Abstract
Myelodysplastic syndromes (MDS) are considered to be diseases associated with splicing defects. A large number of genes involved in the pre-messenger RNA splicing process are mutated in MDS. Deletion of 5q and 7q are of diagnostic value, and those chromosome regions bear the numbers of splicing genes potentially deleted in del(5q) and del(7q)/-7 MDS. In this review, we present the splicing genes already known or suspected to be implicated in MDS pathogenesis. First, we focus on the splicing genes located on chromosome 5 (HNRNPA0, RBM27, RBM22, SLU7, DDX41), chromosome 7 (LUC7L2), and on the SF3B1 gene since both chromosome aberrations and the SF3B1 mutation are the only genetic abnormalities in splicing genes with clear diagnostic values. Then, we present and discuss other splicing genes that are showing a prognostic interest (SRSF2, U2AF1, ZRSR2, U2AF2, and PRPF8). Finally, we discuss the haploinsufficiency of splicing genes, especially from chromosomes 5 and 7, the important amplifier process of splicing defects, and the cumulative and synergistic effect of splicing genes defects in the MDS pathogenesis. At the time, when many authors suggest including the sequencing of some splicing genes to improve the diagnosis and the prognosis of MDS, a better understanding of these cooperative defects is needed.
1. Introduction
Myelodysplastic syndromes (myelodysplasia or MDS) are clonal and acquired myeloid hematologic malignancies characterized by inefficient hematopoiesis. The risk of progression from MDS to acute myeloid leukemia (AML) varies from 15 to 30%, depending on the presence of chromosomal abnormalities and gene mutations. Indeed, MDS result mainly from the accumulation of somatic genetic abnormalities and, in particular, from cytogenetic aberrations in MDS that are clonal and recurrent. They are found in about 50% of de novo MDS and in more than 80% of secondary (therapy-related) MDS. They mostly represent unbalanced abnormalities with the loss of chromosomal material, and translocations are rare. Importantly, recurrent cytogenetic aberrations and their number (defining or not a complex karyotype) in MDS are of diagnostic value [1,2,3]. In cases of cytopenias, recurrent chromosome aberrations can be predictive of MDS even in the absence of morphological MDS characteristics [1]. However, so far, in the WHO reference classification of MDS, only one genetic aberration is characteristic of a specific MDS entity: it consists of the partial deletion of chromosome 5 (del(5q)). Deletion of chromosome 7 is also considered in the WHO MDS classification [2] since its presence precludes it from the ‘MDS with isolated del(5q)’ subtype. Additionally, those recurrent cytogenetic abnormalities are strong independent prognostic markers [4,5]. Chromosomes 5 and 7 bear important genes implicated in the splicing of pre-messenger RNA; those genes could be driver genes involved in the emergence or maintenance of MDS. Moreover, mutations in the gene, encoding the splicing factor SF3B1, present a strong diagnostic value when associated with ring sideroblasts. More rarely, MDS can emerge from a constitutive aberration: in particular, in the case of DDX41 mutations, which is a gene that also encodes a splicing factor. Altogether, MDS can be considered ‘splicing diseases’. Highly recurrent mutations in genes, encoding splicing factors, are reported [6,7,8], with some having diagnostic or prognostic interests.
The goal of this review is to present the splicing genes that are already known, or suspected to be implicated, in the pathogenesis of MDS. First, we focus on the splicing genes located on chromosomes 5, 7, and on SF3B1 gene. Indeed, del(5q), del(7q)/-7, and SF3B1 mutations are the only genetic abnormalities in splicing genes with clear diagnostic values. Then, we present and discuss most of the other splicing genes that are showing a prognostic interest.
2. Chromosome 5 and Its Genes Implicated in Splicing
2.1. The Diagnostic Value of Chromosome 5 Deletion (del(5q) MDS)
In 1974, Van den Berghe et al. described the first recurrent chromosomal abnormality associated with MDS: the deletion of the long arm of chromosome 5 [9]. ‘MDS with isolated del(5q)’ was then included in the WHO reference MDS classification, as a distinct subtype, in 2001. In the last WHO 2016 MDS classification [2], the ‘MDS with isolated del(5q)’ is defined as the entity with: 1–3 dysplastic lineages, 1–2 cytopenias, none or any ring sideroblasts, less than 5% and 1% of blasts in the bone marrow and the peripheral blood, respectively, and importantly, with an isolated del(5q) or with one additional abnormality, except -7 or del(7q).
The deletion of 5q is the most frequent chromosomal abnormality reported in MDS (15%) [10]. It is of good prognosis when found alone or associated with another aberration (65% of the cases of del(5q)), and it is of poor prognosis when associated with two or more aberrations (complex karyotype) (33% of the cases of del(5q)) or in the presence of TP53 mutations [3,4,10,11].
The 5q- syndrome is a distinct entity of MDS, with the sole 5q deletion and specific clinical and biological features (macrocytosis, anemia, normal or high platelet counts and hypolobulated megakaryocytes in the bone marrow, as well as peripheral and medullary blast counts inferior to 5%). Prognosis of patients with 5q- syndrome is excellent [12]. The 5q- syndrome is rare, according to Holtan et al. [13]. In 2022, with the evolution of classifications, the use of the term “5q- syndrome” is somehow confusing and potentially outdated.
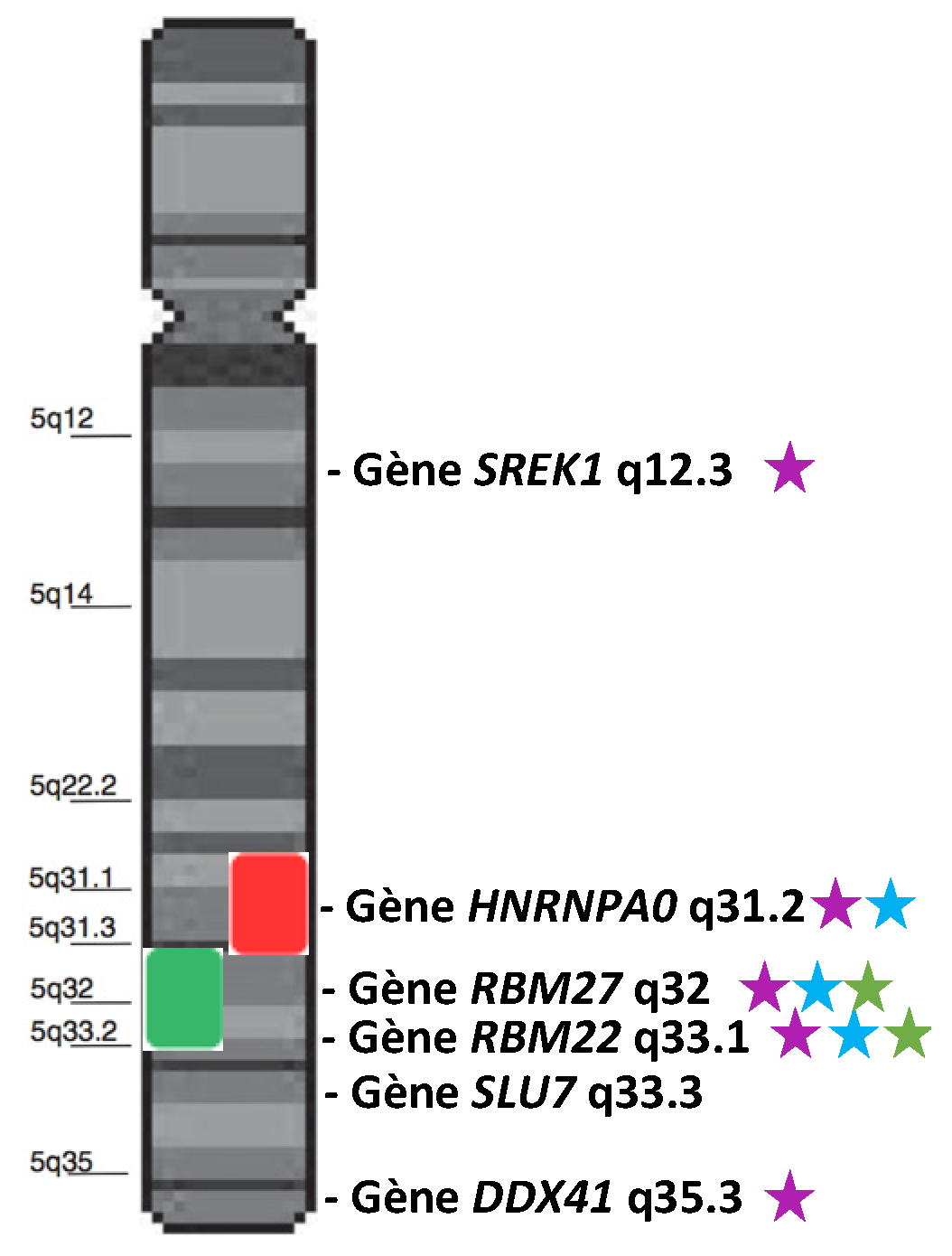
The borders of the commonly deleted region (CDR) of del(5q) slightly differ from one cohort to another [14,15,16,17]. Globally, it encompasses a large region from 5q14.1 to 5q35.1 [17]. Within this CDR, the 5q31.2–5q31.3 sub-region is more associated with higher-risk MDS, while the 5q32–5q33.2 sub-region is characteristic of 5q- syndrome. Several splicing genes are present in the 5q (Figure 1). We will focus on HNRNPA0, RBM27, RBM22, SLU7, and DDX41 because some studies suggest/demonstrate their possible role in the cell cycle, proliferation, hematopoiesis, or in the pathogenesis of MDS.
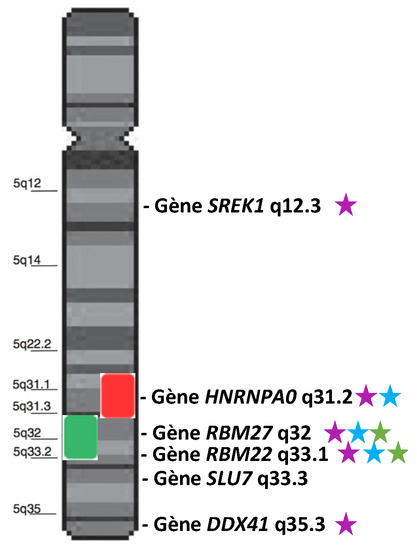
Figure 1.
Schematic representation of chromosome 5 and two characteristic CDRs: the 5q- CDR (green box) and the higher-risk CDR (red box). Genes annotated as “splicing genes” are represented in front of their locus. Purple stars indicate the genes involved/mutated/dysregulated in MDS or hematopoiesis. Blue stars indicate the genes located in the most frequently deleted region. Green stars indicate the most dysregulated genes in the 5q- syndrome.
2.2. Splicing Genes Located in the Commonly Deleted Region of the Deletion of 5q
2.2.1. HNRNPA0 (Heterogeneous Nuclear Ribonucleoprotein A0)
HNRNPA0 is located on chromosome 5, at the locus 5q31.2 in the human genome, and is expressed at reduced levels in CD34+ cells from patients with a del(5q) [18]. Its role is still poorly understood. This intronless gene encodes a minor heterogeneous nuclear ribonucleoprotein (hnRNP) bearing two RNA binding domains and a glycine-rich C-terminus [19]. It binds adenine and uracil (AU)-rich elements (ARE) of nuclear pre-messenger RNAs and stabilizes them. HNRNPA0 is involved in the control of the cell cycle, where it plays a role in DNA damage checkpoints, in particular through the post-transcriptional stabilization of p27(Kip1) and Gadd45α mRNAs [20]. HNRNPA0 is also implicated in the translation control system [21], in mRNA transport, and in nucleic acid metabolism [19].
