Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions
Abstract
:1. Introduction
2. Materials and Methods
- -
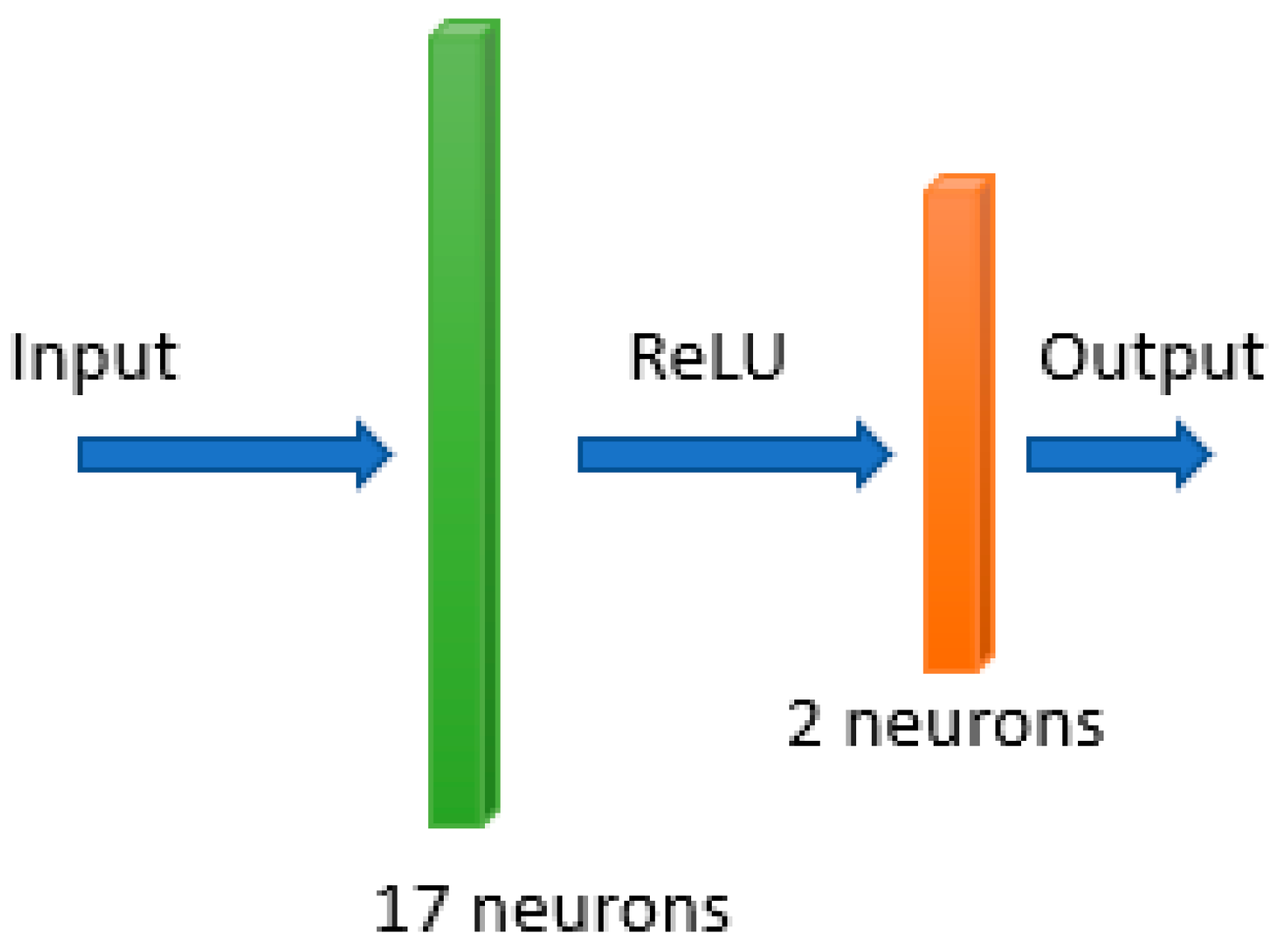
- The input layer receives the input variables.
- -
- The hidden layer is the collection of neurons with activation functions. It is the layer responsible for the extraction of the features from the input data.
- -
- The output layer produces the result for given inputs.
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ECIS—European Cancer Information System. Available online: https://ecis.jrc.ec.europa.eu (accessed on 4 October 2021).
