Diagnostics of COVID-19 Based on CRISPR–Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update
Abstract
:1. Introduction

2. CRISPR–Cas in Diagnostics
3. Nucleic Acid Isothermal Amplification in Diagnostics
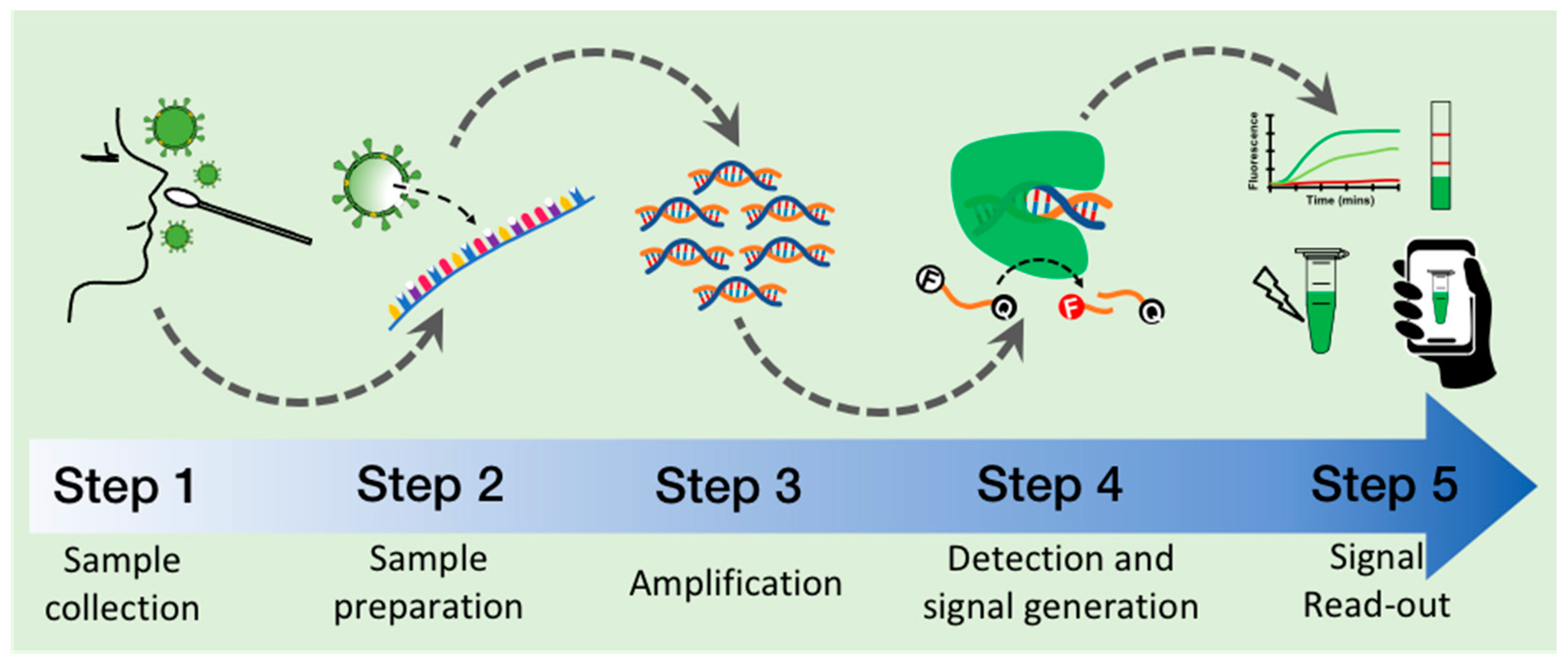
4. General Procedure to Detect SARS-CoV-2 with CRISPR–Cas
4.1. Step 1: Collection of Clinical Sample
4.2. Step 2: RNA Preparation
4.3. Step 3: Isothermal Amplification of Target Sequence
4.4. Step 4: Target Detection and Signal Generation by CRISPR–Cas
4.5. Step 5: Signal Read-Out
5. Key Experimental Parameters
5.1. Type of Sample
5.2. Method of Preparation: Extraction vs. Release of DNA/RNA
5.3. Targeted Genes
5.4. Type of Isothermal Amplification Method
5.5. Time and Temperature of Isothermal Method
5.6. CRISPR–Cas Systems
5.7. Type of Read-Out
5.8. Reporter Probe
5.9. Portable, Lyophilized, and One-Pot Versions
6. Experimental Outputs to Compare between CRISPR-Based Methods
6.1. Total Time to Deliver a Result
6.2. Limit of Detection (LoD)
| Name Method | Acronym | LoD (vc/rx) | LoD (aM) | Iso-thermal | CRISPR–Cas | Read-Out | Total Time Steps 3 and 4 (min) | One-Pot | Targeted Gene(s) | Cas Conc (nM) | gRNA Conc (nM) | RNP Ratio | Probe Conc (nM) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rapid and Sensitive Detection of SARS-CoV-2 Using CRISPR * | NA | 2 | 0.17 | RT-RPA | LbCas12a | F/LF | 60 | Not | M, N, and S | 640 | 640 | 1 | 800 | [63] |
| CRISPR-based Diagnostic for COVID-19 | CRISPR–COVID | 2.5 | 0.17 | RT-RPA | Cas13a | F | 40 | Not | Orf1ab and N | 66.7 | 33.3 | 2.00 | 166 | [25] |
| Cas12a-linked Beam Unlocking Reaction | CALIBURN | 2.5 | 0.42 | RT-RPA | LbCas12a | F | 60 | Not | Orf1ab, S, E, M, and N | NR | 100 | NR | 1250 | [77] |
| ENHanced Analysis of Nucleic acids with CrRNA Extensions | CRISPR–ENHANCE | 3 | 0.10 | RT-LAMP | LbCas12a | F/LF | 35 | Not | N | 60 | 120 | 0.5 | 500 | [102] |
| One-Pot Visual RT-LAMP–CRISPR | opvCRISPR | 5 | 0.13 | RT-LAMP | LbCas12a | F | 45 | Not | N and S | 200 | 600 | 0.33 | 2000 | [94] |
| All-In-One Dual CRISPR–Cas12a Assay | AIOD–CRISPR | 5 | 0.33 | RT-RPA | LbCas12a | F | 40 | Yes | N | 76.8 | 38.4 | 2 | 400 | [35] |
| CRISPR-powered COVID-19 Diagnosis and CRISPR-based Fluorescent Detection System | CRISPR–FDS | 5 | 0.28 | RT-RPA | LbCas12a | F | 40 | Not | N and Orf1a | 33.3 | 30 | 1.11 | 667 | [104] |
| Multiple Cross Displacement Amplification with CRISPR–Cas12a-based Detection | COVID-19 MCCD | 7 | 0.58 | RT-MCDA | LbCas12a | LF | 45 | Not | Orf1ab and N | 75 | 100 | 0.75 | 10,000 | [24] |
| CRISPR-mediated Testing in One-Pot | CRISPR–top | 10 | 0.66 | RT-LAMP | AapCas12b | F/LF | 40 | Yes | Orf1ab and N | 16 | 24 | 0.67 | 2000 | [76] |
| In vitro Specific CRISPR-based Assay for Nucleic Acids Detection | iSCAN | 10 | 0.30 | RT-LAMP | LbCas12a, AacCas12b, AapCas12b | F/LF | 60 | Yes | N and E | 250 | 250 | 1 | 500 | [89] |