HNRNPA0 is considered a tumor suppressor gene due to its overexpression in cancer. In humans, HNRNPA0 is expressed in the hematopoietic tissue in lymphoid and myeloid lineages. In mice, depletion of Hnrnpa0 alters the expression of myeloid specification genes and shifts the hematopoietic cell differentiation from monocytes to granulocytes [22]. In del(5q) MDS patients, different authors suggested a cooperative effect of HNRNPA0 and EGR1 double haploinsufficiency on the dysregulation of the myeloid differentiation [22,23]. Both genes, localized in the CDR of del(5q), are expressed at haploinsufficient levels in del(5q) MDS patients [18].
2.2.2. RBM27 (RNA-Binding Motif 27)
RNA-Binding Motif 27 (RBM27) is an RBM gene located on chromosome 5, at the locus 5q32. It is quite ubiquitously expressed in humans. RBM27 encodes a protein containing an RNA-Binding Motif, which confers its ability to bind RNA. However, RBM27 is still very poorly studied: in particular, in its role in pre-mRNA splicing. RBM27 depletion by RNA interference causes an upregulation of a few alternative splicing events in the HepG2 cell line, such as tandem UTRs, exon skipping, and mutually exclusive exons [24]. The only described function of RBM27 is to be necessary for the Poly(A) Tail eXosome Targeting (PAXT) connection [25].
RBM27, which is located in the CDR of chromosome 5, is very frequently deleted and was identified as one of the most dysregulated genes in CD34+ cells of patients with the 5q- syndrome [14]. Nevertheless, its role in the pathogenesis of MDS remains unknown.
2.2.3. RBM22 (RNA-Binding Motif 22)
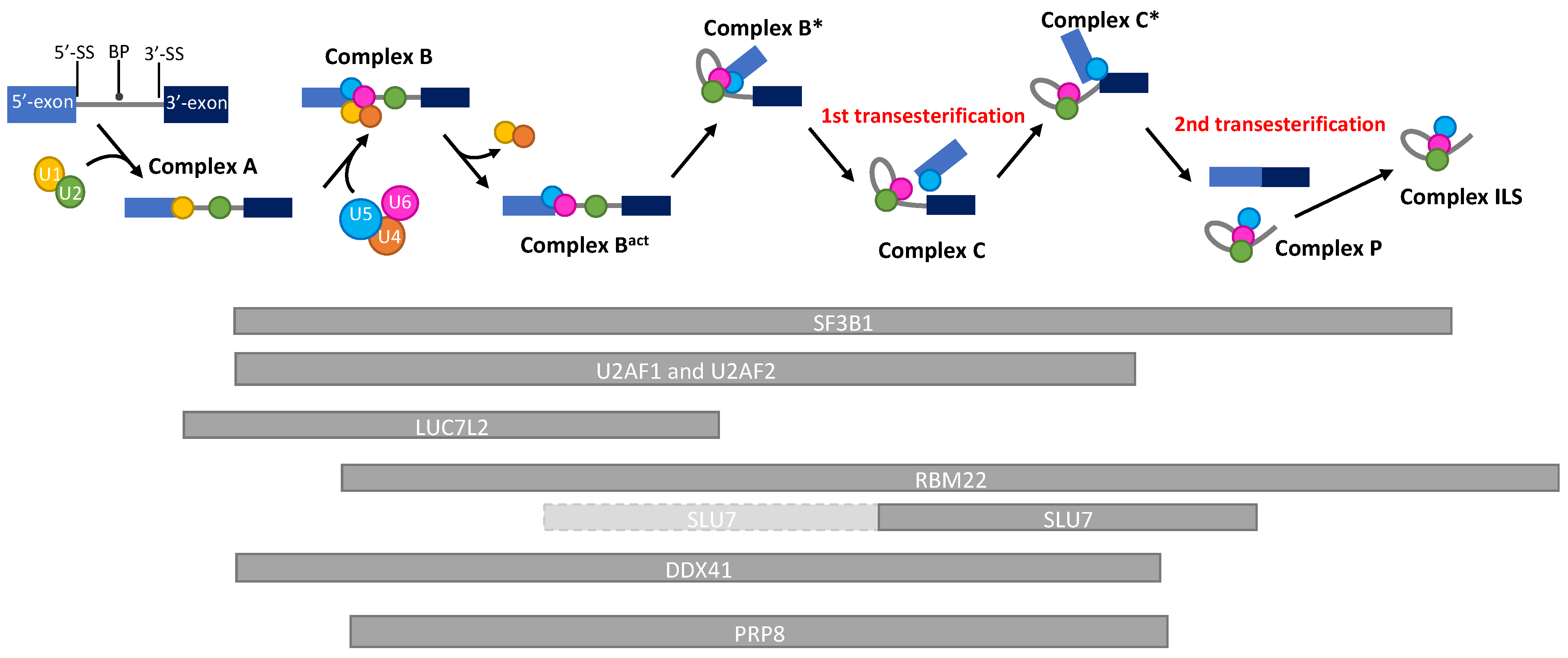
RBM22 is located on chromosome 5 and at the locus 5q33.1 in humans. As RBM27, it is lost in almost all del(5q) MDS since it is localized in the del(5q) CDR (Figure 1). Belonging to the RNA-Binding Motif family of genes, RBM22 encodes a protein containing an RNA-Recognition Motif (RRM) and a Zinc–Finger motif that allow it to bind RNA [26,27]. Its main role is to stabilize the catalytic core of the spliceosome (Figure 2). Indeed, RBM22 binds the RNA-maturing complex before the first transesterification (i.e., the separation of the intron from the 5′-exon, also called branching reaction) during the remodeling of the complex by Brr2 [28]. Mainly involved in the first step of splicing, RBM22 will tether several components together: contacting the ACAGA box and the Internal Stem Loop (ISL) of U6-snRNA, a few nucleotides of the intron downstream of the 5′-splice site and, most likely, as well as a few nucleotides of U2-snRNA [27,29,30]. It also interacts with other proteins, such as PRP19, Aquarius, and PRP8, which contribute to stabilizing the conformation of the catalytic core [29,31,32]. Of note; PRPF8 (encoding PRP8) is also a gene mutated in MDS (see Section 5.2 below). Although its role is not clearly identified in the following steps of splicing, RBM22 stays in the complex during the whole process and leaves it after the intron release in the ILS complex [33,34]. Its presence in the spliceosome is essential, as its depletion leads to the failure of the branching reaction in yeast and several alternative splicing events in yeast, drosophila, or human cell lines [24,26,35]. Interestingly, RBM22 could potentially be involved in the calcium-dependent regulation of splicing, as it interacts with the calcium-binding protein ALG-2, and it is involved in the translocation of the latter and of another splicing factor (SLU7) from the cytoplasm to the nucleus and vice versa [36,37].
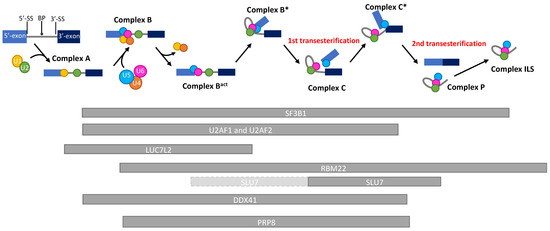
Figure 2.
Schematic representation of the kinetics of the presence of splicing factors during a pre-mRNA splicing reaction. RNA splicing is a process by which introns are removed from pre-mRNAs, and exons are ligated to produce a mature mRNA. By the action of more than 200 proteins and 6 ribonucleoprotein complexes (U1-U6), the intron will be separated from the 5′-exon (1st transesterification) and, subsequently, from the 3′-exon (2nd transesterification), allowing the ligation of both exons. This reaction depends on the recognition of consensus splicing sequences (5′-Splice site, Branch point, 3′-Splice site) by the spliceosome. The 5′ and 3′ exons are represented with light and dark blue boxes, respectively. The U1, U2, U4, U5, and U6-snRNA are, respectively, represented with yellow, green, orange, blue, and pink circles. The kinetics of presence of the proteins discussed in this review are represented by grey bars below the reaction timeline. Light-grey bars with a dashed border line represent a low affinity presence. BP: Branch Point; 5′-SS: 5′-Splice site; 3′-SS: 3′-Splice site.
Furthermore, it was recently shown that RBM22 is also able to interact with DNA and act as a transcription factor, controlling the expression of about 3000 genes [38].
RBM22 is a very conserved gene that is ubiquitously expressed in humans but slightly more in the bone marrow [39]. Though its exact role in hematopoiesis is still unclear, RBM22 is undeniably involved in that process and linked to the occurrence of MDS. Indeed, located in the del(5q) commonly deleted region (CDR) of chromosome 5, RBM22 is one of the most underexpressed genes in patients [14,18]. Its depletion induces significant defects in erythroid differentiation of CD34+ hematopoietic stem cells (HSC), which is one of the features of the 5q- syndrome and, thus, potentially links RBM22 to the pathogenesis of this MDS entity [40]. Moreover, Rbm22 was identified as an essential gene in mouse AML cell lines, and its in vitro downregulation was also associated with a defect of differentiation in the B cell lineage [41,42]. Together, those data suggest an implication of the loss of RBM22 in del(5q) MDS pathogenesis [43].
2.3. Splicing Genes in the Variably Deleted Region of the 5q
2.3.1. SLU7 (Synergistic Lethal with U5 snRNA 7)
Synergistic lethal with U5 snRNA 7 (SLU7) is a gene located at the locus 5q33.3, and it has been identified, in 1992, in a screening of genes that could be synthetically lethal with U5-snRNA in yeast [44]. It encodes the splicing factor SLU7, which is involved in the selection of the 3′-splice site (3′-SS) for the second step of splicing (i.e., the nucleophilic attack of the 3′-SS by the 3′-OH of the 5′-exon, leading to exon ligation) (Figure 2). SLU7 arrives on the complex by low affinity binding on the intron 3′-tail (downstream of the branch point (BP)), during the B* step of splicing, before the first transesterification [45,46]. Then, this affinity becomes stronger before the second step of splicing, facilitated by the stabilization of SLU7 on the spliceosome by Prp22 [47]. For the second transesterification, SLU7 participates in the choice of the 3′-SS, forming a heterodimer with Prp18 [47]. It then facilitates the delocalization of Prp8, which was binding the 5′-SS and BP to make it bind both 5′-SS and 3′-SS, and it stabilizes this new conformation. Thus, SLU7 docks the 3′-SS into the catalytic center of the spliceosome, allowing the exon ligation to occur [45,47,48]. Interestingly, SLU7 is only necessary for the selection of 3′SS that are >7 nucleotides away from the branch point. Under that distance, the second step of splicing is able to occur properly without SLU7 [49]. When the distance is higher than 7 nucleotides, the splicing process stops before exon ligation [48]. Interestingly, the nuclear concentration of SLU7 can be modulated by translocating the protein to the cytoplasm. This process was shown under several stress conditions: calcium-stress, heat shock, or UV-C treatments. Further, the decrease in its nuclear concentration was associated with an effect on the alternative splicing of several genes [36,37,50].
Apart from pre-mRNA splicing, SLU7 is also involved in other biological processes, such as the cell cycle and genome stability. Indeed, the knock-down of SLU7 in several human cell lines induces the formation of R-loops that cause DNA damages; it also affects mitosis by causing a defect in chromatid separation [51].