- Scandurra, C.; Muzii, B.; La Rocca, R.; Di Bello, F.; Bottone, M.; Califano, G.; Longo, N.; Maldonato, N.M.; Mangiapia, F. Social Support Mediates the Relationship between Body Image Distress and Depressive Symptoms in Prostate Cancer Patients. Int. J. Environ. Res. Public Health 2022, 19, 4825. [Google Scholar] [CrossRef]
- Capece, M.; Creta, M.; Calogero, A.; La Rocca, R.; Napolitano, L.; Barone, B.; Sica, A.; Fusco, F.; Santangelo, M.; Dodaro, C.; et al. Does Physical Activity Regulate Prostate Carcinogenesis and Prostate Cancer Outcomes? A Narrative Review. Int. J. Environ. Res. Public Health 2020, 17, 1441. [Google Scholar] [CrossRef] [Green Version]
- Schoentgen, N.; Califano, G.; Manfredi, C.; Romero-Otero, J.; Chun, F.K.H.; Ouzaid, I.; Hermieu, J.-F.; Xylinas, E.; Verze, P. Is it Worth Starting Sexual Rehabilitation Before Radical Prostatectomy? Results From a Systematic Review of the Literature. Front. Surg. 2021, 8, 648345. [Google Scholar] [CrossRef]
- Scandurra, C.; Mangiapia, F.; La Rocca, R.; Di Bello, F.; De Lucia, N.; Muzii, B.; Cantone, M.; Zampi, R.; Califano, G.; Maldonato, N.M.; et al. A cross-sectional study on demoralization in prostate cancer patients: The role of masculine self-esteem, depression, and resilience. Support. Care Cancer 2022, 30, 7021–7030. [Google Scholar] [CrossRef]
- Van Poppel, H.; Hogenhout, R.; Albers, P.; van den Bergh, R.C.N.; Barentsz, J.O.; Roobol, M.J. Early Detection of Prostate Cancer in 2020 and Beyond: Facts and Recommendations for the European Union and the European Commission. Eur. Urol. 2021, 79, 327–329. [Google Scholar] [CrossRef]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Fütterer, J.J.; Briganti, A.; De Visschere, P.; Emberton, M.; Giannarini, G.; Kirkham, A.; Taneja, S.S.; Thoeny, H.; Villeirs, G.; Villers, A. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur. Urol. 2015, 68, 1045–1053. [Google Scholar] [CrossRef]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef]
- Drost, F.-J.H.; Osses, D.; Nieboer, D.; Bangma, C.H.; Steyerberg, E.W.; Roobol, M.J.; Schoots, I.G. Prostate Magnetic Resonance Imaging, with or Without Magnetic Resonance Imaging-targeted Biopsy, and Systematic Biopsy for Detecting Prostate Cancer: A Cochrane Systematic Review and Meta-analysis. Eur. Urol. 2020, 77, 78–94. [Google Scholar] [CrossRef]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- de Rooij, M.; Israël, B.; Tummers, M.; Ahmed, H.U.; Barrett, T.; Giganti, F. ESUR/ESUI consensus statements on multi-parametric MRI for the detection of clinically significant prostate cancer: Quality requirements for image acquisition, interpretation and radiologists’ training. Eur. Radiol. 2020, 30, 5404–5416. [Google Scholar] [CrossRef]
- Stabile, A.; Giganti, F.; Kasivisvanathan, V.; Giannarini, G.; Moore, C.M.; Padhani, A.; Panebianco, V.; Rosenkrantz, A.; Salomon, G.; Turkbey, B.; et al. Factors Influencing Variability in the Performance of Multiparametric Magnetic Resonance Imaging in Detecting Clinically Significant Prostate Cancer: A Systematic Literature Review. Eur. Urol. Oncol. 2020, 3, 145–167. [Google Scholar] [CrossRef]
- Castillo, T.J.M.; Arif, M.; Niessen, W.J.; Schoots, I.G.; Veenland, J.F. Automated Classification of Significant Prostate Cancer on MRI: A Systematic Review on the Performance of Machine Learning Applications. Cancers 2020, 12, 1606. [Google Scholar] [CrossRef]
- Greer, M.D.; Lay, N.; Shih, J.H.; Barrett, T.; Bittencourt, L.K.; Borofsky, S.; Kabakus, I.; Law, Y.M.; Marko, J.; Shebel, H.; et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: An international multi-reader study. Eur. Radiol. 2018, 28, 4407–4417. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Romeo, V.; Green, R.; Cantoni, V.; Ponsiglione, A.; Ugga, L.; Imbriaco, M. Machine learning for the identification of clinically significant prostate cancer on MRI: A meta-analysis. Eur. Radiol. 2020, 30, 6877–6887. [Google Scholar] [CrossRef]
- Ferro, M.; Crocetto, F.; Bruzzese, D.; Imbriaco, M.; Fusco, F.; Longo, N.; Napolitano, L.; La Civita, E.; Cennamo, M.; Liotti, A.; et al. Prostate Health Index and Multiparametric MRI: Partners in Crime Fighting Overdiagnosis and Overtreatment in Prostate Cancer. Cancers 2021, 13, 4723. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forest. In Machine Learning; Robert, E., Ed.; Schapire: Berlin/Heidelberg, Germany, 2001; Volume 45, pp. 5–32. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, 1–8 June 2008; IEEE: Piscataway, NJ, USA, 2008; pp. 1322–1328. Available online: http://ieeexplore.ieee.org/document/4633969/ (accessed on 22 June 2022).
