Abstract
Nasopharyngeal swab (NPS) and oropharyngeal swab (OPS) are the most widely used upper respiratory tract specimens for diagnosis of SARS-CoV-2 using RT-qPCR. In contrast, nasal swab (NS) and saliva (SS), recently recommended by the WHO, are rarely used, and their test accuracy is limited. The method for direct RT-PCR detection of SARS-CoV-2 does not require an RNA extraction and is faster and easier than standard RT-PCR tests with RNA extraction. This study aimed to compare the diagnostic performance of upper respiratory tract samples for SARS-CoV-2 detection using the direct RT-PCR without preliminary heat inactivation. Here we report the application and validation of direct RT-PCR SARS-CoV-2 RNA on 165 clinical specimens of NPS/OP, and 36 samples of NS/NPS and 37 saliva samples (for the latter with prior deproteinization). The overall sensitivity estimates were 95.9%, 94.2%, 88.9%, and 94.6% for NPS/OPS/NS/SS samples, respectively, and the specificity was 100% against standard RT-PCR with RNA extraction. Overall, NS and SS testing by direct RT-PCR had sufficient sensitivity to detect SARS-CoV-2. They can be acceptable alternative to NPS/OPS for rapid detection of SARS-CoV-2 infections in future.
1. Introduction
The pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) still puts a tremendous strain on public health worldwide. The availability of accurate COVID-19 diagnostic tests is one of the critical steps of tackling the pandemic [1].
Real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is the current gold standard for the diagnosis of COVID-19 from NPS and/or OPS specimens, collected in viral transport medium, and is included in the protocols recommended by the WHO and other international organizations [2,3].
The commonly used protocols require a ribonucleic acid (RNA) extraction step, which may be seen as a critical aspect in the massive laboratory testing process, especially in the SARS-CoV-2 era [4]. The huge demand for RT-PCR testing of SARS-CoV-2 has led to a worldwide shortage of laboratory reagents, including RNA isolation kits, which remains a major problem. To overcome the shortage of reagents and expand the capacity for SARS-CoV-2 testing, approaches skipping the RNA extraction step have been developed [5,6,7].
Several groups have shown the feasibility of extraction-free RT-PCR for SARS-CoV-2 mainly on NPS with a preliminary heat inactivation step at 95 °C for 5 min or 70–75 °C for 10–15 min [7,8,9,10]. It has also been suggested that the absence of the preliminary heat inactivation step in direct RT-PCR may lead to insufficient release of RNA from the virion and a decrease in the efficiency of RNA detection by direct RT-PCR [10]. On the other hand, the heat treatment step (95 °C for 5 min), a prior direct RT-PCR aimed to lyse epithelial cells, may, however, reduce the detection rate of the samples with low viral load due to the inverse effect of thermolysis on RNA integrity as was noted by others [11,12,13].
For initial diagnostic testing for current SARS-CoV-2 infections, CDC recommended collecting and testing upper respiratory tract specimens such as NPS and OPS as well mid-turbinate NS and saliva [14]. The FDA also states that that “more data are necessary to better understand the performance when using specific saliva collection or other specimen types for COVID-19 testing” [15]. Moreover, self-collected nasal swabs and saliva are less invasive and easy to collect than NPS/OPS and thus more suitable for mass screening than NPS sampling [16,17].
OPS and NPS have been a preferred specimen type in Russia for SARS-CoV-2 molecular diagnosis since the beginning of the pandemic. However, the invasiveness and painful of this sampling are of its disadvantages. Additionally, variable collection quality and demand for supervised collection by trained personnel create obstacles for the scalability of this testing [18,19]. Recently recommended non-invasive sampling such as saliva [20,21] or nasal swabs [22] combined with rapid direct COVID-19 RT-PCR assays [23] could potentially enable broader SARS-CoV-2 testing of at-risk populations. Standard RT-PCR assays with RNA extraction using NS and SS have been rarely applied in Russia, data about the diagnostic sensitivity for direct RT-PCR is still limited.
The aim of this study was to evaluate the diagnostic performance of the direct multiplex RT-PCR method, without RNA extraction, on clinical specimens of upper respiratory tract: NPS, OPS, NS, and saliva, and to compare with the performance of the traditional method based on RNA extraction, in a real-world setting.