SLU7 is rather ubiquitously expressed, with a sharp stronger expression in the brain, thyroid, testis, and bone marrow, as well as a lower expression in the pancreas, salivary glands, and liver [39]. So far, no link between SLU7 and hematopoiesis has been made, and no correlation with hemopathies has ever been made. However, being located on the 5q chromosome, SLU7 is susceptible to be deleted in del(5q) MDS, but it is not comprised of the so-called CDR (Table 1). Importantly, as already mentioned, RBM22 interacts with SLU7. Of note, apart from their interaction during the translocation of SLU7, they more importantly interact within the same complex of the spliceosome during the second step of splicing [52]. Thereby, RBM22 happens to be co-deleted with SLU7 in some cases. It would, thus, be interesting to question the effect of their co-deletion on the phenotype of del(5q) MDS.

Table 1.
Most important pathogenic mutations in splicing genes potentially involved in MDS.
2.3.2. DDX41 (DEAD-Box Helicase 41)
DDX41 (DEAD-Box Helicase 41) is a gene located on the 5q35.3 cytogenetic band and belongs to DEAD box proteins with a putative RNA helicase function, and it seems to be a tumor suppressor gene. Involved in many cellular processes, such as splicing, rRNA processing, and innate immunity, DDX41 interacts with several spliceosome components, including SF3B proteins, PRP8 (PRPF8 gene) scaffold proteins, as well as U2 and U5 complexes (Figure 2). Mutations of the DDX41 gene disrupt the splicing progress and induce aberrant exon skippings or exon retentions [65].
Germline monoallelic (frameshifts) and somatic DDX41 mutations in other alleles (located in DEAD box and helicase domain) are described in MDS and AML [66] (Table 1). First reported by Polprasert et al. in 2015 [54], these mutations are described as common mutations in adult MDS and AML, without known family history and with frequent preexisting cytopenia, normal karyotype, and favorable outcomes, according to Sebert et al. [67]. Moreover, Alkhateeb and colleagues reported a low incidence of TP53 and splicing factor gene co-mutations in mutated DDX41 patients [68].
DDX41 is quite ubiquitously expressed. Low expression is described in 5q deletion [8]. Located on the 5q chromosome, DDX41 is susceptible to being retained in del(5q) MDS and is rarely deleted in more complex karyotypes with five chromosome abnormalities. Some studies have described a responsiveness to lenalidomide in patients with DDX41 mutations in myeloid neoplasms [69,70].
3. Chromosome 7 and Splicing Genes
3.1. Diagnostic and Prognostic Values
Deletion of 7q and monosomy 7 are among the most frequent chromosomal abnormalities reported in MDS (5%, respectively) [10]. They are mainly found in MDS-EB2, but can be retrieved also in the other MDS entities [11]. Deletion of 7q is associated with an intermediate MDS cytogenetic prognostic score. When associated with other chromosomal aberrations, monosomy 7 or del(7q) is of poor prognosis [4].
Several splicing genes are present in the chromosome 7, including RBM28, RBM33, LSM5, TRA2A, and LUC7L2. We will focus on LUC7L2 because of the existing literature on the role of this gene in the evolution of MDS to AML.
3.2. LUC7L2 Gene
LUC7L2 (Lethal unless CBC 7-Like 2)
LUC7L2 gene is located on chromosome 7 at the locus 7q34. LUC7L2 encodes a pre-mRNA splicing factor bearing Zinc–Finger domains and a serine/arginine-rich (SR) domain in its C-terminal region, which is a domain that is often found in splicing factors. LUC7L2 protein is homologous to the yeast protein Luc7p—where it is essential for splicing—particularly for the recognition of non-consensus 5′ splice donor sites [71]. Two major spliced variants of LUC7L2 can be expressed [72]. In mouse, Luc7l2 interacts with sodium channel modifier 1 (Scnm1) [72]. Yeast Luc7p is found in the early spliceosome pre-B complex, where it recognizes the duplex between the 5′ splice site and the 5′ end of U1 snRNA [73,74,75] (Figure 2). Similar results have been described in humans and mouse cells where LUC7L2 colocalizes with the U1 snRNP spliceosomal subunit: in particular, with U1 snRNP-specific protein U1-70K (SNRNP70) [71,72,73,76,77]. LUC7L2 preferentially binds to exonic sequences near, or at, the 5′SS, rather than to intronic sequences [76]. LUC7L2 either binds directly or not to exonic splicing enhancer (ESE) sequences (AAGAAG sequences) [76]. Alternative splicing events regulated by LUC7L2 have been recently published [57,76,77,78]. In particular, depletion of LUC7L2 increases the expression of multiple spliceosomal factors that will result, directly or not, in the repression of oxidative phosphorylation and the promotion of glycolysis [76,77].
Loss of function of LUC7L2 alters the hematopoietic differentiation of induced pluripotent stem cells (iPSCs) [79].
In MDS, LUC7L2 is located in the most commonly deleted region of -7/del7q MDS: 7q34. This region is deleted in 85% of -7/del7q MDS patients [80,81]. Loss of chromosome 7 in MDS has been associated with high-risk. The 7q deletion including LUC7L2 and EZH2 genes was also associated with mutations in TP53, KRAS and IDH1 [80].
In 2012, a first report described a somatic heterozygous mutation (R27X) in LUC7L2 in secondary AML [55] (Table 1). In 2013, the association between LUC7L2 mutation (giving rise to the truncated LUC7L2 R279X variant) and the evolution from MDS to acute myeloid leukemia (AML) was reported [56]. LUC7L2 mutations/deletions in MDS significantly affect the overall survival [7]. Mutations in LUC7L2 gene are far less frequent than in SF3B1, SRSF2, and U2AF1 genes [55,57,80]. Mutations in LUC7L2 are described as hemizygous, heterozygous, and homozygous [57,81,82].
A lower expression of LUC7L2 is reported in bone marrow samples of myeloid neoplasm patients (including MDS patients) compared to controls [57]. This low expression of LUC7L2 dysregulates 5′SS selection and increases the splicing of normally retained introns (RIs) [57].
In particular, a 40% decreased expression of LUC7L2 and other genes from the long arm of chromosome 7, compared to healthy controls., is reported in the CD34+ HSC from MDS patients, including some with monosomy 7 or del(7q) demonstrating the haploinsufficiency of this gene in MDS with monosomy 7 or del(7q) [83]. MDS samples with mutations in splicing factors other than LUC7L2 (e.g., SRSF2) also harbor aberrant splicing of LUC7L2 transcripts [57,84]. LUC7L2 mutations in MDS -7/del7q patients have been associated with shorter overall survival relative to patients with normal LUC7L2 expression [81].
4. The Only Gene with, So Far, a Diagnostic Value Is a Splicing Gene: SF3B1
SF3B1 (Splicing Factor 3B Subunit 1)
The SF3B1 gene, which is located on chromosome 2 (2q33.1), encodes an essential component of the U2 small nuclear ribonucleoprotein particle (snRNP) that is involved in the recognition of the branch point sequence close to the 3′ splice site during pre-mRNA splicing, thus participating in the definition of intron/exon junctions (Figure 2). SF3B1, which is the largest subunit of the heptameric protein complex Sf3b, contains a highly conserved HEAT domain composed of 20 tandem repeats structured as a superhelix, in C-terminus. SF3B1 interacts with other proteins outside the U2 complex, such as Histone proteins [85] or DNA-repair proteins (FANCI, FANCD2) [86], suggesting U2-independent functions of SF3B1. SF3B1 is a ubiquitous gene, found to be essential for hematopoiesis in zebrafish models [87]. SF3B1 is the most frequently mutated gene in MDS (15–30%) [7,59,60,62], especially in the group of MDS with ring sideroblasts (RS) for which approx. 80% of cases harbor an SF3B1 somatic mutation [6,88,89]. Those are heterozygous missense gain-of-function mutations that affect amino acids at restricted sites in the H4 to H8 repeats of the HEAT domain [90,91]. K700E variant occurs in more than 50% of SF3B1-mutant MDS cases (Table 1). Importantly, cells mutated for splicing factors, including SF3B1, require the wild type allele for survival [92]. SF3B1 mutations mainly alter 3′ splice site selection through the recognition of cryptic branch points, leading to hundreds of novel alternative/aberrant transcripts [93] whose contribution in the MDS pathogenesis is still poorly understood. More than 50% of these transcripts would be recognized and eliminated by the NonSense Mediated mRNA Decay mechanism [93], while others would lead to novel protein isoforms, as exemplified by a variant erythroferrone ERFE [94]. SF3B1 mutations can also affect the splicing of its own pre-mRNA [95]. SF3B1 mutations appear to occur early in the MDS pathogenesis, preceding other known genetic lesions [96,97]. Mutations in SF3B1 are strongly associated with the presence of medullar ring sideroblasts (RS) [59], which represent erythroid precursors with abnormal iron accumulation in mitochondria. In the 2016 update of WHO classification, the criteria to define MDS with RS include the presence of SF3B1 mutations, when as few as 5% of RS are at least detected [2]. Patients with SF3B1 mutations showed significantly better overall survival and lower cumulative incidence of disease progression [89]. Recently, based on the analysis of a comprehensive data set of 3479 patients, the International Working Group for the Prognosis of MDS suggested to create an SF3B1-mutant MDS subtype as a distinct disease subtype, characterized by (i) cytopenia, (ii) somatic SF3B1 mutation, (iii) isolated erythroid or multilineage dysplasia (for which RS will no longer be required for the diagnosis), (iv) bone marrow blasts <5% and peripheral blood blasts <1%, and (v) with selected concomitant genetic lesions as exclusion criteria [98]. This new SF3B1-mutant MDS entity has relatively good prognosis, and a potential response to luspatercept treatment. Nevertheless, all the SF3B1 variants are unlikely to be equivalent. For instance, K666N was associated with an increased progression of MDS, as illustrated by an enrichment in high-risk MDS and AML, and a shorter overall survival compared to non-K666N SF3B1-mutant MDS cases [99]. Comparison of the clinical features of K700E and non-K700E SF3B1-mutant MDS patients shows that only K700E mutation independently predicts overall survival in MDS [100]. SF3B1 mutations are mutually exclusive with SRSF2 or U2AF1 mutations in MDS, suggesting a synthetic lethality interaction, which has been confirmed in different models [92]. SF3B1 physically interacts with SUGP1, whose genetic alterations, which are rare in cancer, mimics SF3B1-mutant splice pattern [101]. Small molecules inhibitors against SF3B1 have demonstrated some effects on tumor cell death [102,103].
5. Other Mutated Splicing Genes with a Prognostic Value: SRSF2, U2AF1, ZRSR2, U2AF2 and PRPF8
5.1. The Most Frequently Mutated Splicing Genes with a Prognostic Value: SRSF2, U2AF1, ZRSR2
5.1.1. SRSF2 (Serine and Arginine Rich Splicing Factor 2)
The SRSF2 gene, which is located on chromosome 17 (17q25.1), encodes an essential member of the serine- and arginine-rich (SR) protein family that plays an important role in constitutive and alternative splicing. SRSF2 promotes exon recognition by binding mRNA exonic splicing enhancer (ESE) motifs through its RNA recognition motif domain (RRM), located in the N-terminus. The SR domain facilitates the interaction between different SR splicing factors. SRSF2 plays an essential role in hematopoiesis during embryonic development. SRSF2 mutations occur in 20–30% of MDS [6]. Those are heterozygous missense mutations, mostly affecting P95 residue (P95H, P95L, P95R), located in an intervening sequence between the RRM and the SR domain (Table 1). Conditional expression of SRSF2P95H impairs hematopoietic differentiation and promotes myelodysplasia in mice [61]. SRSF2 mutations result in genome-wide alterations in ESE preference [61]. In contrast to SRSF2 loss, SRSF2 mutations alters the recognition of specific ESE motifs, leading to recurrent missplicing of key hematopoietic regulators, including the promotion of a poison exon of EZH2 that undergoes NMD, resulting in reduced EZH2 protein level. Expression of hot-spot SRSF2 mutation (P95H, P95R) in CD34+ cells leads to a dramatic inhibition of proliferation via a G2-M phase arrest and an induction of apoptosis [104]. Non-canonical functions of SRSF2, shared with other SR proteins, have been described in the mRNA life cycle, including the regulation of transcription elongation and the NMD mechanism [105,106]. SRSF2 plays also a role in the control of genomic instability [107], together with SF3B1 and U2AF1.
