Author Contributions
Conceptualization, H.A. and M.A.; methodology, H.A., Y.A.-I. and M.A.; software, H.A., M.A. and A.Z.; validation, H.A., Y.A.-I., W.A.M. and M.A.; formal analysis, Y.A.-I., H.A. and M.A.; writing—original draft preparation, Y.A.-I. and H.A.; writing—review and editing, H.A., W.A.M., M.A., Y.A.-I. and I.A.Q. and visualization, H.A. and A.Z.; supervision, H.A. and W.A.M.; project administration, H.A. and Y.A.-I. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Flow chart of the proposed methodology.
Figure 1.
Flow chart of the proposed methodology.
Figure 2.
(A) point-like, (B) point-flaky, (C) and flaky corneal.
Figure 2.
(A) point-like, (B) point-flaky, (C) and flaky corneal.
Figure 3.
(A) Grade 0, (B) Grade 1, (C) Grade 2, and (D) Grade 4.
Figure 3.
(A) Grade 0, (B) Grade 1, (C) Grade 2, and (D) Grade 4.
Figure 4.
(A) Type 0, (B) Type 1, (C) Type 2, (D) Type 4, and (E) Type 4.
Figure 4.
(A) Type 0, (B) Type 1, (C) Type 2, (D) Type 4, and (E) Type 4.
Figure 5.
(A) Original image, (B) cornea area, (C) enhanced gray image, and (D) final enhanced image.
Figure 5.
(A) Original image, (B) cornea area, (C) enhanced gray image, and (D) final enhanced image.
Figure 6.
Residual learning: a building block [
31].
Figure 6.
Residual learning: a building block [
31].
Figure 7.
SVM models for general pattern classification.
Figure 7.
SVM models for general pattern classification.
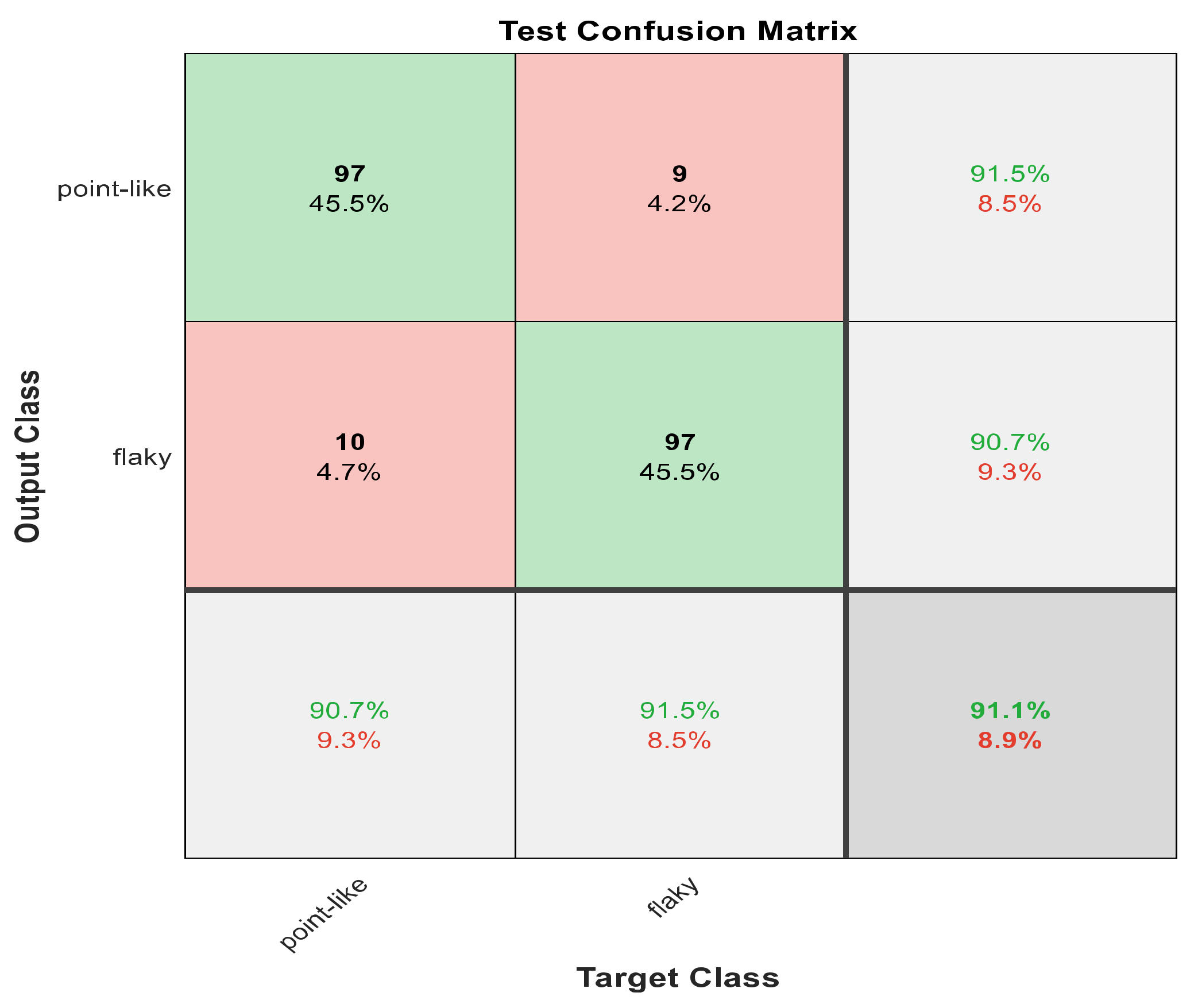
Figure 8.
