Abstract
The human microbiome encodes more than three million genes, outnumbering human genes by more than 100 times, while microbial cells in the human microbiota outnumber human cells by 10 times. Thus, the human microbiota and related microbiome constitute a vast source for identifying disease biomarkers and therapeutic drug targets. Herein, we review the evidence backing the exploitation of the human microbiome for identifying diagnostic biomarkers for human disease. We describe the importance of the human microbiome in health and disease and detail the use of the human microbiome and microbiota metabolites as potential diagnostic biomarkers for multiple diseases, including cancer, as well as inflammatory, neurological, and metabolic diseases. Thus, the human microbiota has enormous potential to pave the road for a new era in biomarker research for diagnostic and therapeutic purposes. The scientific community needs to collaborate to overcome current challenges in microbiome research concerning the lack of standardization of research methods and the lack of understanding of causal relationships between microbiota and human disease.
1. Introduction
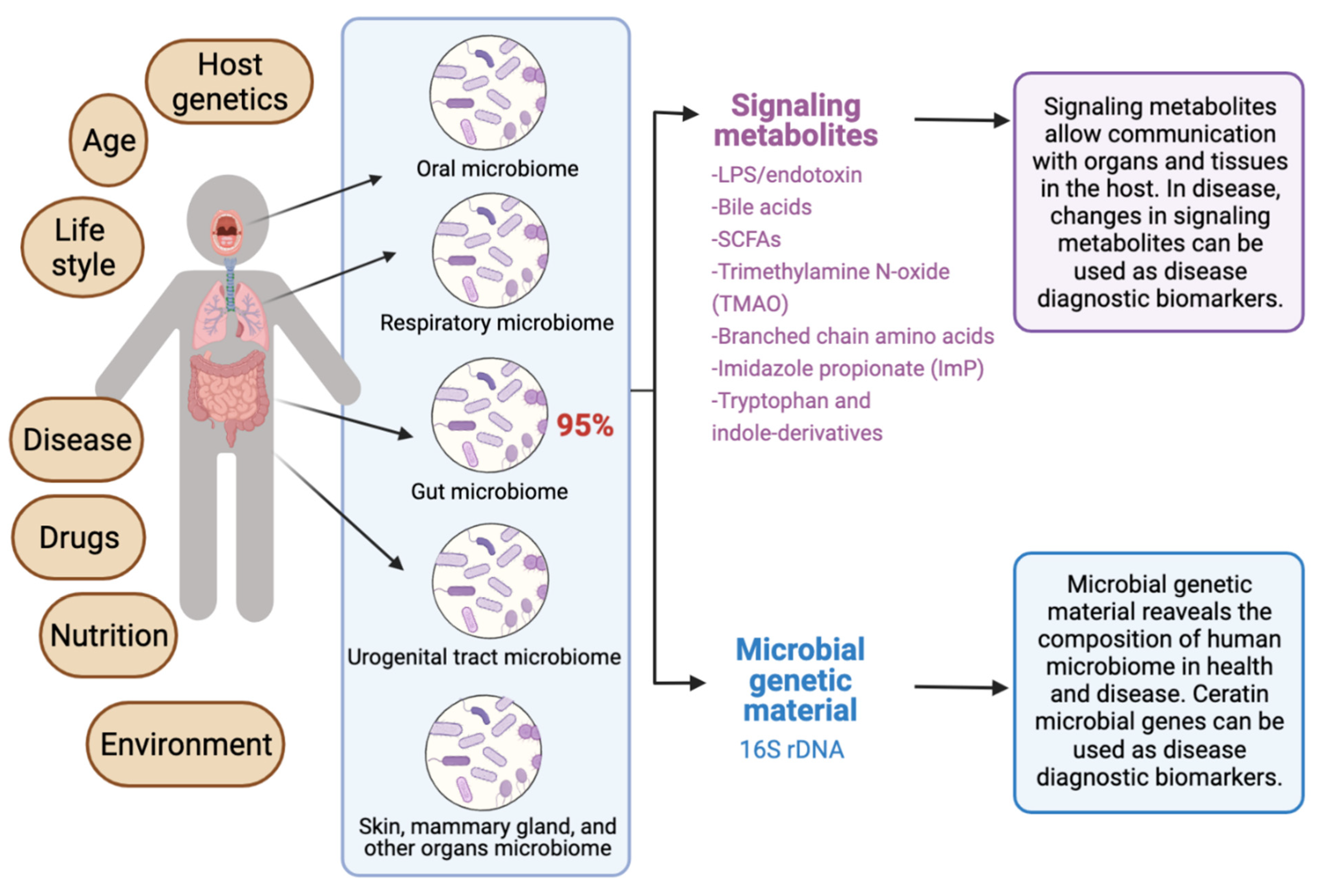
The human microbiota comprises 10–100 trillion symbiotic microbial cells constituting over 10,000 microbial species residing in the human body and outnumbering human cells by 10 times [1]. It consists primarily of bacteria, in addition to viruses, fungi, protozoa, and helminths residing in and on human body organs, such as the skin, mammary glands, mucosa, gastrointestinal (GI), respiratory, and urogenital tracts [2,3,4]. The largest percentage of the human microbiota (95%) resides in the GI tract, and every human being has a unique microbiota composition which could potentially serve as a unique fingerprint. The human microbiome consists of the genes of prokaryotic and eukaryotic cells, and it is often viewed as our “other genome”, which consists of more than three million genes, in comparison with our 23,000 human genes. Hence, the human microbiome has gained increased interest recently with regard to identifying novel drug targets and biomarkers for human disease.