Mutations of SRSF2 in MDS predicted shorter overall survival and more frequent AML progression compared with wild type SRSF2 [60]. A meta-analysis performed on 10 cohort studies, covering 1864 de novo MDS patients, confirmed that SRSF2 mutations had an adverse prognostic impact on overall survival and AML transformation [108]. Mutation status of SRSF2 in patients with lower risk MDS was associated to shorter overall survival in several studies [108,109]. Importantly, targeting exon sequencing of 96 genes performed in 648 cytopenic patients, among which 212 were diagnosed with MDS, showed that variant alleles frequency for seven genes, including SF3B1, U2AF1, and SRSF2, could correctly re-classify subjects as either MDS or other in 74% of cases that were misclassified. This suggests that targeted sequencing of these genes could improve MDS diagnosis [110].
5.1.2. U2AF1 (U2 Small Nuclear RNA Auxiliary Factor 1)
The gene U2AF1 (also known as U2AF35), which is located on chromosome 21 (21q22.3), encodes an essential accessory factor of U2 snRNP, which, together with U2AF2 (also known as U2AF65), binds to the AG dinucleotide at 3′ splice sites, thus ensuring an essential role in defining intron/exon junctions during the initial steps of pre-mRNA splicing (Figure 2). U2AF1 interacts with U2AF2 as a heterodimer, to form the U2 auxiliary factor (U2AF) complex. U2AF1 protein consists of two Zinc–Finger domains, as well as an UHM (U2AF homology motif) domain and an SR domain in C-terminal, which are both involved in the interaction with SRSF2. U2AF1 is required for survival and function of hematopoietic cells [111]. U2AF1 mutations occur in 11–16% of de novo MDS [6,63,112]. They are mainly heterozygous missense hot-spot mutations affecting amino acids S34 (S34F, S34Y) and Q157 (Q157R, Q157P) located in the N-terminal and C-terminal Zinc–Finger domains, respectively. Wild type allele of U2AF1 is required for cell survival [111]. RNAseq analysis performed in CD34+ hematopoietic cells expressing U2AF1S34F (vs. wild type) showed exon-skipping events and alternative splice site usage in a restricted number of genes (affecting less than 1% of expressed junctions) [113], suggesting that mutant U2AF1 alter splicing in a context of specific RNA sequences. Importantly, mutations in SF3B1, SRSF2 and U2AF1 result in different splicing alterations, largely affecting different genes, but with an enrichment in RNA splicing and transport, protein synthesis, mitochondrial dysfunction and signaling cascades, suggesting common mechanisms of action in MDS [84,114]. U2AF1 hot-spot mutations were associated with inferior survival in several studies [62,115], but not in all studies [60]. VAF > 40% of U2AF1 was shown to be an independent factor of short overall survival in MDS patients, but the presence of co-mutated genes (such as ASXL1, RUNX1, TET2) with U2AF1 can affect disease progression and prognosis [63]. A meta-analysis covering 3038 patients from 13 studies showed that U2AF1 mutations were associated with poor survival in MDS patients, and patients with U2AF1Q157 had a worse OS than those with U2AF1S34 (Li et al., 2020). RNAseq analysis performed in myeloid malignancies harboring U2AF1S34 and U2AF1Q157 reveals that only 4% of the genes were commonly misspliced by both mutations [116], emphasizing the necessity to study more deeply the specific effect of each hot spot mutation and their respective incidence for prognosis in MDS.
5.1.3. ZRSR2 (Zinc Finger CCCH-Type, RNA Binding Motif and Serine/Arginine Rich 2)
The gene ZRSR2, which is located on chromosome X (Xp22.2), encodes an essential component of the U12-dependent (minor) spliceosome [117,118], which excises a rare group of introns characterized by highly conserved 5′ splice sites and branch point sequences. More specifically, ZRSR2 is involved in the recognition of 3′SS during the early stages of spliceosomal assembly. Knockdown of ZRSR2 in TF-1 cells leads to retention of U12-type introns, without affecting U2-type introns [117,118]. The protein ZRSR2, which shares structural similarities with U2AF35, consists in two Zinc–Finger domains flanking an UHM domain, with an SR domain at C-terminus. ZRSR2 physically interacts with U2AF2, as well as with SRSF1 and SRSF2 [119]. ZRSR2 somatic mutations, which occur in approx. 3–7% of MDS [6,7,60] are widely distributed along the entire coding region, and correspond to nonsense or frameshift changes or alteration of splicing donor/acceptor sites, consistent with loss of function. ZRSR2 mutations impair splicing of U12-type introns by the minor-spliceosome pathway, mainly through increased intron retention [117,118], affecting several crucial genes involved in cell cycle, signaling, RNA binding and transport. Induced deletion of Zrsr2 in mice revealed enhanced self-renewal of Zrsr2-deficient male and female hematopoietic cells, in contrast to the effects of hot spot mutations in Sf3b1 or Srsf2 [120]. In MDS ZRSR2-mutant samples, 48% of minor introns exhibited significantly increased retention [120]. In particular, minor intron retention in LZTR1 (a cullin-3 adaptor for ubiquitin-mediated suppression of RAS-related GTPases), which correlated with reduced LZTR1 protein in MDS patients, resulted in cytokine independence. Patients with ZRSR2 mutations were not found to have a higher transformation rate to AML [60]. Prognostic impact of ZRSR2 mutations in MDS is currently unknown.
5.2. Other Mutated Splicing Genes with a Prognostic Value: U2AF2, PRPF8
5.2.1. U2AF2 Gene (U2 Small Nuclear RNA Auxiliary Factor 2)
The U2AF2 gene (U2 small nuclear RNA auxiliary factor 2), also known as U2AF65 is located at 19q13.42 band. It is a heterodimeric partner of U2AF1 pre-mRNA splicing factor. U2AF2 mutations mainly affect RNA recognition motifs (RRM1 and RRM2) [64]. These motifs recognize a polypyrimidine tract preceding the major class of 3′ splice sites (Figure 2). U2AF2 protein interacts with other splicing factors involved in MDS, such as SF3B complex in U2 complex [121], ZRSR2 [122] and SF1 [123]. It recruits PRP19 protein (PRPF19 gene), which regulates MDM4-mediated p53 activation inducing cellular senescence [124].
Acquired U2AF2 mutations are involved in gene expression dysregulation in leukemia and in solid tumors. In MDS, contrary to U2AF1 mutations, U2FA2 mutations are rare and recurrent events in myeloid pathologies, with a frequency of 1% or less [125]. They have a prognostic value [7,126], as they are associated with high-risk MDS and AML [127].
5.2.2. PRPF8 (Pre-MRNA Processing Factor 8)
PRPF8 is located on chromosome 17 at the locus 17p13.3. This gene encodes a U5 snRNP protein (PRP8), which is a major component of the catalytic core of the spliceosome. It specifically recognizes the 5′ splice site (5′SS) within the spliceosome complex B [128,129,130]. PRP8 facilitates the folding of the U2/U6 catalytic RNA [131]. Both in yeast and humans, Prp8 interacts with the 5′SS, the branch point, the polypyrimidine tract, and the 3′SS during splicing [128,129]. It is also found in the human C* spliceosome, concomitantly to RBM22 and SLU7 [32,46] (Figure 2).
The implication of PRPF8 in the inherited human disease Retinitis Pigmentosa has been extensively studied since 2005 [132]. However, the first article demonstrating the role of Prpf8 in the myeloid lineage differentiation was published in 2013 in a zebrafish model [133]. In those zebrafish mutants for Prpf8, accumulation of aberrantly spliced transcripts retaining both U2- and U12-type introns was observed [133].
From a cohort of 875 MDS cases, Haferlach et al. demonstrated that PRPF8 mutations/deletions in MDS significantly affect the overall survival [7]. Somatic PRPF8 mutations and hemizygous were reported [58], with half of the cases (del(17p) or PRPF8 variants) that showed a poor prognosis (Table 1). Alterations of PRPF8 increase the proliferation of CD34+ primary bone marrow cells and of a myeloid cell line, as well as resulting in defects of the second step of the RNA splicing process [58]. PRPF8 aberrations were associated with increased myeloblasts and ring sideroblasts in the absence of SF3B1 mutations [58]. Missplicing defects were reported in primary cells from patients with PRPF8 aberrations [58]. Of note, CD34+ cells from MDS patients with SF3B1 mutations exhibit differential expression profile of PRPF8 [134].
6. Perspectives and Conclusions
MDS are a heterogeneous group of myeloid malignancies. For years, the research efforts have been driven by the intention to categorize and stratify those malignancies to group them into homogeneous entities in order to set up proper management of these diseases and decide on the therapeutic options. Successful approaches to diagnose MDS, then categorize them for the prognostic evaluation, have consisted in their genetic characterizations in association with morphological/cytological data. Those approaches have also unveiled some understanding on the molecular pathogenesis of MDS and have led to the emergence of the concept of MDS as malignancies of the RNA splicing. The most obvious examples are the identification of recurrent genetic mutations of MDS that are involved in RNA splicing. They include, for the somatic mutations, SF3B1, SRSF2, U2AF1, ZRSR2, LUC7L2, and, for the constitutional and somatic mutations, DDX41. Taken alone, each gene can play its own distinct role in the molecular pathogenesis of MDS and triggers its own specific landscape of splicing alterations. Most of these genes are included in the recently proposed molecular IPSS-M [126]. Scientific literature accumulates on that subject but more research needs to be done (review in [135]).
Haploinsufficiency of splicing genes is however less studied. Many genes implicated in RNA splicing are located in frequently deleted chromosomes, such as in del(5q) (e.g., HNRNPA0, RBM27, RBM22, SLU7) or in del(7q) (e.g., LUC7L2). Those genes may be mutated or not, and they can also be lost or expressed at a sub-optimal levels for their proper function. Their roles have been described and discussed in this review. Further investigations are required to decipher their exact implication in MDS pathogenesis, their diagnostic and prognostic added values.
The RNA splicing process is highly complex, with sequential steps, multiple actors, some being early-acting spliceosomal proteins (e.g., SF3B1, U2AF1, SRSF2, and ZRSR2), and other later-acting splicing factors (e.g., PRPF8, DDX41) (Figure 2). The consequences of their mutations/deletions can be distinct or convergent. Interesting data have already been brought by several groups in this direction [84], with the identification of RNA splicing signatures associated with each splicing factor mutation. For example, different authors characterized shared sets and also distinct sets of alternative spliced RNA and the common affected biological processes in the different splicing gene mutant in MDS [84,114]. Importantly, several studies also unveiled that many genes involved in splicing are, themselves, dysregulated (up or down) or misspliced, suggesting an important amplifier process of splicing defects in the MDS pathogenesis [76,77,84,95,114,134,136].