The confusion matrix with ECFS-reduced features for Model 1.
Figure 8.
The confusion matrix with ECFS-reduced features for Model 1.
Figure 9.
The confusion matrix with ECFS-reduced features for Model 2.
Figure 9.
The confusion matrix with ECFS-reduced features for Model 2.
Figure 10.
The confusion matrix with ECFS-reduced features for the whole cascading system.
Figure 10.
The confusion matrix with ECFS-reduced features for the whole cascading system.
Figure 11.
The confusion matrix with ECFS-reduced features for type grading.
Figure 11.
The confusion matrix with ECFS-reduced features for type grading.
Figure 12.
The confusion matrix with PCA-reduced features for grade grading.
Figure 12.
The confusion matrix with PCA-reduced features for grade grading.
Figure 13.
The AROC with ECFS-reduced features for Model 1.
Figure 13.
The AROC with ECFS-reduced features for Model 1.
Figure 14.
The AROC with ECFS-reduced features for Model 2.
Figure 14.
The AROC with ECFS-reduced features for Model 2.
Figure 15.
The AROC with ECFS-reduced features for the whole cascading system.
Figure 15.
The AROC with ECFS-reduced features for the whole cascading system.
Figure 16.
The AROC with ECFS-reduced features for type grading.
Figure 16.
The AROC with ECFS-reduced features for type grading.
Figure 17.
The AROC with PCA-reduced features for grade grading.
Figure 17.
The AROC with PCA-reduced features for grade grading.
Figure 18.
The confusion matrix for Model 1 using 1000 deep learning features.
Figure 18.
The confusion matrix for Model 1 using 1000 deep learning features.
Figure 19.
The confusion matrix for Model 2 using 1000 deep learning features.
Figure 19.
The confusion matrix for Model 2 using 1000 deep learning features.
Figure 20.
The confusion matrix for whole cascading system using 1000 deep learning features.
Figure 20.
The confusion matrix for whole cascading system using 1000 deep learning features.
Figure 21.
The confusion matrix with for type grading using 1000 deep learning features.
Figure 21.
The confusion matrix with for type grading using 1000 deep learning features.
Figure 22.
The confusion matrix with for grade grading using 1000 deep learning features.
Figure 22.
The confusion matrix with for grade grading using 1000 deep learning features.
Figure 23.
The AROC for Model 1 using 1000 deep learning features.
Figure 23.
The AROC for Model 1 using 1000 deep learning features.
Figure 24.
The AROC for Model 2 using 1000 deep learning features.
Figure 24.
The AROC for Model 2 using 1000 deep learning features.
Figure 25.
The AROC for cascading model using 1000 deep learning features.
Figure 25.
The AROC for cascading model using 1000 deep learning features.
Figure 26.
The AROC for type grading using 1000 deep learning features.
Figure 26.
The AROC for type grading using 1000 deep learning features.
Figure 27.
The AROC for grade grading using 1000 deep learning features.
Figure 27.
The AROC for grade grading using 1000 deep learning features.
Figure 28.
Accuracy for the 30 most significant features for type grading in both automated and hand-crafted features.
Figure 28.
Accuracy for the 30 most significant features for type grading in both automated and hand-crafted features.
Figure 29.
Accuracy for the 30 most significant features for severity grading in both automated and hand-crafted features.
Figure 29.
Accuracy for the 30 most significant features for severity grading in both automated and hand-crafted features.
Figure 30.
Accuracy for the 30 most significant features for severity grading and type grading employing automatic features.
Figure 30.