2. Materials and Methods
2.1. Upper Respiratory Clinical Sample Collection and Preparation
Upper respiratory tract specimens, including nasopharyngeal, oropharyngeal, and nasal swabs as well as saliva were collected and were a part of routine diagnostics to detect SARS-CoV-2. Informed voluntary consent was obtained from all involved patients. The study was carried out according to the guidelines of the World Medical Association’s Declaration of Helsinki.
We were able to collect 122 OPS/NPS, 36 NS/NPS, and 37 saliva/OPS paired SARS-CoV-2 positive samples, and 43 SARS-CoV-2 negative samples. Symptom status of patients was not recorded at the time of the study swab. OPS collection involved swabbing of the palatine arches and the back wall of the oropharynx. The NPS sampling was performed through one nostril. For NS collection, one nostril was swabbed to a depth of at least 2 cm and rotated 3 times. Saliva specimens were diluted with 3-fold low saline buffer for an endpoint volume of 1.0 mL to reduce the viscosity of the sample. Saliva suspension was centrifuged at 2000× g for 5 min at 4 °C, and the supernatant was treated with Proteinase K at 55 °C for 15 min following heat inactivation at 95 °C for 5 min. Finally, 10 µL of saliva suspension was used in direct RT-PCR. One oropharyngeal and one nasopharyngeal swab from each patient or one nasopharynx and one nasal swab from each patient or oropharyngeal and saliva swab were placed into 0.5 mL of STORE-F transport medium (DNA-technologies, Moscow, Russia) to obtain a sample to set either direct (without RNA extraction) or standard RT-PCR. All types of samples were stored at +4 °C for a maximum of 5 days until processing. The mean number of days from diagnostic swab to study swabs was 3.1 (range 1–5).
2.2. Viral RNA Extraction
All specimens were collected from patients in Moscow and the Moscow region, Russia. Samples were centrifuged at high speed (>2000× g) for 5 min to pellet cellular debris and then stored at +4 °C until qPCR was performed. RNA extraction was performed using manual extraction protocol with a triazole based PREP-NA kit (DNA Technologies, Moscow, Russia) according to the manufacturer’s manual. Viral RNA extract was eluted in 100 μL of the elution buffer, and an RT-PCR assay was performed.
2.3. SARS-CoV-2 RNA Detection Using Standard RT-PCR
In addition, 10 μL of the RNA elute were mixed with 40 μL of PCR master mix SARS-CoV-2 Lite Real-Time PCR Detection Kit (DNA-Technologies, Moscow, Russia) to make a total of 50 μL of reaction volume. The master mix contained components as following PCR buffer with magnesium salt, primers, and probes. Ferment mix included reverse transcriptase, RNAsin, and DNA polymerase. RT-PCR applying SARS-CoV-2 Lite Real-Time PCR Detection Kit was able to target the SARS-CoV-2 E and RdRp genes with detection in the FAM channel. The analytical sensitivity (LOD) reported was 10 copies/reaction. Automatic analysis was performed on DTprime Real-Time Thermal Cycler using the DTmaster software for the DNA Technologies kit, included in the DT Lite system (DNA Technologies, Moscow, Russia). The result was interpreted as a positive if threshold value (Ct value) <40 for all two target genes. The PCR conditions consisted of 1 cycle of 5 min at 60 °C, 2 min at 94 °C and followed by 45 cycles of 5 s at 94 °C, 10 s at 58 °C and 10 s at 64 °C.
2.4. Detection of Viral RNA by Direct RT-PCR Method without RNA Extraction
Direct RT-qPCR without RNA extraction was performed using the same SARS-CoV-2 Lite Real-Time PCR Detection Kit (DNA Technologies, Moscow, Russia) according to the manufacturers’ instructions. Before analysis, the swabs were spin centrifugated at >2000× g for 5 min and 10 µL were used in RT-PCR without prior heat inactivation. In addition, 10 μL of the sample in transport medium STORE-F were mixed with 40 μL of PCR master mix SARS-CoV-2 Lite Real-Time PCR Detection Kit to make a total of 50 μL of reaction volume. Direct RT-PCR applying SARS-CoV-2 Lite Real-Time PCR Detection Kit was conducted with the same protocol as the standard RT-PCR. A positive result, indicating the presence of SARS-CoV-2 RNA, was determined according to the Ct value. Ct values of <40 with the target primer/probe set for SARS-CoV-2 indicated virus presence. Positive (SARS-CoV-2 Lite RNA Positive Control) and negative controls (nuclease-free water) were used to control and exclude possible false-negative results. MS2 phage served as an internal control for control reverse transcription and the amplification of material collected in each sample.
