterations were detected in about a half (52.2%). Intriguingly, only two patients who died within 24 months had a PDAC without KRAS and/or TP53 alterations. Moreover, patients with PDAC harboring the concomitant mutations in KRAS and TP53 have a significantly worse OS if compared to those with PDAC, with only one of the two genes mutated, or with both KRAS and TP53 wild-type (Figure 3).
Analyzing the clinicopathological features, we see that the only statistically significant variable in a logistic regression multivariate analysis was represented by the lymph-nodal status. The other histological parameters that the literature reported to be predictive of recurrence and to be related to survival in PDCA patients (i.e., the tumor stage, the presence of perineural or vascular invasion, and the margin resection status) did not demonstrate a significant prognostic value in term of OS in our patient cohort.
Combining the KRAS and TP53 double-mutated tumor phenotype with the abovementioned lymph-nodal stage at the time of surgery, we observed a dramatic prognosis in the double-mutated PDAC patients with an N2 stage compared to all the other patients (wild-type phenotype or tumors harboring just one mutation, without lymph-nodal spread, N0, or with limited ones, N1) (Figure 4).
Based on our NGS analysis, the current study confirms the data reported by the literature about the main driver genes (i.e., KRAS and TP53) found mutated in PDAC.
The KRAS p.G12C variant is a mutation that could be targeted by specific inhibitor molecules [36]. However, we have not found the KRAS p.G12C mutation in our cohort of patients.
In 2020, the ESMO outlined the indications for the use of NGS in the characterization of several metastatic cancers, including pancreatic ductal adenocarcinoma. According to these guidelines, it is not currently recommended to perform multigene NGS in daily practice, even if multigene sequencing is encouraged in order to get access to innovative drugs [34]. Moreover, NGS can be an alternative technique to PCR-based assays if it is not associated with extra costs for the public healthcare system and if the patient is informed about the putative benefits of this analysis [34]. Our results confirm that, in daily practice testing, KRAS and TP53 alone may be sufficient to improve the clinical management of the patients, and that NGS multigene panels can be used in clinical research centers to increase access to innovative clinical study. Moreover, our data highlight that PDACs with KRAS and TP53 wild-type may represent a distinct molecular subtype of pancreatic cancer that could benefit from tailored treatments [37]. This double-negative PDAC may also be investigated for the presence of HRR genes alterations, detecting a putative cohort of patients that could benefit from PARP inhibitors treatment. On the contrary, the analysis of KRAS and TP53 genes in PDAC patients would help to identify those subjects with worse overall survival (i.e., KRAS and TP53 mutated PDAC).
5. Conclusions
In conclusion, our work highlights that PDCAs harboring both KRAS and TP53 mutation have a shorter OS compared to wild-type tumors or PDCAs harboring only one mutation. The presence of a high lymph-nodal patient’s stage at the time of surgery, combined with the presence of a double mutated tumor phenotype, is indicative of a dramatically short OS.
A clinical implication that can be gathered from our study is the possibility to translate the knowledge of KRAS and TP53 mutational status pre-operatively from cytological material and fine-needle biopsy tissue, conditioning the future clinical management of patients with PDAC. It should be considered that the recent improvement in the needles for endoscopic ultrasound-guided fine-needle biopsy (EUS-FNB) has allowed us to obtain a large amount of material also in preoperative procedures, providing enough cells for molecular analysis in pancreatic lesions [38,39]. Moreover, a good concordance between EUS-FNB and surgical (both formalin-fixed paraffin-embedded) specimens has been demonstrated [40]. For these reasons, the analysis of KRAS and TP53, using NGS or other molecular techniques, could be easily used also in preoperative specimens. The molecular analyses performed on preoperative material of pancreatic lesions, together with the lymph node status, may help to hypothesize patients’ outcomes and influence the decision-making process, for example, pushing toward neoadjuvant chemotherapy.