Finally, the cumulative and synergistic effect of the splicing genes on MDS pathogenesis is still not completely known (Figure 2). Obviously, some genetic defects are exclusive and other co-occurring. The synergy of the splicing factors is less studied. For example, each protein of the trio PRP8, RBM22, and SLU7 are, together, present in the C* spliceosome complex. The effect of the presence of PRPF8 mutation associated with RBM22 deletion (i.e., in del(5q)), associated or not with SLU7 deletion could be investigated. Other genes involved in a component of splicing machinery, such as SF1, PRPF40B, and SF3A1 could be explored in MDS patients.
Many authors advocate for the inclusion of sequencing some of these splicing genes to improve the diagnosis and the prognosis of MDS, so a better understanding of these cooperations, including all types of genetic defects (mutations, deletions, gains, dysregulation of gene expression, spliced variants) will be a future step for MDS management. This could lead to new diagnostic classifications for MDS and more refined prognostic scores, for the direct benefit of MDS patients.
Author Contributions
Conceptualization, M.-B.T. and N.D.-G.; writing—original draft preparation and editing, N.D.-G., B.S., D.G.B. and M.-B.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ligue Régionale contre le cancer (comités 16, 22, 29, 35, 56, 41 and 85), Association Halte au Cancer, Association Gaétan Saleün, Association connaître et combattre la myélodysplasie, le Collectif Agora de Guilers, Association Fondation de l’Avenir and fonds INNOVEO Brest.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A.; et al. The 2008 Revision of the World Health Organization (WHO) Classification of Myeloid Neoplasms and Acute Leukemia: Rationale and Important Changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Khac, F.; Bidet, A.; Daudignon, A.; Lafage-Pochitaloff, M.; Ameye, G.; Bilhou-Nabéra, C.; Chapiro, E.; Collonge-Rame, M.A.; Cuccuini, W.; Douet-Guilbert, N.; et al. The Complex Karyotype in Hematological Malignancies: A Comprehensive Overview by the Francophone Group of Hematological Cytogenetics (GFCH). Leukemia 2022, 36, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised International Prognostic Scoring System for Myelodysplastic Syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef]
- Schanz, J.; Tüchler, H.; Solé, F.; Mallo, M.; Luño, E.; Cervera, J.; Granada, I.; Hildebrandt, B.; Slovak, M.L.; Ohyashiki, K.; et al. New Comprehensive Cytogenetic Scoring System for Primary Myelodysplastic Syndromes (MDS) and Oligoblastic Acute Myeloid Leukemia after MDS Derived from an International Database Merge. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 820–829. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent Pathway Mutations of Splicing Machinery in Myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of Genetic Lesions in 944 Patients with Myelodysplastic Syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Hosono, N.; Makishima, H.; Mahfouz, R.; Przychodzen, B.; Yoshida, K.; Jerez, A.; LaFramboise, T.; Polprasert, C.; Clemente, M.J.; Shiraishi, Y.; et al. Recurrent Genetic Defects on Chromosome 5q in Myeloid Neoplasms. Oncotarget 2017, 8, 6483–6495. [Google Scholar] [CrossRef]
- Van Den Berghe, H.; Cassiman, J.-J.; David, G.; Fryns, J.-P.; Michaux, J.-L.; Sokal, G. Distinct Haematological Disorder with Deletion of Long Arm of No. 5 Chromosome. Nature 1974, 251, 437–438. [Google Scholar] [CrossRef]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 Allelic State for Genome Stability, Clinical Presentation and Outcomes in Myelodysplastic Syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef]
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 1223–1233. [Google Scholar] [CrossRef]
- Giagounidis, A.A.N.; Aul, C. The 5q- Syndrome. Cancer Treat. Res. 2008, 142, 133–148. [Google Scholar]
- Holtan, S.G.; Santana-Davila, R.; Dewald, G.W.; Khetterling, R.P.; Knudson, R.A.; Hoyer, J.D.; Chen, D.; Hanson, C.A.; Porrata, L.; Tefferi, A.; et al. Myelodysplastic Syndromes Associated with Interstitial Deletion of Chromosome 5q: Clinicopathologic Correlations and New Insights from the Pre-Lenalidomide Era. Am. J. Hematol. 2008, 83, 708–713. [Google Scholar] [CrossRef]
- Boultwood, J.; Pellagatti, A.; Cattan, H.; Lawrie, C.H.; Giagounidis, A.; Malcovati, L.; Porta, M.G.D.; Jädersten, M.; Killick, S.; Fidler, C.; et al. Gene Expression Profiling of CD34+ Cells in Patients with the 5q− Syndrome. Br. J. Haematol. 2007, 139, 578–589. [Google Scholar] [CrossRef]
- Eisenmann, K.M.; Dykema, K.J.; Matheson, S.F.; Kent, N.F.; DeWard, A.D.; West, R.A.; Tibes, R.; Furge, K.A.; Alberts, A.S. 5q- Myelodysplastic Syndromes: Chromosome 5q Genes Direct a Tumor-Suppression Network Sensing Actin Dynamics. Oncogene 2009, 28, 3429–3441. [Google Scholar] [CrossRef]
- Douet-Guilbert, N.; De Braekeleer, E.; Basinko, A.; Herry, A.; Gueganic, N.; Bovo, C.; Trillet, K.; Dos Santos, A.; Le Bris, M.J.; Morel, F.; et al. Molecular Characterization of Deletions of the Long Arm of Chromosome 5 (Del(5q)) in 94 MDS/AML Patients. Leukemia 2012, 26, 1695–1697. [Google Scholar] [CrossRef]
- Adema, V.; Palomo, L.; Walter, W.; Mallo, M.; Hutter, S.; Framboise, T.L.; Arenillas, L.; Meggendorfer, M.; Radivoyevitch, T.; Xicoy, B.; et al. Pathophysiologic and Clinical Implications of Molecular Profiles Resultant from Deletion 5q. eBioMedicine 2022, 80, 104059. [Google Scholar] [CrossRef]
- Pellagatti, A.; Cazzola, M.; Giagounidis, A.A.N.; Malcovati, L.; Porta, M.G.D.; Killick, S.; Campbell, L.J.; Wang, L.; Langford, C.F.; Fidler, C.; et al. Gene Expression Profiles of CD34+ Cells in Myelodysplastic Syndromes: Involvement of Interferon-Stimulated Genes and Correlation to FAB Subtype and Karyotype. Blood 2006, 108, 337–345. [Google Scholar] [CrossRef]
- Myer, V.E.; Steitz, J.A. Isolation and Characterization of a Novel, Low Abundance HnRNP Protein: A0. RNA 1995, 1, 171–182. [Google Scholar]
- Cannell, I.G.; Merrick, K.A.; Morandell, S.; Zhu, C.-Q.; Braun, C.J.; Grant, R.A.; Cameron, E.R.; Tsao, M.-S.; Hemann, M.T.; Yaffe, M.B. A Pleiotropic RNA-Binding Protein Controls Distinct Cell Cycle Checkpoints to Drive Resistance of P53-Defective Tumors to Chemotherapy. Cancer Cell 2015, 28, 623–637. [Google Scholar] [CrossRef]
- Zhang, J.; Kong, L.; Guo, S.; Bu, M.; Guo, Q.; Xiong, Y.; Zhu, N.; Qiu, C.; Yan, X.; Chen, Q.; et al. HnRNPs and ELAVL1 Cooperate with UORFs to Inhibit Protein Translation. Nucleic Acids Res. 2017, 45, 2849–2864. [Google Scholar] [CrossRef] [PubMed]
- Young, D.J.; Stoddart, A.; Nakitandwe, J.; Chen, S.-C.; Qian, Z.; Downing, J.R.; Beau, M.M.L. Knockdown of Hnrnpa0, a Del(5q) Gene, Alters Myeloid Cell Fate in Murine Cells through Regulation of AU-Rich Transcripts. Haematologica 2014, 99, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Joslin, J.M.; Tennant, T.R.; Reshmi, S.C.; Young, D.J.; Stoddart, A.; Larson, R.A.; Le Beau, M.M. Cytogenetic and Genetic Pathways in Therapy-Related Acute Myeloid Leukemia. Chem. Biol. Interact. 2010, 184, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.-Y.; Cody, N.A.L.; Dominguez, D.; et al. A Large-Scale Binding and Functional Map of Human RNA-Binding Proteins. Nature 2020, 583, 711–719. [Google Scholar] [CrossRef]
- Silla, T.; Schmid, M.; Dou, Y.; Garland, W.; Milek, M.; Imami, K.; Johnsen, D.; Polak, P.; Andersen, J.S.; Selbach, M.; et al. The Human ZC3H3 and RBM26/27 Proteins Are Critical for PAXT-Mediated Nuclear RNA Decay. Nucleic Acids Res. 2020, 48, 2518–2530. [Google Scholar] [CrossRef]
- Rasche, N.; Dybkov, O.; Schmitzová, J.; Akyildiz, B.; Fabrizio, P.; Lührmann, R. Cwc2 and Its Human Homologue RBM22 Promote an Active Conformation of the Spliceosome Catalytic Centre. EMBO J. 2012, 31, 1591–1604. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, C.; Hang, J.; Finci, L.I.; Lei, J.; Shi, Y. An Atomic Structure of the Human Spliceosome. Cell 2017, 169, 918–929.e14. [Google Scholar] [CrossRef]
- Hogg, R.; McGrail, J.C.; O’Keefe, R.T. The Function of the NineTeen Complex (NTC) in Regulating Spliceosome Conformations and Fidelity During Pre-MRNA Splicing. Biochem. Soc. Trans. 2010, 38, 1110–1115. [Google Scholar] [CrossRef]
- Kastner, B.; Will, C.L.; Stark, H.; Lührmann, R. Structural Insights into Nuclear Pre-MRNA Splicing in Higher Eukaryotes. Cold Spring Harb. Perspect. Biol. 2019, 11, a032417. [Google Scholar] [CrossRef]
- van der Feltz, C.; Nikolai, B.; Schneider, C.; Paulson, J.C.; Fu, X.; Hoskins, A.A. Saccharomyces Cerevisiae Ecm2 Modulates the Catalytic Steps of Pre-MRNA Splicing. RNA 2021, 27, 591–603. [Google Scholar] [CrossRef]
- McGrail, J.C.; Krause, A.; O’Keefe, R.T. The RNA Binding Protein Cwc2 Interacts Directly with the U6 SnRNA to Link the Nineteen Complex to the Spliceosome during Pre-MRNA Splicing. Nucleic Acids Res. 2009, 37, 4205–4217. [Google Scholar] [CrossRef]