2.5. Statistical Analysis
The correlation between the Ct values for both direct and standard methods was calculated by Pearson’s rank correlation using the Excel Analysis ToolPak. p-values of <0.05 were considered statistically significant. The sensitivity and specificity values and corresponding confidence intervals were calculated with assistance of an online tool (Available online: https://www.medcalc.org/calc/diagnostic_test.php (accessed on 18 March 2022)).
3. Results
3.1. Sensitivity Assessment of Direct RT-PCR on Clinical Oropharyngeal and Nasopharyngeal Swabs
A total of 165 tested samples were stored in STORE-F at +4 °C before PCR as recommended by the manufacturer. To evaluate the ability of direct RT-PCR to detect SARS-CoV-2 RNA without RNA isolation, 122 clinical OPS/NPS pair samples from patients with SARS-CoV-2 and 43 SARS-CoV-2 negative ones were performed. The standard RT-PCR protocol served as a reference to evaluate the performance of the direct PCR protocol. Threshold cycle values ranging from 18.3 to 40.5 were previously obtained by standard RT-qPCR using SARS-CoV-2/SARS-CoV Multiplex Real-Time PCR Detection Kit (DNA Technologies, Moscow, Russia) with RNA isolation from a 200 µL sample in transport medium and concentrating in 50 µL of the TE, and subdivided into samples with high viral load (Ct < 24), medium (Ct 25–34), and low (Ct > 35). For direct RT-PCR, 10 µL of swab specimen stored in STORE-F transport medium were used. In parallel, a 100 µL aliquot of each sample was used for RNA isolation without subsequent concentration and the equivalent of 10 µL of the original swab diluent used for RT-PCR, allowing a one-to-one comparison with direct RT-PCR. The overall diagnostic sensitivity of direct RT-PCR on OPS and NPS samples relative to the standard with RNA isolation was 94.2% (95% CI: 86.1–97.6%) and 95.9% (95% CI: 91.2–98.4%) (Table 1). Ct (mean ± standard deviation) values for both targets E (envelope) and RdRP (RNA-dependent RNA polymerase) genes by OPS (27.2 ± 5.1) and NPS (26.8 ± 4.9) testing were similar. Differences in mean Ct values for the target genes in direct RT-PCR on both OPS and NPS were not statistically significant (p = 0.59). The direct RT-PCR protocol using OPS showed 95% concordance with the standard protocol and mean difference in Ct values of 1.69 for both E gene and the RdRp gene targets. The direct RT-PCR protocol using NPS showed 98% concordance with the standard protocol and mean difference in Ct values of 1.62 for both E gene and the RdRp gene targets.

Table 1.
Detection sensitivity of direct RT-qPCR versus standard RT-qPCR on oropharyngeal and nasopharyngeal swabs containing a range of SARS-CoV-2 viral RNA loads.
Of the 122 positive samples, 6 and 5 were false negatives by the direct RT-PCR on OPS and NPS samples, respectively, compared to the standard protocol with RNA extraction. All but one OPS and NPS samples not detected by direct RT-PCR corresponded to Ct = 39.0 and Ct = 40.5 in standard RT-PCR with RNA isolation, i.e., were treated as samples with an extremely low viral load. Only one OPS sample appears to be a false negative with a corresponding Ct value of 33 in standard RT-PCR. Triple dilution and deproteinization with Proteinase K before repeated direct RT-PCR allowed the detection of SARS-CoV-2 in this sample (Ct = 34.4). The presence of saliva inhibitory proteins and salts in this OPS sample assumed to cause a failure to detect SARS-CoV-2 RNA in the first setting.