Microbiota affect human health and disease by modulating important metabolic and immunomodulatory processes [3,5]. The interactions between the human body and microbiota form a complex, distinct, and harmonized bionetwork that defines the relationship between the host and its microbiota as commensal, symbiotic, or pathogenic. The human microbiota is continually developing and changing throughout life by responding to host factors such as age, genes, hormonal changes, nutrition, predisposing disease, lifestyle, and many environmental factors [6,7,8,9]. Harmonized microbiota contribute substantially to healthy livelihood [7], while a disruption in microbiota hemostasis (dysbiosis) might contribute to life-threatening diseases [10]. The significant contribution of the human microbiome in health and disease has been recently described in the biomedical literature [11,12,13,14,15,16,17,18,19,20,21,22] delineating gastrointestinal [10,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37], urinary tract [4,38], and skin [3] microbiota. Evidence from the biomedical literature indicates that alterations in host immunity might be closely related to the compositional and functional changes of gut flora [24,39].
Thus, the human microbiota can potentially lead to the discovery of effective disease diagnostic biomarkers. According to the National Institute of Health (NIH), a biomarker is “a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” [40]. A diagnostic biomarker is simply a biomarker that “detects or confirms the presence of a disease or condition of interest, or identifies an individual with a subtype of the disease” [41]. The most frequently used biomarkers are derived from either biological materials or imaging data. More recently, machine learning (ML) and artificial intelligence (AI) have enabled the identification of highly predictive, disease-specific biomarkers [42].
In fact, microflora disturbances have been linked to many human diseases, including GI tract diseases [10,43], cardiovascular disease [13,44,45], allergies [39,46], inflammation [44,45,47], neuro-disease, stubborn bacterial infections [48,49,50,51], and cancer [37,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68]. Aberrations in the human microbiome are linked to several cancers, including breast, colorectal, gastric, pancreatic, and hepatic cancers [69,70]. Additionally, cancer could be provoked by viruses, fungi, helminths, and bacteria [69,70]. Microbiota might also contribute to cancer development by disrupting the balance between the growth and death of host cells after altering the immune system and affecting metabolism [58,71,72]. Furthermore, Microbiota affects cancer prognosis by several mechanisms, including genotoxicity, inflammation, and metabolism [73].
Recent reviews indicated that microbiome signatures can be exploited as disease diagnostic biomarkers [71,72,74,75,76,77,78,79]. Herein, we review the available evidence supporting the use of the human microbiome- and microbiota-derived metabolites for the purposes of disease diagnosis. A graphical summary of the concept in provided in Figure 1. We detail potential microbiota-derived biomarkers for the diagnosis of a variety of diseases, including complex diseases like diabetes, neuro-diseases, and cancer.
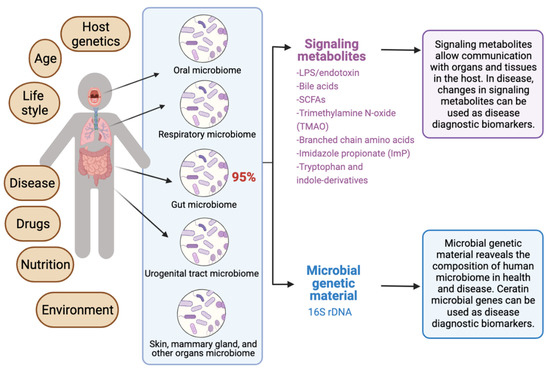
Figure 1.
Exploiting the human microbiome for diagnostic disease biomarkers.
2. The Rationale for Microbiome-Based Disease Biomarkers
The identification of “ideal biomarkers” is considered a daunting task for many diseases, including some cancer types. Most of the current sampling techniques for cancer tissues cannot identify individuals who will lack response to therapy, and they fall short in classifying cancer types correctly, owing to the inter- and intra-tumor heterogeneity of tumors [80]. A biomarker should be easily measurable, non-invasive, and cost-effective. The human microbiome, particularly the gut microbiome, can be considered as a non-invasive approach to identify disease biomarkers that can detect many diseases in the early stages [71,81]. Additionally, the identification of microbiome-based biomarkers can increase the accuracy of disease classification when it is combined with clinical information and other biomarkers. For example, some microbes are known to contribute to the adenoma-carcinoma transition in some cancers, such as colorectal carcer (CRC). Such microbes can be exploited as disease and immunotherapy efficacy biomarkers for CRC [71,81].