- Bertram, K.; Agafonov, D.E.; Liu, W.-T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Lührmann, R. Cryo-EM Structure of a Human Spliceosome Activated for Step 2 of Splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef]
- Liu, S.; Li, X.; Zhang, L.; Jiang, J.; Hill, R.C.; Cui, Y.; Hansen, K.C.; Zhou, Z.H.; Zhao, R. Structure of the Yeast Spliceosomal Postcatalytic P Complex. Science 2017, 358, 1278–1283. [Google Scholar] [CrossRef]
- Wan, R.; Yan, C.; Bai, R.; Lei, J.; Shi, Y. Structure of an Intron Lariat Spliceosome from Saccharomyces Cerevisiae. Cell 2017, 171, 120–132.e12. [Google Scholar] [CrossRef]
- Park, J.W.; Parisky, K.; Celotto, A.M.; Reenan, R.A.; Graveley, B.R. Identification of Alternative Splicing Regulators by RNA Interference in Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 15974–15979. [Google Scholar] [CrossRef]
- Montaville, P.; Dai, Y.; Cheung, C.Y.; Giller, K.; Becker, S.; Michalak, M.; Webb, S.E.; Miller, A.L.; Krebs, J. Nuclear Translocation of the Calcium-Binding Protein ALG-2 Induced by the RNA-Binding Protein RBM22. Biochim. Biophys. Acta 2006, 1763, 1335–1343. [Google Scholar] [CrossRef]
- Janowicz, A.; Michalak, M.; Krebs, J. Stress Induced Subcellular Distribution of ALG-2, RBM22 and HSlu7. Biochim. Biophys. Acta BBA Mol. Cell Res. 2011, 1813, 1045–1049. [Google Scholar] [CrossRef]
- Xiao, R.; Chen, J.-Y.; Liang, Z.; Luo, D.; Chen, G.; Lu, Z.J.; Chen, Y.; Zhou, B.; Li, H.; Du, X.; et al. Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription. Cell 2019, 178, 107–121.e18. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-Specific Expression by Genome-Wide Integration of Transcriptomics and Antibody-Based Proteomics. Mol. Cell. Proteom. MCP 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Ebert, B.L.; Pretz, J.; Bosco, J.; Chang, C.Y.; Tamayo, P.; Galili, N.; Raza, A.; Root, D.E.; Attar, E.; Ellis, S.R.; et al. Identification of RPS14 as a 5q- Syndrome Gene by RNA Interference Screen. Nature 2008, 451, 335–339. [Google Scholar] [CrossRef]
- Yamauchi, T.; Masuda, T.; Canver, M.C.; Seiler, M.; Semba, Y.; Shboul, M.; Al-Raqad, M.; Maeda, M.; Schoonenberg, V.A.C.; Cole, M.A.; et al. Genome-Wide CRISPR-Cas9 Screen Identifies Leukemia-Specific Dependence on a Pre-MRNA Metabolic Pathway Regulated by DCPS. Cancer Cell 2018, 33, 386–400.e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ahmed, T.; Krysiak, K.; Shirai, C.L.; Shao, J.; Nunley, R.; Bucala, R.; McKenzie, A.; Ndonwi, M.; Walter, M.J. Haploinsufficiency of Multiple Del(5q) Genes Induce B Cell Abnormalities in Mice. Leuk. Res. 2020, 96, 106428. [Google Scholar] [CrossRef] [PubMed]
- Soubise, B.; Jiang, Y.; Douet-Guilbert, N.; Troadec, M.-B. RBM22, a Key Player of Pre-MRNA Splicing and Gene Expression Regulation, Is Altered in Cancer. Cancers 2022, 14, 643. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.; Patterson, B.; Guthrie, C. Synthetic Lethal Mutations Suggest Interactions between U5 Small Nuclear RNA and Four Proteins Required for the Second Step of Splicing. Mol. Cell. Biol. 1992, 12, 5197–5205. [Google Scholar] [CrossRef]
- Ohrt, T.; Odenwälder, P.; Dannenberg, J.; Prior, M.; Warkocki, Z.; Schmitzová, J.; Karaduman, R.; Gregor, I.; Enderlein, J.; Fabrizio, P.; et al. Molecular Dissection of Step 2 Catalysis of Yeast Pre-MRNA Splicing Investigated in a Purified System. RNA 2013, 19, 902–915. [Google Scholar] [CrossRef][Green Version]
- Chung, C.-S.; Tseng, C.-K.; Lai, Y.-H.; Wang, H.-F.; Newman, A.J.; Cheng, S.-C. Dynamic Protein-RNA Interactions in Mediating Splicing Catalysis. Nucleic Acids Res. 2019, 47, 899–910. [Google Scholar] [CrossRef]
- James, S.-A.; Turner, W.; Schwer, B. How Slu7 and Prp18 Cooperate in the Second Step of Yeast Pre-MRNA Splicing. RNA 2002, 8, 1068–1077. [Google Scholar] [CrossRef][Green Version]
- Chua, K.; Reed, R. Human Step II Splicing Factor HSlu7 Functions in Restructuring the Spliceosome between the Catalytic Steps of Splicing. Genes Dev. 1999, 13, 841–850. [Google Scholar] [CrossRef]
- Brys, A.; Schwer, B. Requirement for SLU7 in Yeast Pre-MRNA Splicing Is Dictated by the Distance between the Branchpoint and the 3′ Splice Site. RNA 1996, 2, 707–717. [Google Scholar]
- Shomron, N.; Alberstein, M.; Reznik, M.; Ast, G. Stress Alters the Subcellular Distribution of HSlu7 and Thus Modulates Alternative Splicing. J. Cell Sci. 2005, 118, 1151–1159. [Google Scholar] [CrossRef]
- Jiménez, M.; Urtasun, R.; Elizalde, M.; Azkona, M.; Latasa, M.U.; Uriarte, I.; Arechederra, M.; Alignani, D.; Bárcena-Varela, M.; Álvarez-Sola, G.; et al. Splicing Events in the Control of Genome Integrity: Role of SLU7 and Truncated SRSF3 Proteins. Nucleic Acids Res. 2019, 47, 3450–3466. [Google Scholar] [CrossRef]
- Xu, D.; Friesen, J.D. Splicing Factor Slt11p and Its Involvement in Formation of U2/U6 Helix II in Activation of the Yeast Spliceosome. Mol. Cell. Biol. 2001, 21, 1011–1023. [Google Scholar] [CrossRef][Green Version]
- Quesada, A.E.; Routbort, M.J.; DiNardo, C.D.; Bueso-Ramos, C.E.; Kanagal-Shamanna, R.; Khoury, J.D.; Thakral, B.; Zuo, Z.; Yin, C.C.; Loghavi, S.; et al. DDX41 Mutations in Myeloid Neoplasms Are Associated with Male Gender, TP53 Mutations and High-Risk Disease. Am. J. Hematol. 2019, 94, 757–766. [Google Scholar] [CrossRef]
- Polprasert, C.; Schulze, I.; Sekeres, M.A.; Makishima, H.; Przychodzen, B.; Hosono, N.; Singh, J.; Padgett, R.A.; Gu, X.; Phillips, J.G.; et al. Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell 2015, 27, 658–670. [Google Scholar] [CrossRef]
- Makishima, H.; Visconte, V.; Sakaguchi, H.; Jankowska, A.M.; Abu Kar, S.; Jerez, A.; Przychodzen, B.; Bupathi, M.; Guinta, K.; Afable, M.G.; et al. Mutations in the Spliceosome Machinery, a Novel and Ubiquitous Pathway in Leukemogenesis. Blood 2012, 119, 3203–3210. [Google Scholar] [CrossRef]
- Singh, H.; Lane, A.A.; Correll, M.; Przychodzen, B.; Sykes, D.B.; Stone, R.M.; Ballen, K.K.; Amrein, P.C.; Maciejewski, J.; Attar, E.C. Putative RNA-Splicing Gene LUC7L2 on 7q34 Represents a Candidate Gene in Pathogenesis of Myeloid Malignancies. Blood Cancer J. 2013, 3, e117. [Google Scholar] [CrossRef]
- Hershberger, C.E.; Moyer, D.C.; Adema, V.; Kerr, C.M.; Walter, W.; Hutter, S.; Meggendorfer, M.; Baer, C.; Kern, W.; Nadarajah, N.; et al. Complex Landscape of Alternative Splicing in Myeloid Neoplasms. Leukemia 2021, 35, 1108–1120. [Google Scholar] [CrossRef]
- Kurtovic-Kozaric, A.; Przychodzen, B.; Singh, J.; Konarska, M.M.; Clemente, M.J.; Otrock, Z.K.; Nakashima, M.; Hsi, E.D.; Yoshida, K.; Shiraishi, Y.; et al. PRPF8 Defects Cause Missplicing in Myeloid Malignancies. Leukemia 2015, 29, 126–136. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 Mutation in Myelodysplasia with Ring Sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Thol, F.; Kade, S.; Schlarmann, C.; Löffeld, P.; Morgan, M.; Krauter, J.; Wlodarski, M.W.; Kölking, B.; Wichmann, M.; Görlich, K.; et al. Frequency and Prognostic Impact of Mutations in SRSF2, U2AF1, and ZRSR2 in Patients with Myelodysplastic Syndromes. Blood 2012, 119, 3578–3584. [Google Scholar] [CrossRef]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.-W.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.-B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Mudireddy, M.; Finke, C.M.; Nicolosi, M.; Lasho, T.L.; Hanson, C.A.; Patnaik, M.M.; Pardanani, A.; Gangat, N. U2AF1 Mutation Variants in Myelodysplastic Syndromes and Their Clinical Correlates. Am. J. Hematol. 2018, 93, E146–E148. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, Y.; Dong, Z.; Li, T.; Xie, X.; Wan, D.; Jiang, Z.; Yu, J.; Guo, R. Differential U2AF1 Mutation Sites, Burden and Co-Mutation Genes Can Predict Prognosis in Patients with Myelodysplastic Syndrome. Sci. Rep. 2020, 10, 18622. [Google Scholar] [CrossRef] [PubMed]
- Maji, D.; Glasser, E.; Henderson, S.; Galardi, J.; Pulvino, M.J.; Jenkins, J.L.; Kielkopf, C.L. Representative Cancer-Associated U2AF2 Mutations Alter RNA Interactions and Splicing. J. Biol. Chem. 2020, 295, 17148–17157. [Google Scholar] [CrossRef]
- Antony-Debré, I.; Steidl, U. Functionally Relevant RNA Helicase Mutations in Familial and Sporadic Myeloid Malignancies. Cancer Cell 2015, 27, 609–611. [Google Scholar] [CrossRef][Green Version]
- Kadono, M.; Kanai, A.; Nagamachi, A.; Shinriki, S.; Kawata, J.; Iwato, K.; Kyo, T.; Oshima, K.; Yokoyama, A.; Kawamura, T.; et al. Biological Implications of Somatic DDX41 p.R525H Mutation in Acute Myeloid Leukemia. Exp. Hematol. 2016, 44, 745–754.e4. [Google Scholar] [CrossRef]
- Sébert, M.; Passet, M.; Raimbault, A.; Rahmé, R.; Raffoux, E.; Sicre de Fontbrune, F.; Cerrano, M.; Quentin, S.; Vasquez, N.; Da Costa, M.; et al. Germline DDX41 Mutations Define a Significant Entity within Adult MDS/AML Patients. Blood 2019, 134, 1441–1444. [Google Scholar] [CrossRef]
- Alkhateeb, H.B.; Nanaa, A.; Viswanatha, D.; Foran, J.M.; Badar, T.; Sproat, L.; He, R.; Nguyen, P.; Jevremovic, D.; Salama, M.E.; et al. Genetic Features and Clinical Outcomes of Patients with Isolated and Comutated DDX41-Mutated Myeloid Neoplasms. Blood Adv. 2022, 6, 528–532. [Google Scholar] [CrossRef]