3.2. Sensitivity Assessment of Direct RT-PCR on Clinical Nasal and Nasopharyngeal Swabs
Next, we tested NS and NPS samples in parallel followed by direct RT-qPCR versus standard RT-qPCR with RNA extraction on a collection of 68 patients. Of these subjects, 36 (52.9%) were found to be positive and 32 (47.1%) negative for SARS-CoV-2 RNA using the standard RT-qPCR with RNA extraction from NPS. The sensitivity of SARS-CoV-2 RNA detection by direct RT-PCR from 36 nasal samples was 88.9% (95% CI: 81.3–94.1%), and specificity was 100.0% (95% CI: 94.6–100.0%) (Table 2). Thus, we found that the % positive NS (88.9% (95% CI: 76.3–93.2%)) was lower than % positive NPS swab (94.4% (95% CI: 86.1–97.8%)). Upon comparison of direct RT-qPCR with standard RT-qPCR, NS showed an agreement of 94.1% for both positive and negative samples. The direct RT-PCR protocol using NPS showed 97% concordance with the standard protocol. Upon comparison of direct RT-qPCR using NS versus NPS, an agreement was 97.0%. Ct (mean ± standard deviation) values for both targets E and RdRP genes by NPS (26.9 ± 4.9) and NS (27.6 ± 5.2) were similar. Differences in Ct values for the target genes were not statistically significant (p = 0.59).

Table 2.
Detection sensitivity of direct RT-qPCR versus standard RT-qPCR on nasal swabs and nasopharyngeal swabs containing a range of SARS-CoV-2 viral RNA loads.
Of the 36 positive results from direct RT-PCR protocol on NS, 9 samples (25.0%) exhibited low viral loads (Ct > 35). Only 4 and 2 NS and NPS samples failed to be detected by direct RT-PCR. However, one NS sample was false negative by direct RT-PCR, while it has been detected in paired NPS by direct RT-PCR (Ct = 32.6) and by standard RT-PCR (Ct = 31). This result probably indicated either the uneven virus distribution in the NS/NPS or inhibitory effect of mucus in the NS sample.
Nasal swab sampling with direct RT-PCR correlated with the standard RT-PCR across the whole range of Ct values as shown in Figure 1. The correlation coefficient (R2 = 0.893) may be associated with the analysis of normalized Ct values.
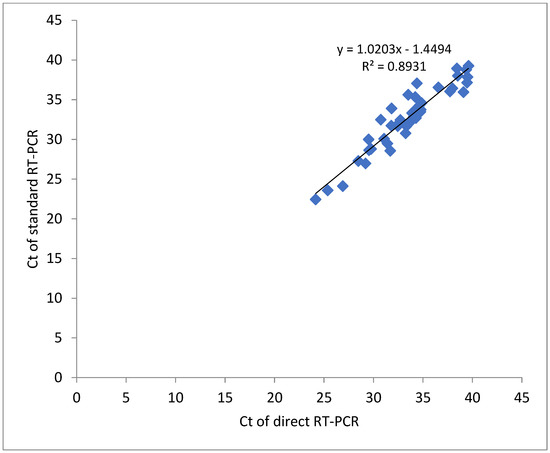
Figure 1.
Correlation of Ct values detected for NS samples in direct and standard SARS-CoV-2 RT-qPCR methods.
3.3. Direct RT-PCR on Saliva Samples
Next, the effectiveness of direct multiplex RT-PCR using saliva samples was tested. Previously, we have found that, to reduce the inhibitory effect of salivary proteins before performing direct RT-PCR on saliva samples, it is necessary to add Proteinase K (+ProK) to the samples and deproteinize for 15 min at 55 °C, followed by proteinase inactivation at 95 °C for 5 min. Using only heat treatment (95 °C for 5–10 min) and fold dilution of the original saliva sample resulted in a significant number of samples non-detectable or invalid due to the lack of signal for both target targets and internal control. In addition to deproteinization, the second factor improving the efficiency of direct RT-PCR was at least 3-fold dilution of the original saliva sample to reduce viscosity as recommended [24].
We compared the effectiveness of direct RT-PCR on saliva samples with Proteinase K treatment (Saliva/ProK) and with Proteinase K and 3-fold dilution (saliva/ProK+:3) against standard RT-PCR on 37 positive and 24 negative saliva samples. The sensitivity of SARS-CoV-2 RNA detection by direct RT-PCR on both variants was 91.9% (95% CI: 84.1–95.2%) and 94.6 (95% CI: 86.2–97.9%), respectively. Specificity was 100.0% (95% CI: 94.6–100.0%) (Table 3). A few false-negative saliva samples in direct RT-PCR corresponded to samples with a Ct value ranging of 39.2 to 40.0 in standard RT-PCR, indicating a viral load close to the LOD level (10 copies/reaction). Upon comparing direct RT-qPCR with standard RT-qPCR, SS with dilution showed an agreement of 97.0%, while SS without dilution –95.0%. Ct (mean ± standard deviation) values for both targets E and RdRP genes for SS without dilution (27.3 ± 4.8) and SS with dilution (28.1 ± 5.2) were similar. Thus, we found that the % positive saliva samples (94.6%) were similar to % positive OPS swab (94.2%).