| CRISPR/Cas12a-based Detection with Naked-Eye Read-Out | CRISPR/Cas12a–NER | 10 | 0.83 | RT-RAA | LbCas12a | F | 45 | Not | Orf1ab, N, and E | 70 | 1000 | 0.07 | NR | [95] |
| Synthetic Mismatch Integrated crRNA-Guided Cas12a Detection | symRNA–Cas12a | 10 | 0.83 | RT-RPA | LbCas12a | F | 45 | Not | E and S | 70 | 1000 | 0.07 | 0.025 | [105] |
6.3. Clinical Sensitivity and Specificity
| Name Method | Acronym | Sensitivity (%) | Specificity (%) | Clinical Samples (Number) | Positive Samples (Number) | Negative Samples (Number) | Type of Samples | Iso-Thermal | CRISPR–Cas | Read-Out | Total Time Steps 3 and 4 (min) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRISPR-based Diagnostic for COVID-19 | CRISPR–COVID | 100 | 100 | 114 | 61 | 53 | NP and BALF | RT-RPA | Cas13a | F | 40 | [25] |
| All-In-One Dual CRISPR–Cas12a Assay | AIOD–CRISPR | 100 | 100 | 28 | 8 | 20 | Clinical swabs | RT-RPA | LbCas12a | F | 40 | [35] |
| One-Pot Visual RT-LAMP–CRISPR | opvCRISPR | 100 | 100 | 26 | NR | NR | NP | RT-LAMP | LbCas12a | F | 45 | [94] |
| Multiple Cross Displacement Amplification with CRISPR–Cas12a-based Detection | COVID-19 MCCD | 100 | 100 | 114 | 37 | 77 | NP | RT-MCDA | LbCas12a | LF | 45 | [24] |
| In vitro Specific CRISPR-based Assay for Nucleic Acids Detection | iSCAN | 100 | 100 | 7 | 5 | 2 | NP | RT-LAMP | LbCas12a, AacCas12b, AapCas12b | F/LF | 60 | [89] |
| CRISPR/Cas12a-based Detection with Naked-Eye Read-Out | CRISPR/Cas12a–NER | 100 | 100 | 31 | 16 | 15 | NP | RT-RAA | LbCas12a | F | 45 | [95] |
| Digitization-Enhanced CRISPR/Cas-Assisted One-Pot Virusdetection | deCOViD | 100 | 100 | 4 | 2 | 2 | NP | RT-RPA | LbCas12a | F | 15 | [28] |
| Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a * | NA | 100 | 100 | 10 | 7 | 3 | NP and OP | RT-LAMP | LbCas12a | F | 45 | [109] |
| SHERLOCK | SHERLOCK | 100 | 100 | 534 | 81 | 380 | Surgery | RT-RPA | LwaCas13a | F/LF | 55 | [21] |
| SHERLOCK Testing in One Pot | STOPCovid | 100 | 100 | 17 | 12 | 5 | NP | RT-LAMP | AapCas12b | F/LF | 40 | [83] |
| Autonomous lab-on-paper platform | NA | 100 | 100 | 21 | 8 | 13 | Clinical swabs | RT-RPA | LbCas12a | F | 40 | [99] |
| Manganese-enhanced Cas12a | MeCas12a | 100 | 100 | 24 | 13 | 11 | NP and saliva | RT-RAA | LbCas12a, AsCas12a | F | 45 | [78] |
| CRISPR Optical Detection of Anisotropy | CODA | 100 | 100 | 20 | 10 | 10 | Clinical swabs | RT-RPA | LbCas12a | FA | 20 | [92] |
| CRISPR–Csm-based Detection of SARS-CoV-2 | NA | 100 | 100 | 56 | 46 | 10 | NP | RT-LAMP | Cas10 | F/C | 30 | [22] |
7. Cost and Manufacturing
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins Coronavirus Resource Center COVID-19 Map. Available online: https://coronavirus.jhu.edu/map.html (accessed on 29 March 2022).