- Negoro, E.; Radivoyevitch, T.; Polprasert, C.; Adema, V.; Hosono, N.; Makishima, H.; Przychodzen, B.; Hirsch, C.; Clemente, M.J.; Nazha, A.; et al. Molecular Predictors of Response in Patients with Myeloid Neoplasms Treated with Lenalidomide. Leukemia 2016, 30, 2405–2409. [Google Scholar] [CrossRef]
- Abou Dalle, I.; Kantarjian, H.; Bannon, S.A.; Kanagal-Shamanna, R.; Routbort, M.; Patel, K.P.; Hu, S.; Bhalla, K.; Garcia-Manero, G.; DiNardo, C.D. Successful Lenalidomide Treatment in High Risk Myelodysplastic Syndrome with Germline DDX41 Mutation. Am. J. Hematol. 2020, 95, 227–229. [Google Scholar] [CrossRef]
- Fortes, P.; Bilbao-Cortés, D.; Fornerod, M.; Rigaut, G.; Raymond, W.; Séraphin, B.; Mattaj, I.W. Luc7p, a Novel Yeast U1 SnRNP Protein with a Role in 5′ Splice Site Recognition. Genes Dev. 1999, 13, 2425–2438. [Google Scholar] [CrossRef]
- Howell, V.M.; Jones, J.M.; Bergren, S.K.; Li, L.; Billi, A.C.; Avenarius, M.R.; Meisler, M.H. Evidence for a Direct Role of the Disease Modifier SCNM1 in Splicing. Hum. Mol. Genet. 2007, 16, 2506–2516. [Google Scholar] [CrossRef]
- Puig, O.; Bragado-Nilsson, E.; Koski, T.; Séraphin, B. The U1 SnRNP-Associated Factor Luc7p Affects 5′ Splice Site Selection in Yeast and Human. Nucleic Acids Res. 2007, 35, 5874–5885. [Google Scholar] [CrossRef]
- Bai, R.; Wan, R.; Yan, C.; Lei, J.; Shi, Y. Structures of the Fully Assembled Saccharomyces Cerevisiae Spliceosome before Activation. Science 2018, 360, 1423–1429. [Google Scholar] [CrossRef]
- Plaschka, C.; Lin, P.-C.; Charenton, C.; Nagai, K. Prespliceosome Structure Provides Insights into Spliceosome Assembly and Regulation. Nature 2018, 559, 419–422. [Google Scholar] [CrossRef]
- Daniels, N.J.; Hershberger, C.E.; Gu, X.; Schueger, C.; DiPasquale, W.M.; Brick, J.; Saunthararajah, Y.; Maciejewski, J.P.; Padgett, R.A. Functional Analyses of Human LUC7-like Proteins Involved in Splicing Regulation and Myeloid Neoplasms. Cell Rep. 2021, 35, 108989. [Google Scholar] [CrossRef]
- Jourdain, A.A.; Begg, B.E.; Mick, E.; Shah, H.; Calvo, S.E.; Skinner, O.S.; Sharma, R.; Blue, S.M.; Yeo, G.W.; Burge, C.B.; et al. Loss of LUC7L2 and U1 SnRNP Subunits Shifts Energy Metabolism from Glycolysis to OXPHOS. Mol. Cell 2021, 81, 1905–1919.e12. [Google Scholar] [CrossRef]
- Li, C.; Feng, L.; Luo, W.-W.; Lei, C.-Q.; Li, M.; Shu, H.-B. The RNA-Binding Protein LUC7L2 Mediates MITA/STING Intron Retention to Negatively Regulate Innate Antiviral Response. Cell Discov. 2021, 7, 46. [Google Scholar] [CrossRef]
- Kotini, A.G.; Chang, C.-J.; Boussaad, I.; Delrow, J.J.; Dolezal, E.K.; Nagulapally, A.B.; Perna, F.; Fishbein, G.A.; Klimek, V.M.; Hawkins, R.D.; et al. Functional Analysis of a Chromosomal Deletion Associated with Myelodysplastic Syndromes Using Isogenic Human Induced Pluripotent Stem Cells. Nat. Biotechnol. 2015, 33, 646–655. [Google Scholar] [CrossRef]
- Shen, W.; Szankasi, P.; Sederberg, M.; Schumacher, J.; Frizzell, K.A.; Gee, E.P.; Patel, J.L.; South, S.T.; Xu, X.; Kelley, T.W. Concurrent Detection of Targeted Copy Number Variants and Mutations Using a Myeloid Malignancy next Generation Sequencing Panel Allows Comprehensive Genetic Analysis Using a Single Testing Strategy. Br. J. Haematol. 2016, 173, 49–58. [Google Scholar] [CrossRef]
- Hershberger, C.E.; Hosono, N.; Singh, J.; Dietrich, R.C.; Gu, X.; Makishima, H.; Saunthararajah, Y.; Maciejewski, J.P.; Padgett, R.A. The Role of LUC7L2 in Splicing and MDS. Blood 2016, 128, 5504. [Google Scholar] [CrossRef]
- Visconte, V.; O Nakashima, M.; J Rogers, H. Mutations in Splicing Factor Genes in Myeloid Malignancies: Significance and Impact on Clinical Features. Cancers 2019, 11, 1844. [Google Scholar] [CrossRef] [PubMed]
- Jerez, A.; Sugimoto, Y.; Makishima, H.; Verma, A.; Jankowska, A.M.; Przychodzen, B.; Visconte, V.; Tiu, R.V.; O’Keefe, C.L.; Mohamedali, A.M.; et al. Loss of Heterozygosity in 7q Myeloid Disorders: Clinical Associations and Genomic Pathogenesis. Blood 2012, 119, 6109–6117. [Google Scholar] [CrossRef]
- Madan, V.; Li, J.; Zhou, S.; Teoh, W.W.; Han, L.; Meggendorfer, M.; Malcovati, L.; Cazzola, M.; Ogawa, S.; Haferlach, T.; et al. Distinct and Convergent Consequences of Splice Factor Mutations in Myelodysplastic Syndromes. Am. J. Hematol. 2020, 95, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kfir, N.; Lev-Maor, G.; Glaich, O.; Alajem, A.; Datta, A.; Sze, S.K.; Meshorer, E.; Ast, G. SF3B1 Association with Chromatin Determines Splicing Outcomes. Cell Rep. 2015, 11, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Moriel-Carretero, M.; Ovejero, S.; Gérus-Durand, M.; Vryzas, D.; Constantinou, A. Fanconi Anemia FANCD2 and FANCI Proteins Regulate the Nuclear Dynamics of Splicing Factors. J. Cell Biol. 2017, 216, 4007–4026. [Google Scholar] [CrossRef] [PubMed]
- De La Garza, A.; Cameron, R.C.; Nik, S.; Payne, S.G.; Bowman, T.V. Spliceosomal Component Sf3b1 Is Essential for Hematopoietic Differentiation in Zebrafish. Exp. Hematol. 2016, 44, 826–837.e4. [Google Scholar] [CrossRef]
- Malcovati, L.; Papaemmanuil, E.; Bowen, D.T.; Boultwood, J.; Della Porta, M.G.; Pascutto, C.; Travaglino, E.; Groves, M.J.; Godfrey, A.L.; Ambaglio, I.; et al. Clinical Significance of SF3B1 Mutations in Myelodysplastic Syndromes and Myelodysplastic/Myeloproliferative Neoplasms. Blood 2011, 118, 6239–6246. [Google Scholar] [CrossRef]
- Malcovati, L.; Karimi, M.; Papaemmanuil, E.; Ambaglio, I.; Jädersten, M.; Jansson, M.; Elena, C.; Gallì, A.; Walldin, G.; Della Porta, M.G.; et al. SF3B1 Mutation Identifies a Distinct Subset of Myelodysplastic Syndrome with Ring Sideroblasts. Blood 2015, 126, 233–241. [Google Scholar] [CrossRef]
- Cretu, C.; Schmitzová, J.; Ponce-Salvatierra, A.; Dybkov, O.; De Laurentiis, E.I.; Sharma, K.; Will, C.L.; Urlaub, H.; Lührmann, R.; Pena, V. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol. Cell 2016, 64, 307–319. [Google Scholar] [CrossRef]
- Seiler, M.; Peng, S.; Agrawal, A.A.; Palacino, J.; Teng, T.; Zhu, P.; Smith, P.G.; Buonamici, S.; Yu, L.; Caesar-Johnson, S.J.; et al. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep. 2018, 23, 282–296.e4. [Google Scholar] [CrossRef]
- Lee, S.C.-W.; North, K.; Kim, E.; Jang, E.; Obeng, E.; Lu, S.X.; Liu, B.; Inoue, D.; Yoshimi, A.; Ki, M.; et al. Synthetic Lethal and Convergent Biological Effects of Cancer-Associated Spliceosomal Gene Mutations. Cancer Cell 2018, 34, 225–241.e8. [Google Scholar] [CrossRef]
- Darman, R.B.; Seiler, M.; Agrawal, A.A.; Lim, K.H.; Peng, S.; Aird, D.; Bailey, S.L.; Bhavsar, E.B.; Chan, B.; Colla, S.; et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015, 13, 1033–1045. [Google Scholar] [CrossRef]
- Bondu, S.; Alary, A.-S.; Lefèvre, C.; Houy, A.; Jung, G.; Lefebvre, T.; Rombaut, D.; Boussaid, I.; Bousta, A.; Guillonneau, F.; et al. A Variant Erythroferrone Disrupts Iron Homeostasis in SF3B1-Mutated Myelodysplastic Syndrome. Sci. Transl. Med. 2019, 11, eaav5467. [Google Scholar] [CrossRef]
- Bergot, T.; Lippert, E.; Douet-Guilbert, N.; Commet, S.; Corcos, L.; Bernard, D.G. Human Cancer-Associated Mutations of SF3B1 Lead to a Splicing Modification of Its Own RNA. Cancers 2020, 12, 652. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and Biological Implications of Driver Mutations in Myelodysplastic Syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef]
- Mian, S.A.; Rouault-Pierre, K.; Smith, A.E.; Seidl, T.; Pizzitola, I.; Kizilors, A.; Kulasekararaj, A.G.; Bonnet, D.; Mufti, G.J. SF3B1 Mutant MDS-Initiating Cells May Arise from the Haematopoietic Stem Cell Compartment. Nat. Commun. 2015, 6, 10004. [Google Scholar] [CrossRef]
- Malcovati, L.; Stevenson, K.; Papaemmanuil, E.; Neuberg, D.; Bejar, R.; Boultwood, J.; Bowen, D.T.; Campbell, P.J.; Ebert, B.L.; Fenaux, P.; et al. SF3B1 -Mutant MDS as a Distinct Disease Subtype: A Proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136, 157–170. [Google Scholar] [CrossRef]
- Dalton, W.B.; Helmenstine, E.; Pieterse, L.; Li, B.; Gocke, C.D.; Donaldson, J.; Xiao, Z.; Gondek, L.P.; Ghiaur, G.; Gojo, I.; et al. The K666N Mutation in SF3B1 Is Associated with Increased Progression of MDS and Distinct RNA Splicing. Blood Adv. 2020, 4, 1192–1196. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R.; Montalban-Bravo, G.; Sasaki, K.; Darbaniyan, F.; Jabbour, E.; Bueso-Ramos, C.; Wei, Y.; Chien, K.; Kadia, T.; Ravandi, F.; et al. Only SF3B1 Mutation Involving K700E Independently Predicts Overall Survival in Myelodysplastic Syndromes. Cancer 2021, 127, 3552–3565. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, J.; Sun, Y.; Perea-Chamblee, T.E.; Manley, J.L.; Rabadan, R. Pan-Cancer Analysis Identifies Mutations in SUGP1 That Recapitulate Mutant SF3B1 Splicing Dysregulation. Proc. Natl. Acad. Sci. USA 2020, 117, 10305–10312. [Google Scholar] [CrossRef]