Table 3.
Detection sensitivity of direct RT-qPCR versus standard RT-qPCR on saliva and 3-fold diluted saliva samples containing a range of SARS-CoV-2 viral RNA loads.
For the Ct value correlation between the direct method and the standard method for saliva samples, the correlation coefficient of the E gene and RdRP genes was 0.987. Thus, Ct values of the detected genes were highly correlated between the two methods (Figure 2).
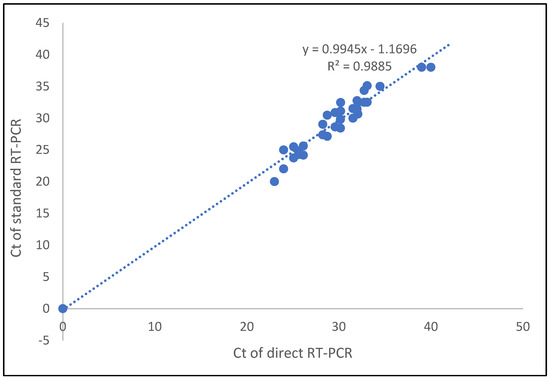
Figure 2.
Correlation of Ct values detected for saliva samples in direct and standard SARS-CoV-2 RT-qPCR methods.
4. Discussion
In this study, we demonstrated that the direct RT-PCR method for detection of SARS-CoV-2 may be a valuable alternative to a standard RT-PCR method with RNA extraction. Our data describe quite sensitive direct detection of SARS-CoV-2 RNA by multiplex RT-qPCR without heat shock compared to standard RT-PCR on real-world upper respiratory tract samples including NPS, OPS, nasal (mid-turbinate) swabs, and saliva. Regarding COVID-19, all of these upper respiratory tract samples are recommended for SARS-CoV-2 RT-qPCR detection [2].
All four sample types tested by the direct RT-PCR appeared to capture slightly lower % positives: NPS (95.9% (95% CI: 91.2–98.4%)) and OPS (94.2% (95% CI: 86.1–97.6%)), nasal swabs (88.9% (95% CI 76.3–93.2%)), and saliva (94.6 (95% CI: 86.2–97.9%)) in comparison to standard RT-PCR with RNA extraction. We found that the detection rate for NPS seemed to be slightly higher compared to OPS, but it was estimated on a small number of positive patients.
Previous studies have reported opposing conclusions regarding whether nasal sampling is concordant or discordant with NPS [25,26]. Here, we report that the molecular detection of SARS-CoV-2 by direct RT-PCR on using NS was slightly lower for the detection using NPS. We observed that the discordant samples in pair NPS+/NS- had low viral load on the NPS. This may well be uneven distribution of the virus through the upper respiratory tracts (nasal and nasopharyngeal) or an inhibitory effect of mucosal secretion in the NS sample. Our results strongly suggest that the difference in NS and NPS/OPS performance depends on the viral load in positive samples, with high concordance for medium-to-high viral loads (Ct < 35) and low concordance for very low viral loads, which may lie close to the LOD of the direct RT-PCR assay. On the other hand, sequential sampling (NPS first, then NS or NPS first, then OPS or OPS first, then saliva) may have also affected performance with subsequent swabs. Overall, SARS-CoV-2 detection using direct RT-PCR from nasal samples showed appropriate sensitivity and high specificity.
Using direct RT-PCR with saliva specimens, we found similar performance compared to NPSs and very similar to OPS (94.6 (95% CI: 86.2–97.9%)) versus 95.9% (95% CI: 91.2–98.4%) and 94.2% (95% CI: 86.1–97.6%), respectively, and only the absence of sufficient dilution of saliva sample resulted in a noticeably lower rate of detection (91.9% (95% CI, 84.1–95.2%)). Several previous studies reported that saliva specimens show similar sensitivity to NPS and OPS and support our data [16,24,27,28,29].