Author Contributions
Conceptualization, D.d.B., D.M. and F.V.; methodology, D.d.B., T.M. and V.S.; formal analysis, D.d.B., D.M. and F.V.; data curation, A.D.L., A.F., M.M., C.B., R.L., M.F.O., S.F., M.D.M. and M.R.; writing—original draft preparation, D.d.B. and D.M.; writing—review and editing, D.d.B. and D.M.; visualization, A.D., E.F., A.A.B., E.J. and G.T.; project administration, D.d.B.; funding acquisition, G.T. and D.d.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All information regarding the human material was managed by using anonymous numerical codes, and the study was carried out in accordance with the ethical principles of the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/, accessed on 27 August 2021). The study was approved by the local review board committee (Azienda USL Bologna, protocol number 15035, Bologna Italy).
Data Availability Statement
All available data are contained within the article.
Conflicts of Interest
D.d.B. has received personal fees (as consultant and/or speaker bureau) from Boehringer Ingelheim and Eli Lilly that are unrelated to the current work.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Huang, C.; Cui Zhou, D.; Hu, Y.; Lih, T.M.; Savage, S.R.; Krug, K.; Clark, D.J.; Schnaubelt, M.; Chen, L.; et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 2021, 184, 5031–5052.e5026. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M. Pancreatic cancer. N. Engl. J. Med. 2010, 362, 1605–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.J.; Goldstein, D.; Hamm, J.; Figer, A.; Hecht, J.R.; Gallinger, S.; Au, H.J.; Murawa, P.; Walde, D.; Wolff, R.A.; et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007, 25, 1960–1966. [Google Scholar] [CrossRef]
- Qian, Y.; Gong, Y.; Fan, Z.; Luo, G.; Huang, Q.; Deng, S.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2020, 13, 130. [Google Scholar] [CrossRef]
- Sinn, M.; Bahra, M.; Liersch, T.; Gellert, K.; Messmann, H.; Bechstein, W.; Waldschmidt, D.; Jacobasch, L.; Wilhelm, M.; Rau, B.M.; et al. CONKO-005: Adjuvant Chemotherapy With Gemcitabine Plus Erlotinib Versus Gemcitabine Alone in Patients After R0 Resection of Pancreatic Cancer: A Multicenter Randomized Phase III Trial. J. Clin. Oncol. 2017, 35, 3330–3337. [Google Scholar] [CrossRef]
- Han, S.S.; Jang, J.Y.; Kim, S.W.; Kim, W.H.; Lee, K.U.; Park, Y.H. Analysis of long-term survivors after surgical resection for pancreatic cancer. Pancreas 2006, 32, 271–275. [Google Scholar] [CrossRef]
- Ferrone, C.R.; Brennan, M.F.; Gonen, M.; Coit, D.G.; Fong, Y.; Chung, S.; Tang, L.; Klimstra, D.; Allen, P.J. Pancreatic adenocarcinoma: The actual 5-year survivors. J. Gastrointest Surg. 2008, 12, 701–706. [Google Scholar] [CrossRef]
- Adham, M.; Jaeck, D.; Le Borgne, J.; Oussoultzouglou, E.; Chenard-Neu, M.P.; Mosnier, J.F.; Scoazec, J.Y.; Mornex, F.; Partensky, C. Long-term survival (5-20 years) after pancreatectomy for pancreatic ductal adenocarcinoma: A series of 30 patients collected from 3 institutions. Pancreas 2008, 37, 352–357. [Google Scholar] [CrossRef]
- Ferrone, C.R.; Pieretti-Vanmarcke, R.; Bloom, J.P.; Zheng, H.; Szymonifka, J.; Wargo, J.A.; Thayer, S.P.; Lauwers, G.Y.; Deshpande, V.; Mino-Kenudson, M.; et al. Pancreatic ductal adenocarcinoma: Long-term survival does not equal cure. Surgery 2012, 152, S43–S49. [Google Scholar] [CrossRef] [Green Version]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, C.A.; Lawrence, S.A.; Richards, A.L.; Chou, J.F.; Wong, W.; Capanu, M.; Berger, M.F.; Donoghue, M.T.A.; Yu, K.H.; Varghese, A.M.; et al. Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer 2020, 126, 3939–3949. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Pentheroudakis, G.; Committee, E.G. Recent eUpdates to the ESMO Clinical Practice Guidelines on hepatocellular carcinoma, cancer of the pancreas, soft tissue and visceral sarcomas, cancer of the prostate and gastric cancer. Ann. Oncol. 2019, 30, 1395–1397. [Google Scholar] [CrossRef]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goere, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. 5), v56–v68. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Raphael, B.J.; Hruban, R.H.; Aguirre, A.J.; Moffitt, R.A.; JenYeh, J.; Stewart, C.; GordonRobertson, A.; Cherniack, A.D.; Gupta, M. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e113. [Google Scholar] [CrossRef] [Green Version]
- Bernard, V.; Semaan, A.; Huang, J.; San Lucas, F.A.; Mulu, F.C.; Stephens, B.M.; Guerrero, P.A.; Huang, Y.; Zhao, J.; Kamyabi, N.; et al. Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clin. Cancer Res. 2019, 25, 2194–2205. [Google Scholar] [CrossRef] [Green Version]
- Fischer, C.G.; Wood, L.D. From somatic mutation to early detection: Insights from molecular characterization of pancreatic cancer precursor lesions. J. Pathol. 2018, 246, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Maitra, A.; Adsay, N.V.; Argani, P.; Iacobuzio-Donahue, C.; De Marzo, A.; Cameron, J.L.; Yeo, C.J.; Hruban, R.H. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod. Pathol. 2003, 16, 902–912. [Google Scholar] [CrossRef] [Green Version]
- Murphy, S.J.; Hart, S.N.; Lima, J.F.; Kipp, B.R.; Klebig, M.; Winters, J.L.; Szabo, C.; Zhang, L.; Eckloff, B.W.; Petersen, G.M.; et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 2013, 145, 1098–1109.e1091. [Google Scholar] [CrossRef] [Green Version]
- Visani, M.; Acquaviva, G.; De Leo, A.; Sanza, V.; Merlo, L.; Maloberti, T.; Brandes, A.A.; Franceschi, E.; Di Battista, M.; Masetti, M.; et al. Molecular alterations in pancreatic tumors. World J. Gastroenterol. 2021, 27, 2710–2726. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Brosens, L.A.A.; Wood, L.D.; Chatterjee, D.; Shin, J.I.; Sciammarella, C.; Fiadone, G.; Malleo, G.; Salvia, R.; Kryklyva, V.; et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: Histology, molecular pathology and clinical implications. Gut 2021, 70, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [Green Version]
- Masetti, M.; Acquaviva, G.; Visani, M.; Tallini, G.; Fornelli, A.; Ragazzi, M.; Vasuri, F.; Grifoni, D.; Di Giacomo, S.; Fiorino, S.; et al. Long-term survivors of pancreatic adenocarcinoma show low rates of genetic alterations in KRAS, TP53 and SMAD4. Cancer Biomark 2018, 21, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Oshima, M.; Okano, K.; Muraki, S.; Haba, R.; Maeba, T.; Suzuki, Y.; Yachida, S. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann. Surg. 2013, 258, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.M.; Hwang, H.K.; Park, J.; Kim, C.; Cho, S.K.; Yun, M.; Lee, W.J. Maximum Standard Uptake Value as a Clinical Biomarker for Detecting Loss of SMAD4 Expression and Early Systemic Tumor Recurrence in Resected Left-Sided Pancreatic Cancer. Medicine 2016, 95, e3452. [Google Scholar] [CrossRef]
- Amin, M.B.; American Joint Committee on Cancer; American Cancer Society. AJCC Cancer Staging Manual, 8th ed.; American Joint Committee on Cancer, Springer: Chicago IL, USA, 2017; p. xvii. 1024p. [Google Scholar]
- de Biase, D.; Acquaviva, G.; Visani, M.; Sanza, V.; Argento, C.M.; De Leo, A.; Maloberti, T.; Pession, A.; Tallini, G. Molecular Diagnostic of Solid Tumor Using a Next Generation Sequencing Custom-Designed Multi-Gene Panel. Diagnostics 2020, 10, 250. [Google Scholar] [CrossRef]