Accuracy for the 30 most significant features for severity grading and type grading employing automatic features.
Figure 31.
Accuracy for the 50 most significant features for severity grading and type grading employing automatic features.
Figure 31.
Accuracy for the 50 most significant features for severity grading and type grading employing automatic features.
Table 1.
The distribution of the images used in the system.
Table 1.
The distribution of the images used in the system.
| General Pattern | Point-Like | Point-Flaky Mixed | Flaky | | |
|---|
| Number of images | 358 | 263 | 91 | | |
| Type grading (specific pattern) | Type 0 | Type 1 | Type 2 | Type 3 | Type 4 |
| Number of images | 36 | 98 | 203 | 273 | 102 |
| Grade grading (severity degree) | Grade 0 | Grade 1 | Grade 2 | Grade 3 | Grade 4 |
| Number of images | 36 | 78 | 50 | 50 | 548 |
Table 2.
Augmentation Process for General Grading Dataset.
Table 2.
Augmentation Process for General Grading Dataset.
| General Grading Class | Point Like | Flaky | Point-Flaky Mixed |
|---|
| Before Augmentation | 358 | 91 | 263 |
| After Augmentation | 358 | 182 | 263 |
| Augmentation Multiplier | 0 | 2 | 0 |
Table 3.
Augmentation Process for Type Grading Classes.
Table 3.
Augmentation Process for Type Grading Classes.
| Type Grading Classes | Type 0 | Type 1 | Type 2 | Type 3 | Type 4 |
|---|
| Before Augmentation | 36 | 98 | 203 | 273 | 102 |
| After Augmentation | 288 | 294 | 203 | 273 | 306 |
| Augmentation Multiplier | 8 | 3 | 0 | 0 | 3 |
Table 4.
Augmentation Process for Grade Grading Classes.
Table 4.
Augmentation Process for Grade Grading Classes.
| Grade Grading Classes | Grade 0 | Grade 1 | Grade 2 and 3 | Grade 4 |
|---|
| Before Augmentation | 36 | 78 | 50 | 548 |
| After Augmentation | 540 | 624 | 550 | 548 |
| Augmentation Multiplier | 15 | 8 | 11 | 0 |
Table 5.
The stricter of ResNet101 [
31].
Table 5.
The stricter of ResNet101 [
31].
| Layer Name | Output Size | ResNet101 |
|---|
| Conv1 | 112 112 | 7 7, 64, stride 2 |
| Conv2_x | 56 56 | 3 3 max pool, stride 2 |
|
| Conv3_x | 28 28 | |
| Conv4_x | 14 14 | |
| Conv5_x | 7 7 | |
| | 1 1 | Average pool, 1000-d FC, softmax |
Table 6.
Testing accuracy results using 30 most significant hand-crafted features.
Table 6.
Testing accuracy results using 30 most significant hand-crafted features.
Image
Categorization | General Pattern | Type Grading | Grade Grading |
|---|
| Model 1 | Model 2 |
|---|
| 60 features | 84.5% | 89% | 60.9% | 74.5% |
| PCA | 85.2% | 94% | 64.5% | 82.2% |
| ECFS | 91.1% | 95.6% | 65.8% | 77.7% |
| ILFS | 88% | 90% | 62.3% | 78.2% |
| Fisher | 87% | 93.6% | 62.5% | 76.3% |
Table 7.
Testing accuracy results using 30 most significant automatic features.
Table 7.
Testing accuracy results using 30 most significant automatic features.
| Image Categorization | General Pattern | Type Grading | Grade Grading |
|---|
| Model 1 | Model 2 |
|---|
| 1000 features | 88.3% | 93.9% | 72.2% | 83.9% |
| PCA | 72.3% | 80.7% | 60.3% | 74.0% |
| ECFS | 69.1% | 75.1% | 61.7% | 72.3% |
| ILFS | 69.5% | 79.3% | 55.2% | 70.6% |
| Fisher | 65.2% | 72.9% | 58.7% | 65.2% |
Table 8.
Testing accuracy results using 50 most significant automatic features.
Table 8.
Testing accuracy results using 50 most significant automatic features.
| Image Categorization | General Pattern | Type Grading | Grade Grading |
|---|
| Model 1 | Model 2 |
|---|
| 1000 features | 88.3% | 93.9% | 72.2% | 83.9% |
| PCA | 86.4% | 91.2% | 72.2% | 81.2% |
| ECFS | 75.9% | 86.5% | 68.4% | 80.0% |
| ILFS | 70.6% | 79.3% | 66.1% | 78.5% |
| Fisher | 74.6% | 84.2% | 65.7% | 75.6% |