Our results provide data that the direct detection of SARS-CoV-2 without heat pretreatment might be an acceptable alternative to the standard RT-PCR with RNA extraction for SARS-CoV-2 detection, although very low viral loads samples would not be detected. Several direct RT-PCR protocols have been described and validated using a pre-heat step at high temperatures above 95 °C for direct RT-qPCR without RNA extraction [8,9,10,30]. The heat pretreatment procedure introduced in order to lyse epithelial cells may, however, reduce sensitivity and increase false negative rate as it was noted for temperature regimen 95 °C [12,13,31] as well as 70–75 °C [9]. In our study, the direct RT-qPCR was in complete agreement with the standard RT-PCR method with all four types of upper respiratory tract samples. However, the detection accuracy with NS samples highly depends on the viral load in the sample, and several false negative results are possible in the samples with very low viral load close to the limit of detection.
This study has several limitations, especially regarding the sample collection conditions for SARS-CoV-2 RT-qPCR. We did not have any information regarding whether the participants were symptomatic at the time of swabbing, which could impact viral shedding dynamics for different types of samples. The collectors did not provide sufficient information about the collection day of infection and other key issues (asymptomatic or symptomatic, early or late testing from symptom onset, severity or mild status). We also were not able to collect all samples in all four types for each patient. Secondly, we provided evidence about the sensitivity of NS and saliva samples on a limited number of patients with SARS-CoV-2 positive and negative diagnosis. We did not have enough participants to say with 95% certainty that NS was 8% less sensitive than NPS or that saliva was as sensitive as OPS. We cannot exclude that sensitivity values would be lower on aa larger group of participants. Another limitation to the saliva sampling is the fact that some patients failed to produce enough saliva. Food or drug contaminants in saliva might also deteriorate the quality of the assays. As for NS samples, it is not always possible to evaluate the influence of variable concomitant inhibitory factors to yield sensitivity of direct RT-PCR. Such inhibitory factor is excessive mucus secretion caused by rhinitis. An additional limitation of our method is safety concerns for healthcare professionals being potentially exposed to the SARS-CoV-2 virus shortly before PCR set up, although now direct RT-PCR is a routinely used method by trained personnel in the special facilities in Russia.
Several groups recently reported about sufficient sensitivity of standard and direct RT-PCR on self-collected NS and saliva samples in SARS-CoV-2 diagnosis among symptomatic patients [23,32]. Authors described the opportunity of self-collection sampling as a vivid alternative to routine collection by trained personnel. Due to current guidelines of the Russian Ministry of Health on diagnosis and treatment of novel coronavirus infection (COVID-19) [33] to make sample collection by health-care-workers only, we did not make an attempt to evaluate the performance of our direct RT-PCR on self-collected samples.
5. Conclusions
In conclusion, we have shown that a direct RT-qPCR method without RNA extraction can effectively detect SARS-CoV-2 RNA from upper respiratory tract specimens including nasal swabs and saliva. The direct RT-PCR method had sufficient sensitivity to detect SARS-CoV-2 with viral loads that correlate with the presence of an infectious virus. We propose that the use of non-invasive nasal swabs and saliva combined with direct RT-qPCR has a potential to provide the diagnostic utility of this approach for future SARS-CoV-2 testing.
Author Contributions
Conceptualization, S.A.K.; Methodology, S.A.K. and T.A.L.; Software, M.V.K. and T.A.L.; Validation, S.A.K., T.A.L. and V.V.K.; Formal analysis, S.A.K., T.A.L. and M.V.K.; Investigation, T.A.L. and V.V.K.; Resources, S.A.K., D.Y.T. and A.P.S.; Data curation, S.A.K. and T.A.L.; Writing—original draft preparation, S.A.K. and M.V.K.; Writing—review and editing, S.A.K. and M.V.K.; Visualization, S.A.K. and M.V.K.; Supervision, A.P.S. and D.Y.T.; Project administration, S.A.K.; Funding acquisition, A.P.S. and D.Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Borkakoty, B.; Jakharia, A.; Bali, N.K.; Sarmah, M.D.; Hazarika, R.; Baruah, G.; Bhattacharya, C.; Biswas, D. A preliminary evaluation of normal saline as an alternative to viral transport medium for COVID-19 diagnosis. Indian J. Med. Res. 2021, 153, 684–688. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance; WHO: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/handle/10665/331329 (accessed on 18 March 2022).
- European Centre for Disease Prevention and Control. COVID-19 Testing Strategies and Objectives; ECDC: Stockholm, Sweden, 2020; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/TestingStrategy_Objective-Sept-2020.pdf (accessed on 5 March 2021).