- Venkataramany, A.S.; Schieffer, K.M.; Lee, K.; Cottrell, C.E.; Wang, P.Y.; Mardis, E.R.; Cripe, T.P.; Chandler, D.S. Alternative RNA Splicing Defects in Pediatric Cancers: New Insights in Tumorigenesis and Potential Therapeutic Vulnerabilities. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2022, 33, 578–592. [Google Scholar] [CrossRef]
- Effenberger, K.A.; Urabe, V.K.; Jurica, M.S. Modulating Splicing with Small Molecular Inhibitors of the Spliceosome. Wiley Interdiscip. Rev. RNA 2017, 8, e1381. [Google Scholar] [CrossRef]
- Bapat, A.; Keita, N.; Martelly, W.; Kang, P.; Seet, C.; Jacobsen, J.R.; Stoilov, P.; Hu, C.; Crooks, G.M.; Sharma, S. Myeloid Disease Mutations of Splicing Factor SRSF2 Cause G2-M Arrest and Skewed Differentiation of Human Hematopoietic Stem and Progenitor Cells. Stem Cells 2018, 36, 1663–1675. [Google Scholar] [CrossRef]
- Slišković, I.; Eich, H.; Müller-McNicoll, M. Exploring the Multifunctionality of SR Proteins. Biochem. Soc. Trans. 2022, 50, 187–198. [Google Scholar] [CrossRef]
- Rahman, M.A.; Lin, K.-T.; Bradley, R.K.; Abdel-Wahab, O.; Krainer, A.R. Recurrent SRSF2 Mutations in MDS Affect Both Splicing and NMD. Genes Dev. 2020, 34, 413–427. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.-Y.; Huang, Y.-J.; Gu, Y.; Qiu, J.; Qian, H.; Shao, C.; Zhang, X.; Hu, J.; Li, H.; et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol. Cell 2018, 69, 412–425.e6. [Google Scholar] [CrossRef]
- Zheng, X.; Zhan, Z.; Naren, D.; Li, J.; Yan, T.; Gong, Y. Prognostic Value of SRSF2 Mutations in Patients with de Novo Myelodysplastic Syndromes: A Meta-Analysis. PLoS ONE 2017, 12, e0185053. [Google Scholar] [CrossRef]
- Jiang, L.; Luo, Y.; Zhu, S.; Wang, L.; Ma, L.; Zhang, H.; Shen, C.; Yang, W.; Ren, Y.; Zhou, X.; et al. Mutation Status and Burden Can Improve Prognostic Prediction of Patients with Lower-risk Myelodysplastic Syndromes. Cancer Sci. 2020, 111, 580–591. [Google Scholar] [CrossRef]
- Goll, J.B.; Jensen, T.L.; Lindsley, R.C.; Bejar, R.; Walker, J.; Fulton, R.; Abel, G.A.; Al Baghdadi, T.; Deeg, H.J.; DeZern, A.E.; et al. Targeted Sequencing of 7 Genes Can Help Reduce Pathologic Misclassification of MDS. Blood 2020, 136, 32–33. [Google Scholar] [CrossRef]
- Dutta, A.; Yang, Y.; Le, B.T.; Zhang, Y.; Abdel-Wahab, O.; Zang, C.; Mohi, G. U2af1 Is Required for Survival and Function of Hematopoietic Stem/Progenitor Cells. Leukemia 2021, 35, 2382–2398. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Lasho, T.L.; Patnaik, M.M.; Saeed, L.; Mudireddy, M.; Idossa, D.; Finke, C.; Ketterling, R.P.; Pardanani, A.; Gangat, N. Targeted Next-Generation Sequencing in Myelodysplastic Syndromes and Prognostic Interaction between Mutations and IPSS-R. Am. J. Hematol. 2017, 92, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Okeyo-Owuor, T.; White, B.S.; Chatrikhi, R.; Mohan, D.R.; Kim, S.; Griffith, M.; Ding, L.; Ketkar-Kulkarni, S.; Hundal, J.; Laird, K.M.; et al. U2AF1 Mutations Alter Sequence Specificity of Pre-MRNA Binding and Splicing. Leukemia 2015, 29, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Armstrong, R.N.; Steeples, V.; Sharma, E.; Repapi, E.; Singh, S.; Sanchi, A.; Radujkovic, A.; Horn, P.; Dolatshad, H.; et al. Impact of Spliceosome Mutations on RNA Splicing in Myelodysplasia: Dysregulated Genes/Pathways and Clinical Associations. Blood 2018, 132, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-J.; Tang, J.-L.; Lin, C.-T.; Kuo, Y.-Y.; Li, L.-Y.; Tseng, M.-H.; Huang, C.-F.; Lai, Y.-J.; Lee, F.-Y.; Liu, M.-C.; et al. Clinical Implications of U2AF1 Mutation in Patients with Myelodysplastic Syndrome and Its Stability during Disease Progression. Am. J. Hematol. 2013, 88, E277–E282. [Google Scholar] [CrossRef] [PubMed]
- Adema, V.; Hershberger, C.E.; Walter, W.; Kerr, C.M.; Hutter, S.; Nagata, Y.; Awada, H.; Kongkiatkamon, S.; Snider, C.; Co, M.; et al. Hotspot U2AF1 Mutations Determine Missplicing Selectivity: Novel Mechanisms Altering Splicing Factors. Blood 2019, 134, 2985. [Google Scholar] [CrossRef]
- Madan, V.; Kanojia, D.; Jia, L.; Okamoto, R.; Sato-Otsubo, A.; Kohlmann, A.; Sanada, M.; Grossmann, V.; Sundaresan, J.; Shiraishi, Y.; et al. ZRSR2 Mutations Cause Dysregulated RNA Splicing in MDS. Blood 2014, 124, 4609. [Google Scholar] [CrossRef]
- Madan, V.; Kanojia, D.; Li, J.; Okamoto, R.; Sato-Otsubo, A.; Kohlmann, A.; Sanada, M.; Grossmann, V.; Sundaresan, J.; Shiraishi, Y.; et al. Aberrant Splicing of U12-Type Introns Is the Hallmark of ZRSR2 Mutant Myelodysplastic Syndrome. Nat. Commun. 2015, 6, 6042. [Google Scholar] [CrossRef]
- Tronchre, H.; Wang, J.; Fu, X.-D. A Protein Related to Splicing Factor U2AF35 That Interacts with U2AF65 and SR Proteins in Splicing of Pre-MRNA. Nature 1997, 388, 397–400. [Google Scholar] [CrossRef]
- Inoue, D.; Polaski, J.T.; Taylor, J.; Castel, P.; Chen, S.; Kobayashi, S.; Hogg, S.J.; Hayashi, Y.; Bello Pineda, J.M.; Penson, A.V.; et al. ZRSR2 Mutation Induced Minor Intron Retention Drives MDS and Diverse Cancer Predisposition Via Aberrant Splicing of LZTR1. Blood 2020, 136, 10–11. [Google Scholar] [CrossRef]
- Warnasooriya, C.; Feeney, C.F.; Laird, K.M.; Ermolenko, D.N.; Kielkopf, C.L. A Splice Site-Sensing Conformational Switch in U2AF2 Is Modulated by U2AF1 and Its Recurrent Myelodysplasia-Associated Mutation. Nucleic Acids Res. 2020, 48, 5695–5709. [Google Scholar] [CrossRef]
- Corsini, L.; Bonnal, S.; Bonna, S.; Basquin, J.; Hothorn, M.; Scheffzek, K.; Valcárcel, J.; Sattler, M. U2AF-Homology Motif Interactions Are Required for Alternative Splicing Regulation by SPF45. Nat. Struct. Mol. Biol. 2007, 14, 620–629. [Google Scholar] [CrossRef]
- Selenko, P.; Gregorovic, G.; Sprangers, R.; Stier, G.; Rhani, Z.; Krämer, A.; Sattler, M. Structural Basis for the Molecular Recognition between Human Splicing Factors U2AF65 and SF1/MBBP. Mol. Cell 2003, 11, 965–976. [Google Scholar] [CrossRef]
- Yano, K.; Takahashi, R.-U.; Shiotani, B.; Abe, J.; Shidooka, T.; Sudo, Y.; Yamamoto, Y.; Kan, S.; Sakagami, H.; Tahara, H. PRPF19 Regulates P53-Dependent Cellular Senescence by Modulating Alternative Splicing of MDM4 MRNA. J. Biol. Chem. 2021, 297, 100882. [Google Scholar] [CrossRef]
- Larsson, C.A.; Cote, G.; Quintás-Cardama, A. The Changing Mutational Landscape of Acute Myeloid Leukemia and Myelodysplastic Syndrome. Mol. Cancer Res. MCR 2013, 11, 815–827. [Google Scholar] [CrossRef]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango, O.J.E.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2022, 1, EVIDoa2200008. [Google Scholar] [CrossRef]
- Stanley, R.F.; Abdel-Wahab, O. Dysregulation and Therapeutic Targeting of RNA Splicing in Cancer. Nat. Cancer 2022, 3, 536–546. [Google Scholar] [CrossRef]
- Reyes, J.L.; Kois, P.; Konforti, B.B.; Konarska, M.M. The Canonical GU Dinucleotide at the 5′ Splice Site Is Recognized by P220 of the U5 SnRNP within the Spliceosome. RNA 1996, 2, 213–225. [Google Scholar]
- Reyes, J.L.; Gustafson, E.H.; Luo, H.R.; Moore, M.J.; Konarska, M.M. The C-Terminal Region of HPrp8 Interacts with the Conserved GU Dinucleotide at the 5′ Splice Site. RNA 1999, 5, 167–179. [Google Scholar] [CrossRef][Green Version]
- MacRae, A.J.; Mayerle, M.; Hrabeta-Robinson, E.; Chalkley, R.J.; Guthrie, C.; Burlingame, A.L.; Jurica, M.S. Prp8 Positioning of U5 SnRNA Is Linked to 5′ Splice Site Recognition. RNA 2018, 24, 769–777. [Google Scholar] [CrossRef]
- Townsend, C.; Leelaram, M.N.; Agafonov, D.E.; Dybkov, O.; Will, C.L.; Bertram, K.; Urlaub, H.; Kastner, B.; Stark, H.; Lührmann, R. Mechanism of Protein-Guided Folding of the Active Site U2/U6 RNA during Spliceosome Activation. Science 2020, 370, eabc3753. [Google Scholar] [CrossRef]
- Grainger, R.J.; Beggs, J.D. Prp8 Protein: At the Heart of the Spliceosome. RNA 2005, 11, 533–557. [Google Scholar] [CrossRef]
- Keightley, M.-C.; Crowhurst, M.O.; Layton, J.E.; Beilharz, T.; Markmiller, S.; Varma, S.; Hogan, B.M.; de Jong-Curtain, T.A.; Heath, J.K.; Lieschke, G.J. In Vivo Mutation of Pre-MRNA Processing Factor 8 (Prpf8) Affects Transcript Splicing, Cell Survival and Myeloid Differentiation. FEBS Lett. 2013, 587, 2150–2157. [Google Scholar] [CrossRef]
- Dolatshad, H.; Pellagatti, A.; Fernandez-Mercado, M.; Yip, B.H.; Malcovati, L.; Attwood, M.; Przychodzen, B.; Sahgal, N.; Kanapin, A.A.; Lockstone, H.; et al. Disruption of SF3B1 Results in Deregulated Expression and Splicing of Key Genes and Pathways in Myelodysplastic Syndrome Hematopoietic Stem and Progenitor Cells. Leukemia 2015, 29, 1092–1103. [Google Scholar] [CrossRef]
- Hosono, N. Genetic Abnormalities and Pathophysiology of MDS. Int. J. Clin. Oncol. 2019, 24, 885–892. [Google Scholar] [CrossRef]
- Shiozawa, Y.; Malcovati, L.; Gallì, A.; Sato-Otsubo, A.; Kataoka, K.; Sato, Y.; Watatani, Y.; Suzuki, H.; Yoshizato, T.; Yoshida, K.; et al. Aberrant Splicing and Defective MRNA Production Induced by Somatic Spliceosome Mutations in Myelodysplasia. Nat. Commun. 2018, 9, 3649. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).