- Barugola, G.; Partelli, S.; Marcucci, S.; Sartori, N.; Capelli, P.; Bassi, C.; Pederzoli, P.; Falconi, M. Resectable pancreatic cancer: Who really benefits from resection? Ann. Surg. Oncol. 2009, 16, 3316–3322. [Google Scholar] [CrossRef]
- Guo, S.W.; Shen, J.; Gao, J.H.; Shi, X.H.; Gao, S.Z.; Wang, H.; Li, B.; Yuan, W.L.; Lin, L.; Jin, G. A preoperative risk model for early recurrence after radical resection may facilitate initial treatment decisions concerning the use of neoadjuvant therapy for patients with pancreatic ductal adenocarcinoma. Surgery 2020, 168, 1003–1014. [Google Scholar] [CrossRef]
- Kimura, K.; Amano, R.; Nakata, B.; Yamazoe, S.; Hirata, K.; Murata, A.; Miura, K.; Nishio, K.; Hirakawa, T.; Ohira, M.; et al. Clinical and pathological features of five-year survivors after pancreatectomy for pancreatic adenocarcinoma. World J. Surg. Oncol. 2014, 12, 360. [Google Scholar] [CrossRef] [Green Version]
- La Torre, M.; Nigri, G.; Lo Conte, A.; Mazzuca, F.; Tierno, S.M.; Salaj, A.; Marchetti, P.; Ziparo, V.; Ramacciato, G. Is a preoperative assessment of the early recurrence of pancreatic cancer possible after complete surgical resection? Gut Liver 2014, 8, 102–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, S.; Shimoda, M.; Shimazaki, J.; Maruyama, T.; Oshiro, Y.; Nishida, K.; Sahara, Y.; Nagakawa, Y.; Tsuchida, A. Predictive Early Recurrence Factors of Preoperative Clinicophysiological Findings in Pancreatic Cancer. Eur. Surg. Res. 2018, 59, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Kohno, T.; Ueno, H.; Hiraoka, N.; Kondo, S.; Saito, M.; Shimada, Y.; Ichikawa, H.; Kato, M.; Shibata, T.; et al. Utility of Assessing the Number of Mutated KRAS, CDKN2A, TP53, and SMAD4 Genes Using a Targeted Deep Sequencing Assay as a Prognostic Biomarker for Pancreatic Cancer. Pancreas 2017, 46, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Gillson, J.; Ramaswamy, Y.; Singh, G.; Gorfe, A.A.; Pavlakis, N.; Samra, J.; Mittal, A.; Sahni, S. Small Molecule KRAS Inhibitors: The Future for Targeted Pancreatic Cancer Therapy? Cancers 2020, 12, 1341. [Google Scholar] [CrossRef]
- Luchini, C.; Paolino, G.; Mattiolo, P.; Piredda, M.L.; Cavaliere, A.; Gaule, M.; Melisi, D.; Salvia, R.; Malleo, G.; Shin, J.I.; et al. KRAS wild-type pancreatic ductal adenocarcinoma: Molecular pathology and therapeutic opportunities. J. Exp. Clin. Cancer Res. 2020, 39, 227. [Google Scholar] [CrossRef]
- Crino, S.F.; Di Mitri, R.; Nguyen, N.Q.; Tarantino, I.; de Nucci, G.; Deprez, P.H.; Carrara, S.; Kitano, M.; Shami, V.M.; Fernandez-Esparrach, G.; et al. Endoscopic Ultrasound-guided Fine-needle Biopsy With or Without Rapid On-site Evaluation for Diagnosis of Solid Pancreatic Lesions: A Randomized Controlled Non-Inferiority Trial. Gastroenterology 2021, 161, 899–909.e895. [Google Scholar] [CrossRef]
- Fabbri, C.; Fornelli, A.; Fuccio, L.; Giovanelli, S.; Tarantino, I.; Antonini, F.; Liotta, R.; Frazzoni, L.; Gusella, P.; La Marca, M.; et al. High diagnostic adequacy and accuracy of the new 20G procore needle for EUS-guided tissue acquisition: Results of a large multicentre retrospective study. Endosc. Ultrasound. 2019, 8, 261–268. [Google Scholar] [CrossRef]
- Larghi, A.; Lawlor, R.T.; Crino, S.F.; Luchini, C.; Rizzatti, G.; Curatolo, M.; Gabbrielli, A.; Inzani, F.; Scarpa, A. Endoscopic ultrasound guided fine needle biopsy samples to drive personalized medicine: A proof of concept study. Pancreatology 2020, 20, 778–780. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).