- Wang, H.; Liu, Q.; Hu, J.; Zhou, M.; Yu, M.Q.; Li, K.Y.; Xu, D.; Xiao, Y.; Yang, J.Y.; Lu, Y.J.; et al. Nasopharyngeal swabs are more sensitive than oropharyngeal swabs for COVID-19 diagnosis and monitoring the SARS-CoV-2 load. Front. Med. 2020, 7, 334. [Google Scholar] [CrossRef] [PubMed]
- Wee, S.K.; Sivalingam, S.P.; Yap, E.P.H. Rapid Direct Nucleic Acid Amplification Test without RNA Extraction for SARS-CoV-2 Using a Portable PCR Thermocycler. Genes 2020, 11, 664. [Google Scholar] [CrossRef] [PubMed]
- Alcoba-Florez, J.; González-Montelongo, R.; Íñigo-Campos, A.; de Artola, D.G.; Gil-Campesino, H.; The Microbiology Technical Support Team; Ciuffreda, L.; Valenzuela-Fernández, A.; Flores, C. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int. J. Infect. Dis. 2020, 97, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Kriegova, E.; Fillerova, R.; Kvapil, P. Direct-RT-qPCR detection of SARS-CoV-2 without RNA extraction as part of a COVID-19 testing strategy: From sample to result in one hour. Diagnostics 2020, 10, 605. [Google Scholar] [CrossRef]
- Bruce, E.A.; Huang, M.L.; Perchetti, G.A.; Tighe, S.; Laaguiby, P.; Hoffman, J.J.; Gerrard, D.L.; Nalla, A.K.; Wei, Y.; Greninger, A.L.; et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol. 2020, 18, e3000896. [Google Scholar] [CrossRef] [PubMed]
- Smyrlaki, I.; Ekman, M.; Lentini, A.; de Sousa, R.; Papanicolaou, N.; Vondracek, M.; Aarum, J.; Safari, H.; Muradrasoli, S.; Rothfuchs, A.G.; et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARSCoV-2 RT-PCR. Nat. Commun. 2020, 11, 4812. [Google Scholar] [CrossRef]
- Byrnes, S.A.; Gallagher, R.; Steadman, A.; Bennett, C.; Rivera, R.; Ortega, C.; Motley, S.T.; Jain, P.; Weigl, B.H.; Connelly, J.T. Multiplexed and extraction-free amplification for simplified SARS-CoV-2 RT-PCR tests. Anal. Chem. 2021, 93, 4160–4165. [Google Scholar] [CrossRef]
- Zou, J.; Zhi, S.; Chen, M.; Su, X.; Kang, L.; Li, C.; Su, X.; Zhang, S.; Ge, S.; Li, W. Heat inactivation decreases the qualitative real-time RT-PCR detection rates of clinical samples with high cycle threshold values in COVID-19. Diagn. Microbiol. Infect. Dis. 2020, 98, 115109. [Google Scholar] [CrossRef]
- Burton, J.; Love, H.; Richards, K.; Burton, C.; Summers, S.; Pitman, J.; Easterbrook, L.; Davies, K.; Spencer, P.; Killip, M.; et al. The effect of heat-treatment on SARS-CoV-2 viability and detection. J. Virol. Methods. 2021, 290, 114087. [Google Scholar] [CrossRef]
- Grant, P.R.; Turner, M.E.; Shin, G.Y.; Nastouli, E.; Levett, L.J. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. BioRxiv 2020. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed on 18 March 2022).
- U.S. Food and Drug Administration, Center for Devices and Radiological Health. FAQs on Testing for SARS-CoV-2. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/faqs-testing-sars-cov-2 (accessed on 12 October 2020).