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (accessed on 29 March 2022).
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wu, Y.; Liu, G.; Gooding, J.J. CRISPR Mediated Biosensing Toward Understanding Cellular Biology and Point-of-Care Diagnosis. Angew. Chemie Int. Ed. 2020, 59, 20754–20766. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2021. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Hoehn, S.; Hardinge, P.; Dighe, S.N.; Ukeri, J.; Pease, D.; Griffin, J.; Warrington, J.I.; Saud, Z.; Hottinger, E.; et al. VarLOCK-sequencing independent, rapid detection of SARS-CoV-2 variants of concern for point-of-care testing, qPCR pipelines and national wastewater surveillance. medRxiv 2022. [Google Scholar] [CrossRef]
- Yuanhao, L.; Hongqing, L.; Lirong, Z.; Jianhui, Z.; Baisheng, L.; Haiying, W.; Jing, L.; Jiufeng, S.; Xingfen, Y.; Xiaoling, D.; et al. CRISPR-Cas12a-Based Detection for the Major SARS-CoV-2 Variants of Concern. Microbiol. Spectr. 2022, 9, e01017-21. [Google Scholar] [CrossRef]
- Chakraborty, D.; Agrawal, A.; Maiti, S. Rapid identification and tracking of SARS-CoV-2 variants of concern. Lancet 2021, 397, 1346–1347. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Frangos, A.; Hall, L.N.; Nemudraia, A.; Nemudryi, A.; Krishna, P.; Wiegand, T.; Wilkinson, R.A.; Snyder, D.T.; Hedges, J.F.; Cicha, C.; et al. Intrinsic signal amplification by type III CRISPR-Cas systems provides a sequence-specific SARS-CoV-2 diagnostic. Cell Rep. Med. 2021, 2, 100319. [Google Scholar] [CrossRef] [PubMed]
- Marsic, T.; Ali, Z.; Tehseen, M.; Mahas, A.; Hamdan, S.; Mahfouz, M. Vigilant: An Engineered VirD2-Cas9 Complex for Lateral Flow Assay-Based Detection of SARS-CoV2. Nano Lett. 2021, 21, 3596–3603. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, X.; Li, S.; Luo, W.; Zhang, X.; Wang, C.; Chen, Q.; Yu, S.; Tai, J.; Wang, Y. Rapid, Ultrasensitive, and Highly Specific Diagnosis of COVID-19 by CRISPR-Based Detection. ACS Sensors 2021, 6, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLOS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 23–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.S.; Hsieh, K.; Chen, L.; Kaushik, A.; Trick, A.Y.; Wang, T.-H. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv. Sci. 2021, 8, 2003564. [Google Scholar] [CrossRef]
- Liu, L.; Li, X.; Ma, J.; Li, Z.; You, L.; Wang, J.; Wang, M.; Zhang, X.; Wang, Y. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 2017, 170, 714–726.e10. [Google Scholar] [CrossRef]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef] [PubMed]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef]
- Baek, Y.H.; Um, J.; Antigua, K.J.C.; Park, J.H.; Kim, Y.; Oh, S.; Kim, Y.I.; Choi, W.S.; Kim, S.O.Y.; Jeong, J.W.; et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 998–1007. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Venzor, A.; Rueda-Zarazua, B.; Marquez-Garcia, E.; Maldonado, V.; Moncada-Morales, A.; Olivera, H.; Lopez, I.; Zuñiga, J.; Melendez-Zajgla, J. SARS-CoV-2 Direct Detection Without RNA Isolation With Loop-Mediated Isothermal Amplification (LAMP) and CRISPR-Cas12. Front. Med. 2021, 8, 125. [Google Scholar] [CrossRef]
- Lu, R.F.; Wu, X.M.; Wan, Z.Z.; Li, Y.X.; Jin, X.; Zhang, C.Y. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 2826. [Google Scholar] [CrossRef] [Green Version]
- Maiti, B.; Anupama, K.P.; Rai, P.; Karunasagar, I.; Karunasagar, I. Isothermal amplification-based assays for rapid and sensitive detection of severe acute respiratory syndrome coronavirus 2: Opportunities and recent developments. Rev. Med. Virol. 2022, 32, e2274. [Google Scholar] [CrossRef]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y.; et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Gadkar, V.J.; Goldfarb, D.M.; Gantt, S.; Tilley, P.A.G. Real-time Detection and Monitoring of Loop Mediated Amplification (LAMP) Reaction Using Self-quenching and De-quenching Fluorogenic Probes. Sci. Rep. 2018, 8, 5548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Martzy, R.; Kolm, C.; Krska, R.; Mach, R.L.; Farnleitner, A.H.; Reischer, G.H. Challenges and perspectives in the application of isothermal DNA amplification methods for food and water analysis. Anal. Bioanal. Chem. 2019, 411, 1695–1702. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Newbigging, A.M.; Tao, J.; Cao, Y.; Peng, H.; Le, C.; Wu, J.; Pang, B.; Li, J.; Tyrrell, D.L.; et al. CRISPR technology incorporating amplification strategies: Molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021, 12, 4683–4698. [Google Scholar] [CrossRef] [PubMed]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Mimura, S.; et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test to Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20. [Google Scholar] [CrossRef] [PubMed]