- van der Leest, M.; Cornel, E.; Israël, B.; Hendriks, R.; Padhani, A.R.; Hoogenboom, M. Head-to-head Comparison of Transrectal Ultrasound-guided Prostate Biopsy Versus Multiparametric Prostate Resonance Imaging with Subsequent Magnetic Resonance-guided Biopsy in Biopsy-naïve Men with Elevated Prostate-specific Antigen: A Large Prospective Multicenter Clinical Study. Eur. Urol. 2019, 75, 570–578. [Google Scholar]
- Venderink, W.; van Luijtelaar, A.; Bomers, J.G.R.; van der Leest, M.; Hulsbergen-van de Kaa, C.; Barentsz, J.O. Results of Targeted Biopsy in Men with Magnetic Resonance Imaging Lesions Classified Equivocal, Likely or Highly Likely to Be Clinically Significant Prostate Cancer. Eur. Urol. 2018, 73, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Hansen, N.; Koo, B.; Warren, A.; Kastner, C.; Barrett, T. Sub-differentiating equivocal PI-RADS-3 lesions in multiparametric magnetic resonance imaging of the prostate to improve cancer detection. Eur. J. Radiol. 2017, 95, 307–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermie, I.; Van Besien, J.; De Visschere, P.; Lumen, N.; Decaestecker, K. Which clinical and radiological characteristics can predict clinically significant prostate cancer in PI-RADS 3 lesions? A retrospective study in a high-volume academic center. Eur. J. Radiol. 2019, 114, 92–98. [Google Scholar] [CrossRef] [PubMed]


| Age (years) | Median | 67 |
| IQR | 58–79 | |
| BMI | Median | 26.8 |
| IQR | 18.2–34.9 | |
| Prostate volume, gr. | Median | 48 |
| IQR | 19–138 | |
| PSA, ng/mL | Median | 6.2 |
| IQR | 0.24–15.43 | |
| PSA density | Median | 0.13 |
| IQR | 0.01–0.8 | |
| Serum Glucose, mg/dL | Median | 95 |
| IQR | 73–196 | |
| Serum Creatinine, mg/dL | Median | 1.03 |
| IQR | 0.79–1.84 | |
| Gleason Score 6 (3 + 3) | N. of patients | 18 |
| Gleason Score 7 (3 + 4) | N. of patients | 25 |
| Gleason Score 7 (4 + 3) | N. of patients | 17 |
| Gleason Score 8 (4 + 4) | N. of patients | 6 |
| Gleason Score 9 (4 + 5) | N. of patients | 3 |
| Method | ACC | SPE | SENS | F1 | AUC |
|---|---|---|---|---|---|
| RF | 77.98% | 71.05% | 81.69% | 82.86% | 83.32% |
| NN | 70.53% | 53.33% | 78.46% | 78.46% | 74.51% |
| Ctree | 74.31% | 73.68% | 74.65% | 79.10% | 74.30% |
| SVM | 72.48% | 73.68% | 71.83% | 77.27% | 72.76% |
| Method | Selected Features |
|---|---|
| RF | BMI-equator-apex-TOT_ZONE-PSA density-ratio-Blood glucose-HDL-Triglycerides-Creatinine - |
| Ctree | TOT_ZONE-prostate volume-Blood glucose-HDL-Triglycerides- |
| NN | BMI-base-equator-apex-transitional-TOT_ZONE-prostate volume-PSA-psa density-Free PSA-ratio-Blood glucose-Total Cholesterol-HDL–LDL-Triglycerides-Creatinine- |
| SVM | BMI-base-TOT_ZONE-PSA-psa density-ratio-Blood glucose-Triglycerides-Creatinine- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gravina, M.; Spirito, L.; Celentano, G.; Capece, M.; Creta, M.; Califano, G.; Collà Ruvolo, C.; Morra, S.; Imbriaco, M.; Di Bello, F.; et al. Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions. Diagnostics 2022, 12, 1565. https://doi.org/10.3390/diagnostics12071565
Gravina M, Spirito L, Celentano G, Capece M, Creta M, Califano G, Collà Ruvolo C, Morra S, Imbriaco M, Di Bello F, et al. Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions. Diagnostics. 2022; 12(7):1565. https://doi.org/10.3390/diagnostics12071565
Chicago/Turabian StyleGravina, Michela, Lorenzo Spirito, Giuseppe Celentano, Marco Capece, Massimiliano Creta, Gianluigi Califano, Claudia Collà Ruvolo, Simone Morra, Massimo Imbriaco, Francesco Di Bello, and et al. 2022. "Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions" Diagnostics 12, no. 7: 1565. https://doi.org/10.3390/diagnostics12071565
APA StyleGravina, M., Spirito, L., Celentano, G., Capece, M., Creta, M., Califano, G., Collà Ruvolo, C., Morra, S., Imbriaco, M., Di Bello, F., Sciuto, A., Cuocolo, R., Napolitano, L., La Rocca, R., Mirone, V., Sansone, C., & Longo, N. (2022). Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions. Diagnostics, 12(7), 1565. https://doi.org/10.3390/diagnostics12071565