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical evaluation of self-collected saliva by quantitative reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, reverse transcription–loop-mediated isothermal amplification, and a rapid antigen test to Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Fujisawa, S.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; Fukumoto, T.; Sato, K.; Oguri, S.; Taki, K.; Senjo, H.; et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020, 81, e145–e147. [Google Scholar] [CrossRef]
- Kinloch, N.N.; Ritchie, G.; Brumme, C.J.; Dong, W.; Dong, W.; Lawson, T.; Jones, R.B.; Montaner, J.S.G.; Leung, V.; Romney, M.G.; et al. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J. Infect. Dis. 2020, 222, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Surkova, E.; Nikolayevskyy, V.; Drobniewski, F. False-positive COVID-19 results: Hidden problems and costs. Lancet Respir. Med. 2020, 8, 1167–1168. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Vaz, S.N.; Santana, D.S.; Netto, E.M.; Pedroso, C.; Wang, W.K.; Santos, F.D.A.; Brites, C. Saliva is a reliable, non-invasive specimen for SARS-CoV-2 detection. Braz. J. Infect. Dis. 2020, 24, 422–427. [Google Scholar] [CrossRef]
- McCulloch, D.J.; Kim, A.E.; Wilcox, N.C.; Logue, J.K.; Greninger, A.L.; Englund, J.A.; Chu, H.Y. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw. Open. 2020, 3, e2016382. [Google Scholar] [CrossRef]
- Kriegova, E.; Fillerova, R.; Raska, M.; Manakova, J.; Dihel, M.; Janca, O.; Sauer, P.; Klimkova, M.; Strakova, P.; Kvapil, P. Excellent option for mass testing during the SARS-CoV-2 pandemic: Painless self-collection and direct RT-qPCR. Virol. J. 2021, 18, 95. [Google Scholar] [CrossRef]
- Ranoa, D.R.E.; Holland, R.L.; Alnaji, F.G.; Green, K.J.; Wang, L.; Brooke, C.B.; Burke, M.D.; Fan, T.M.; Hergenrother, P.J. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. BioRxiv 2020. [Google Scholar] [CrossRef]
- Kojima, N.; Turner, F.; Slepnev, V.; Bacelar, A.; Deming, L.; Kodeboyina, S.; Klausner, J.D. 2020. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for for coronavirus disease 2019 detection. Clin. Infect. Dis. 2021, 73, e3106–e3109. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.A.; Herigon, J.C.; Benedetti, A.; Pollock, N.R.; Denkinger, C.M. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. J. Clin. Microbiol. 2021, 59, e02881-20. [Google Scholar] [CrossRef] [PubMed]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- Teo, A.K.J.; Choudhury, Y.; Tan, I.B.; Cher, C.Y.; Chew, S.H.; Wan, Z.Y.; Cheng, L.T.E.; Oon, L.L.E.; Tan, M.H.; Chan, K.S.; et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci. Rep. 2021, 11, 3134. [Google Scholar] [CrossRef]
- Lai, J.; German, J.; Hong, F.; Tai, S.H.S.; McPhaul, K.M.; Milton, D.K. University of Maryland StopCOVID Research Group for the University of Maryland StopCOVID Research Group. Comparison of Saliva and Midturbinate Swabs for Detection of SARS-CoV-2. Microbiol. Spectr. 2022, e00128-22. [Google Scholar] [CrossRef]
- Beltrán-Pavez, C.; Alonso-Palomares, L.A.; Valiente-Echeverría, F.; Gaggero, A.; Soto-Rifo, R.; Barriga, G.P. Accuracy of a RT-qPCR SARS-CoV-2 detection assay without prior RNA extraction. J. Virol. Methods. 2021, 287, 113969. [Google Scholar] [CrossRef] [PubMed]
- Domnich, A.; De Pace, V.; Pennati, B.M.; Caligiuri, P.; Varesano, S.; Bruzzone, B.; Orsi, A. Evaluation of extraction-free RT-qPCR methods for SARS-CoV-2 diagnostics. Arch. Virol. 2021, 166, 2825–2828. [Google Scholar] [CrossRef]
- Gertler, M.; Krause, E.; van Loon, W.; Krug, N.; Kausch, F.; Rohardt, C.; Rössig, H.; Michel, J.; Nitsche, A.; Mall, M.A.; et al. Self-collected oral, nasal and saliva samples yield sensitivity comparable to professionally collected oro-nasopharyngeal swabs in SARS-CoV-2 diagnosis among symptomatic outpatients. Int. J. Infect. Dis. 2021, 110, 261–266. [Google Scholar] [CrossRef]
- Russian Ministry of Health. Current Guidelines: Prevention, Diagnosis and Treatment of Novel Coronavirus Infection. Version 15 (22.02.22). Available online: https://static-0.minzdrav.gov.ru/system/attachments/attaches/000/059/392/original/BMP_COVID-19_V15.pdf (accessed on 18 March 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).