- Thi, V.L.D.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- Rauch, J.N.; Valois, E.; Solley, S.C.; Braig, F.; Lach, R.S.; Audouard, M.; Ponce-Rojas, J.C.; Costello, M.S.; Baxter, N.J.; Kosik, K.S.; et al. A Scalable, Easy-to-Deploy Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material. J. Clin. Microbiol. 2021, 59, e02402-20. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Teng, P.; Xiao, W.; He, G.; Song, Q.; Zhang, Y.; Peng, B.; Li, G.; Hu, L.; Cao, D.; et al. Application of the amplification-free SERS-based CRISPR/Cas12a platform in the identification of SARS-CoV-2 from clinical samples. J. Nanobiotech. 2021, 19, 273. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, H.; Taguchi, Y.; Nakagawa, R.; Makino, A.; Okazaki, S.; Nakano, M.; Muramoto, Y.; Takahashi, C.; Takahashi, I.; Ando, J.; et al. Amplification-free RNA detection with CRISPR–Cas13. Commun. Biol. 2021, 4, 476. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef] [PubMed]
- Jayamohan, H.; Lambert, C.J.; Sant, H.J.; Jafek, A.; Patel, D.; Feng, H.D.; Beeman, M.; Mahmood, T.; Nze, U.; Gale, B.K. SARS-CoV-2 pandemic: A review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021, 413, 49–71. [Google Scholar] [CrossRef] [PubMed]
- Wurstle, S.; Spinner, C.D.; Voit, F.; Hoffmann, D.; Hering, S.; Weidlich, S.; Schneider, J.; Zink, A.; Treiber, M.; Iakoubov, R.; et al. Self-sampling versus health care professional-guided swab collection for SARS-CoV-2 testing. Infection 2021, 49, 927–934. [Google Scholar] [CrossRef]
- Vlek, A.L.M.; Wesselius, T.S.; Achterberg, R.; Thijsen, S.F.T. Combined throat/nasal swab sampling for SARS-CoV-2 is equivalent to nasopharyngeal sampling. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 193–195. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.P.; Osorio, J.; Videla, M.; Angel, G.; Camponovo, R.; Henríquez-Henríquez, M. Analytical and Clinical Validation for RT-qPCR Detection of SARS-CoV-2 Without RNA Extraction. Front. Med. 2020, 7, 673. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Muller, T.G.; Khalid, D.; Sonntag-Buck, V.; Heuser, A.M.; Glass, B.; Meurer, M.; Morales, I.; Schillak, A.; Freistaedter, A.; et al. SARS-CoV-2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses 2020, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Kriegova, E.; Fillerova, R.; Kvapil, P. Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour. Diagnostics 2020, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Azmi, I.; Faizan, M.I.; Kumar, R.; Raj Yadav, S.; Chaudhary, N.; Kumar Singh, D.; Butola, R.; Ganotra, A.; Datt Joshi, G.; Deep Jhingan, G.; et al. A Saliva-Based RNA Extraction-Free Workflow Integrated With Cas13a for SARS-CoV-2 Detection. Front. Cell. Infect. Microbiol. 2021, 11, 632646. [Google Scholar] [CrossRef] [PubMed]
- Ooi, K.H.; Liu, M.M.; Tay, J.W.D.; Teo, S.Y.; Kaewsapsak, P.; Jin, S.; Lee, C.K.; Hou, J.; Maurer-Stroh, S.; Lin, W.; et al. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021, 12, 1739. [Google Scholar] [CrossRef] [PubMed]
- Curti, L.A.; Primost, I.; Valla, S.; Ibañez Alegre, D.; Olguin Perglione, C.; Repizo, G.D.; Lara, J.; Parcerisa, I.; Palacios, A.; Llases, M.E.; et al. Evaluation of a Lyophilized CRISPR-Cas12 Assay for a Sensitive, Specific, and Rapid Detection of SARS-CoV-2. Viruses 2021, 13, 420. [Google Scholar] [CrossRef] [PubMed]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Tsou, J.-H.; Liu, H.; Stass, S.A.; Jiang, F. Rapid and Sensitive Detection of SARS-CoV-2 Using Clustered Regularly Interspaced Short Palindromic Repeats. Biomedicines 2021, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Lucia, C.; Federico, P.-B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hjelmsø, M.H.; Hellmér, M.; Fernandez-Cassi, X.; Timoneda, N.; Lukjancenko, O.; Seidel, M.; Elsässer, D.; Aarestrup, F.M.; Löfström, C.; Bofill-Mas, S.; et al. Evaluation of Methods for the Concentration and Extraction of Viruses from Sewage in the Context of Metagenomic Sequencing. PLoS ONE 2017, 12, e0170199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, N.; Rampazzo, R.D.C.P.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. BioMed Res. Int. 2017, 2017, 9306564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, L. Viral RNA Isolation Methods Reviewed: Spin vs. Magnetic. Available online: https://www.kbdna.com/publishinglab/viral-RNA-isolation-methods-reviewed-spin-vs-magnetic (accessed on 29 March 2022).
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, R.A.; De Puig, H.; Nguyen, P.Q.; Angenent-Mari, N.M.; Donghia, N.M.; McGee, J.P.; Dvorin, J.D.; Klapperich, C.M.; Pollock, N.R.; Collins, J.J. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. USA 2020, 117, 25722–25731. [Google Scholar] [CrossRef]
- Behrmann, O.; Bachmann, I.; Spiegel, M.; Schramm, M.; Abd El Wahed, A.; Dobler, G.; Dame, G.; Hufert, F.T. Rapid Detection of SARS-CoV-2 by Low Volume Real-Time Single Tube Reverse Transcription Recombinase Polymerase Amplification Using an Exo Probe with an Internally Linked Quencher (Exo-IQ). Clin. Chem. 2020, 66, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.Z.; Zhou, Y.; Ye, J.W.; Al-Maskri, A.A.A.; Kang, Y.; Zeng, S.; Cai, S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. [Google Scholar] [CrossRef] [PubMed]
- James, A.S.; Alawneh, J.I. COVID-19 Infection Diagnosis: Potential Impact of Isothermal Amplification Technology to Reduce Community Transmission of SARS-CoV-2. Diagnostics 2020, 10, 399. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Method of Synthesizing Single-Stranded Nucleic Acid. EP Patent 1333089A1, 16 March 2005. [Google Scholar]
- Piepenburg, O.; Williams, C.H.; Armes, N.A.; Stemple, D.L. Recombinase Polymerase Amplification. U.S. Patent 7270981B2, 18 September.
- Ding, X.; Yin, K.; Li, Z.; Sfeir, M.M.; Liu, C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens. Bioelectron. 2021, 184, 113218. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Huang, J.; Ren, L.; Jiang, W.; Wang, M.; Zhuang, L.; Zheng, Q.; Yang, R.; Zeng, Y.; Luu, L.D.W.; et al. A one-step, one-pot CRISPR nucleic acid detection platform (CRISPR-top): Application for the diagnosis of COVID-19. Talanta 2021, 233, 122591. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, D.; Lin, H.; Chen, D.; Sun, J.; Xie, Y.; Wang, X.; Ma, P.; Nie, Y.; Mei, H.; et al. Development of a Broadly Applicable Cas12a-Linked Beam Unlocking Reaction for Sensitive and Specific Detection of Respiratory Pathogens Including SARS-CoV-2. ACS Chem. Biol. 2021, 16, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Meng, Q.; Sun, B.; Zhao, B.; Dang, L.; Zhong, M.; Liu, S.; Xu, H.; Mei, H.; Liu, J.; et al. MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection. Adv. Sci. 2020, 7, 2001300. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Geng, B.; Muenker, M.C.; Moore, A.J.; Vogels, C.B.F.; et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Majam, M.; Msolomba, V.; Scott, L.; Stevens, W.; Marange, F.; Kahamba, T.; Venter, F.; Conserve, D.F. Self-Sampling for SARS-CoV-2 Diagnostic Testing by Using Nasal and Saliva Specimens: Protocol for Usability and Clinical Evaluation. JMIR Res. Protoc. 2021, 10, e24811. [Google Scholar] [CrossRef] [PubMed]
- Gertler, M.; Krause, E.; van Loon, W.; Krug, N.; Kausch, F.; Rohardt, C.; Rössig, H.; Michel, J.; Nitsche, A.; Mall, M.A.; et al. Self-collected oral, nasal and saliva samples yield sensitivity comparable to professionally collected oro-nasopharyngeal swabs in SARS-CoV-2 diagnosis among symptomatic outpatients. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 110, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Sun, X.; Wang, X.; Liang, C.; Jiang, H.; Gao, Q.; Dai, M.; Qu, B.; Fang, S.; Mao, Y.; et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.-L.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv 2020. [Google Scholar] [CrossRef]
- Azhar, M.; Phutela, R.; Kumar, M.; Ansari, A.H.; Rauthan, R.; Gulati, S.; Sharma, N.; Sinha, D.; Sharma, S.; Singh, S.; et al. Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens. Bioelectron. 2021, 183, 113207. [Google Scholar] [CrossRef] [PubMed]
- Prevention, C. Centers for Disease Control and Prevention and Real-time RT–PCR Panel for Detection 2019-nCoV (US Centers for Disease Control and Prevention, 2020). Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/covid-19-tests.html (accessed on 29 March 2022).
- World Health Organization. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RT–PCR; World Health Organization: Geneva, Switzerland, 2020.
- Kumar, M.; Gulati, S.; Ansari, A.H.; Phutela, R.; Acharya, S.; Azhar, M.; Murthy, J.; Kathpalia, P.; Kanakan, A.; Maurya, R.; et al. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. Elife 2021, 10, e67130. [Google Scholar] [CrossRef]
- Huang, W.; Yu, L.; Wen, D.; Wei, D.; Sun, Y.; Zhao, H.; Ye, Y.; Chen, W.; Zhu, Y.; Wang, L.; et al. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine 2020, 61, 103036. [Google Scholar] [CrossRef]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, K.; Takeshita, K.; Yamayoshi, S.; Shibumura, S.; Yamauchi, Y.; Yamamoto, M.; Yotsuyanagi, H.; Kawaoka, Y.; Mashimo, T. Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3. medRxiv 2020. [Google Scholar] [CrossRef]
- Lee, C.Y.; Degani, I.; Cheong, J.; Lee, J.-H.; Choi, H.-J.; Cheon, J.; Lee, H. Fluorescence polarization system for rapid COVID-19 diagnosis. Biosens. Bioelectron. 2021, 178, 113049. [Google Scholar] [CrossRef] [PubMed]
- Azhar, M.; Phutela, R.; Ansari, A.H.; Sinha, D.; Sharma, N.; Kumar, M.; Aich, M.; Sharma, S.; Rauthan, R.; Singhal, K.; et al. Rapid, field-deployable nucleobase detection and identification using FnCas9. BioRxiv 2021. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, M.; Liu, Y.; Ma, P.; Dang, L.; Meng, Q.; Wan, W.; Ma, X.; Liu, J.; Yang, G.; et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. 2020, 65, 1436–1439. [Google Scholar] [CrossRef]
- Samacoits, A.; Nimsamer, P.; Mayuramart, O.; Chantaravisoot, N.; Sitthi-amorn, P.; Nakhakes, C.; Luangkamchorn, L.; Tongcham, P.; Zahm, U.; Suphanpayak, S.; et al. Machine Learning-Driven and Smartphone-Based Fluorescence Detection for CRISPR Diagnostic of SARS-CoV-2. ACS Omega 2021, 6, 2727–2733. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, E.; Verhagen, H.J.M.P.; van de Laar, T.J.W.; Claas, E.C.J.; Cornelissen, M.; van den Akker, E. Rapid, Sensitive, and Specific Severe Acute Respiratory Syndrome Coronavirus 2 Detection: A Multicenter Comparison Between Standard Quantitative Reverse-Transcriptase Polymerase Chain Reaction and CRISPR-Based DETECTR. J. Infect. Dis. 2021, 223, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Sfeir, M.M.; Ballesteros, E.; Liu, C. Autonomous lab-on-paper for multiplexed, CRISPR-based diagnostics of SARS-CoV-2. Lab Chip 2021, 21, 2730–2737. [Google Scholar] [CrossRef] [PubMed]
- Metsky, H.C.; Freije, C.A.; Kosoko-Thoroddsen, T.-S.F.; Sabeti, P.C.; Myhrvold, C. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A.; et al. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. medRxiv 2020. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Smith, B.M.; Jain, P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020, 11, 4906. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Valois, E.; Ponce-Rojas, J.C.; Aralis, Z.; Lach, R.S.; Zappa, F.; Audouard, M.; Solley, S.C.; Vaidya, C.; Costello, M.; et al. Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Screening Using Reverse Transcriptase-Quantitative Polymerase Chain Reaction or CRISPR-Based Assays in Asymptomatic College Students. JAMA Netw. Open 2021, 4, e2037129. [Google Scholar] [CrossRef]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Meng, Q.; Wang, X.; Wang, Y.; Dang, L.; Liu, X.; Ma, X.; Chi, T.; Wang, X.; Zhao, Q.; Yang, G.; et al. Detection of the SARS-CoV-2 D614G mutation using engineered Cas12a guide RNA. Biotechnol. J. 2021, 16, 2100040. [Google Scholar] [CrossRef]
- Nimsamer, P.; Mayuramart, O.; Rattanaburi, S.; Chantaravisoot, N.; Saengchoowong, S.; Puenpa, J.; Poovorawan, Y.; Payungporn, S. Comparative performance of CRISPR-Cas12a assays for SARS-CoV-2 detection tested with RNA extracted from clinical specimens. J. Virol. Methods 2021, 290, 114092. [Google Scholar] [CrossRef]
- Pu, R.; Liu, S.; Ren, X.; Shi, D.; Ba, Y.; Huo, Y.; Zhang, W.; Ma, L.; Liu, Y.; Yang, Y.; et al. The screening value of RT-LAMP and RT-PCR in the diagnosis of COVID-19: Systematic review and meta-analysis. J. Virol. Methods 2022, 300, 114392. [Google Scholar] [CrossRef] [PubMed]
- Subali, A.D.; Wiyono, L. Reverse Transcriptase Loop Mediated Isothermal Amplification (RT-LAMP) for COVID-19 diagnosis: A systematic review and meta-analysis. Pathog. Glob. Health 2021, 115, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef] [PubMed]
| Key Parameter (Condition/Component) | Options | |
|---|---|---|
| Step 1 | Type of sample | Nasopharyngeal and Oropharyngeal swabs, viral RNA, saliva, sputum, other |
| Step 2 | Type and time of method of preparation | RNA extraction (5–40 min) and RNA release (5–30 min) |
| Step 3 | Targeted Genes | Genes N, Orf1ab, S, E, other |
| Type of Isothermal Amplification Method | RT-RPA, RT-LAMP, other | |
| Temperature | 59–65 °C (RT-LAMP) and 37–42 °C (RT-RPA) | |
| Time | 20 to 40 min (RT-LAMP) and 15 to 30 min (RT-RPA) | |
| Step 4 | Type CRISPR–Cas system | Cas12a, Cas13a, Cas12b, Cas9, Cas10, Cas3, other |
| Cas protein concentration | 2.2–1000 nM | |
| gRNA concentration | 20–1000 nM | |
| RNP ratio (Cas/gRNA) | 0.07–17.5 | |
| Temperature | 25–70 °C | |
| Time | 1–90 min | |
| Step 5 | Type of Read-Out | Fluorescence, Lateral Flow, Fluorescence Anisotropy, Electrophoresis in gel, other |
| Probe Type of Sequence Length Type of fluorophore Type of quencher | Thymine rich, Adenine/Thymine rich, Uracil-rich, other 5–16 nt FAM, HEX, and Alexa Iowa Black and Black Hole | |
| Format | Portable vs. Lab-based Lyophilized vs Solution-based | |
| Two-step vs. One-pot |
| Name Method | Acronym | Total Time Steps 3 and 4 (min) | Isothermal | Isothermal Time (min) | CRISPR–Cas | CRISPR–Cas Time (min) | Read-Out | One-Pot * | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Digitization-Enhanced CRISPR/Cas-Assisted One-Pot Virus detection | deCOViD | 15 | RT-RPA | * | LbCas12a | * | F | Yes | [28] |
| CRISPR Optical Detection of Anisotropy | CODA | 20 | RT-RPA | * | LbCas12a | * | FA | Yes | [92] |
| Isotachophoresis-mediated CRISPR–Cas12 DNA Detection | ITP–CRISPR assay | 25 | RT-LAMP | 20 | LbCas12a | 5 | F | Not | [98] |
| Variant Nucleotide Guard | VaNGuard | 27 | RT-LAMP | 22 | LbCas12a/AsCas12a/enAsCas12a | 5 | F/LF | Not | [60] |
| CRISPR–Csm-based Detection of SARS-CoV-2 | NA | 30 | RT-LAMP | 29 | Cas10 | 1 | F/C | Not | [22] |
| SHERLOCK and HUDSON Integration to Navigate Epidemics | SHINE | 30 | RT-RPA | * | LwaCas13a | * | F/LF | Yes | [62] |
| DNA Endonuclease Targeted CRISPR Trans Reporter | DETECTR ** | 30 | RT-LAMP | 20 | LbCas12a | 10 | F/LF | Not | [101] |
| DNA Endonuclease Targeted CRISPR Trans Reporter | DETECTR ** | 35 | RT-LAMP | 20 | LbCas12a | 15 | F/LF | Not | [97] |
| ENHanced Analysis of Nucleic acids with CrRNA Extensions | CRISPR–ENHANCE | 35 | RT-LAMP | 20 | LbCas12a | 15 | F/LF | Not | [102] |
| VirD2–dCas9 Guided and LFA-coupled Nucleic Acid Test | VIGILANT | 36 | RT-RPA | 25 | SpCas9 | 11 | LF | Not | [23] |
| Name Method | Acronym | Isothermal | CRISPR–Cas | Read-Out | Total Time Steps 3 and 4 (min) | Portable * | Reference |
|---|---|---|---|---|---|---|---|
| Digitization-Enhanced CRISPR/Cas-Assisted One-Pot Virus detection | deCOViD | RT-RPA | LbCas12a | F | 15 | Yes | [28] |
| CRISPR Optical Detection of Anisotropy | CODA | RT-RPA | LbCas12a | FA | 20 | Yes | [92] |
| SHERLOCK and HUDSON Integration to Navigate Epidemics | SHINE | RT-RPA | LwaCas13a | F/LF | 30 | Not | [62] |
| All-In-One Dual CRISPR–Cas12a Assay | AIOD–CRISPR | RT-RPA | LbCas12a | F | 40 | Yes | [35] |
| SHERLOCK Testing in One Pot | STOPCovid | RT-LAMP | AapCas12b | F/LF | 40 | Yes | [83] |
| CRISPR-mediated Testing in One Pot | CRISPR–top | RT-LAMP | AapCas12b | F/LF | 40 | Yes | [76] |
| SHERLOCK Testing in One Pot | STOPCovid.v2 | RT-LAMP | AapCas12b | F/LF | 45 | Yes | [90] |
| In vitro Specific CRISPR-based Assay for Nucleic Acids Detection | iSCAN | RT-LAMP | LbCas12a, AacCas12b, AapCas12b | F/LF | 60 | Yes | [89] |
| Digital Warm-Start CRISPR Assay | dWS–CRISPR | RT-DAMP | AsCas12a | F | 90 | Not | [75] |
| Method | Acronym | STEP 1 | STEP 2 | STEP 3 | STEP 4 | STEP 5 | Experimental Outputs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Sample Preparation | Isothermal | Temp (°C) | Time (min) | CRISPR–Cas | Temp (°C) | Time (min) | Read-Out | One-Pot | Portable? | Total Time (Steps 3 and 4) (min) ** | LoD (c/r) | LoD (aM) | Specificity (%) | Sensitivity (%) | Reference | ||
| Digitization-Enhanced CRISPR/Cas-Assisted One-Pot Virus detection | deCOViD | NP | R | RT-RPA | 42 | 15 | LbCas12a | RT | 15 | F | Yes | Portable | 15 | 15 | 10 | 100 | 100 | [28] |
| CRISPR-based Diagnostic for COVID-19 | CRISPR–COVID | NP and BALF | E | RT-RPA | 42 | 30 | Cas13a | 42 | 10 | F | Not | Lab | 40 | 2.5 | 1 | 100 | 100 | [25] |
| All-In-One Dual CRISPR–Cas12a Assay | AIOD–CRISPR | C | E | RT-RPA | 37 | 40 | LbCas12a | 37 | 40 | F | Yes | Portable | 40 | 5 | 2 | 100 | 100 | [35] |
| One-Pot Visual RT-LAMP-CRISPR | opvCRISPR | NP | NR | RT-LAMP | 65 | 40 | LbCas12a | 37 | 5 | F | Not | Lab | 45 | 5 | 0.8 | 100 | 100 | [94] |
| Multiple Cross Displacement Amplification with CRISPR–Cas12a-based Detection | COVID-19 MCCD | NP | NR | RT-MCDA | 63 | 35 | LbCas12a | 37 | 5 | LF | Not | Portable | 45 | 7 | 3.5 | 100 | 100 | [24] |
| CRISPR/Cas12a-based Detection with Naked-Eye Read-Out | CRISPR/Cas12a–NER | NP | E | RT-RAA | 39 | 30 | LbCas12a | 37 | 15 | F | Not | Portable | 45 | 10 | 5 | 100 | 100 | [95] |
| CRISPR Optical Detection of Anisotropy | CODA | C | E | RT-RPA | 42 | 20 | LbCas12a | 42 | 20 | FA | Yes | Portable | 20 | 150 | 30 | 100 | 100 | [92] |
| Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a * | NA | C | NR | RT-LAMP | 65 | 40 | LbCas12a | 37 | 5 | F | Not | Portable | 45 | 20 | 4.5 | 100 | 100 | [109] |
| In vitro Specific CRISPR-based Assay for Nucleic Acids Detection | iSCAN | NP and OP | E | RT-LAMP | 62 | 60 | LbCas12a, AacCas12b, AapCas12b | 62 | 60 | F/LF | Yes | Portable | 60 | 10 | 1.8 | 100 | 100 | [89] |
| Autonomous lab-on-paper platform * | NA | C | E | RT-RPA | 37 | 15 | LbCas12a | 37 | 25 | F | Not | Portable | 40 | 100 | 40 | 100 | 100 | [99] |
| SHERLOCK Testing in One Pot | STOPCovid | NP | R | RT-LAMP | 60 | 40 | AapCas12b | 70 | 40 | F/LF | Yes | Portable | 40 | 100 | 20 | 100 | 100 | [83] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez-Garcia, A.; Morales-Moreno, M.D.; Valdés-Galindo, E.G.; Jimenez-Nieto, E.P.; Quezada, A. Diagnostics of COVID-19 Based on CRISPR–Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update. Diagnostics 2022, 12, 1434. https://doi.org/10.3390/diagnostics12061434
Hernandez-Garcia A, Morales-Moreno MD, Valdés-Galindo EG, Jimenez-Nieto EP, Quezada A. Diagnostics of COVID-19 Based on CRISPR–Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update. Diagnostics. 2022; 12(6):1434. https://doi.org/10.3390/diagnostics12061434
Chicago/Turabian StyleHernandez-Garcia, Armando, Melissa D. Morales-Moreno, Erick G. Valdés-Galindo, Eric P. Jimenez-Nieto, and Andrea Quezada. 2022. "Diagnostics of COVID-19 Based on CRISPR–Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update" Diagnostics 12, no. 6: 1434. https://doi.org/10.3390/diagnostics12061434
APA StyleHernandez-Garcia, A., Morales-Moreno, M. D., Valdés-Galindo, E. G., Jimenez-Nieto, E. P., & Quezada, A. (2022). Diagnostics of COVID-19 Based on CRISPR–Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update. Diagnostics, 12(6), 1434. https://doi.org/10.3390/diagnostics12061434