In addition to microbiome-based biomarkers, there is also an emerging interest in mast cells (MCs) [82,83,84,85], microRNAs (miRNAs) [86,87], imaging, and machine-learning models [42] as non-invasive disease diagnostic and prognostic biomarkers that promise to shape the future of precision medicine. Sometimes, there is a crosstalk between the human microbiota and other genetic or chemical biomarkers. For example, alterations in fecal small RNA profiles in CRC reflect gut microbiome composition in stool samples [88]. Thus, using multiple connected biomarkers of the network type (i.e., “network biomarkers”) may increase the effectiveness of existing biomarkers.
3. The Significance of Human Microbiota in Health and Disease
The human microbiota plays several important roles in the human body, such as helping in food digestion, producing vitamins, regulating the immune system, and protecting against pathogenic disease-causing microbes. In the following subsections, we review the significance of the human microbiota in health and disease and the importance of classifying healthy microbiomes from unhealthy microbiomes in clinical practice.
3.1. Conservation of Homeostasis
The human microbiota controls the immune system and affects the inflammatory cascade and immune homeostasis in newborn and children [89]. Children developing allergies at advanced ages showed ubiquity of anaerobic bacteria and Bacteroidaceae, as well as a low number of Lactobacillus, Bifidobacterium bifidum, and Bifidobacterium adolescentis [11,27]. Studies reported that these microbes hydrolyze adulterants such as pesticides, plastic particles, heavy metals, polycyclic aromatic hydrocarbons, and organic compounds [23]. Further studies revealed that the urinary tract microbiomes detoxify toxins [90]. Studies showed that female genital tract microbiomes provoke an immune response through secreting antimicrobial peptides, inhibitory compounds, and cytokines [90].
3.2. Involvement in Host Immune System
The symbiosis interaction between the indigenous microbiome and the immune system results in the evolution of immune responses and the development of the immune system to recognize pathogens and beneficial microbiota [91,92]. Indeed, the immune system is shaped by the human microbiome [93]. The lack or alterations in the human microbiome might weaken the immune system and induce type II immunity responses and allergies [39,94]. Aberrations of microbiota induce allergic rhinitis in children [39,94]. The gut microbiome activates the regulatory T-cells (Tregs) and proinflammatory Th17cells in the intestine [95,96]. The older neutrophil decreases the proinflammatory properties in vivo [91]. The microbiota induces the growth of neutrophil through MyD88-mediated and Toll-like receptor (TLR) signaling cascades [91]. Changes in microbial flora decrease the old neutrophils and induce inflammation-mediated tissue injury, such as septic shock and sickle cell disease. Altogether, the microbial flora supervise disease-inducing neutrophil, which is a substantial component in inflammatory diseases [91]. In addition, the gut microbiomes protect the body against harmful pathogens through inducing colonization resistance, as well as synthesizing antimicrobial compounds [97]. A stable intestinal microbiota controls antibodies of CD8+T (killer) and CD4+ (helper) cells that impede the influx of the influenza virus to the respiratory system [89,97]. The gut flora supports and optimizes the functionality of the GIT [98,99]. Activating the regulatory T cells is essential in maintaining the hemostasis of the immune system [89].
3.3. Involvement in Host Nutrition and Metabolism
Gut microbiota provide nutrients to the host by digesting complex dietary elements (e.g., fiber and other complex carbohydrates) in food, permitting their absorption from the gut [100]. Additionally, intestinal microbiota offer essential nutrients that are not available, but are necessary for maintaining GI tract functionality [101]. Furthermore, intestinal microbiota halt cancer prognosis in the GI tract by generating butyrate, which is a product of fermentation complex nutrients [102]. Studies revealed that fruits’ and vegetables’ carbohydrates maintain a healthy GI tract microbiome [97]. In addition, the gut microbiome provide the required vitamins (K and folic acid) for host growth, such as enterobacteria and GI tract bacteria, including Bacteroides and Bifidobacterium species [100]. Moreover, gut microbiota contribute to red and white blood cells (RBC and WBC) synthesis [103]. Live microorganisms (probiotics) are deployed for treating allergic diseases [97]. Probiotics decrease and/or inhibit the activation of T-cells and restrain the tumor necrosis factor (TNF) that participates in systemic inflammation [97]. Gut microbiota produce important vitamins needed for blood coagulation, including B vitamins such as B12, thiamine and riboflavin, and Vitamin K [104,105,106].
3.4. Classifying Healthy and Unhealthy Microbiomes
The identification of microbiome-based biomarkers for disease diagnosis, prognosis, risk profiling, and precision medicine relies on the determination of microbial features associated with health or disease. It is often a daunting task to clearly define what constitutes a healthy microbiome in different human populations, especially because a person’s microbiota can be affected by many factors, including age, lifestyle, diet, smoking, exercise, ethnicity, environmental factors, and other factors. Another challenge in classifying healthy versus unhealthy microbiomes stems from limitations in the current technologies and methodologies that do not provide a high microbial resolution on the strain-level, impeding the functional understanding or relevance for health or disease [10].
4. Metagenomics-Derived Genes as Potential Disease Biomarkers
There is emerging evidence highlighting important functional links between microbiota dysbiosis and disease. Cataloging the types of organisms and the numbers of each type is extremely helpful in studying microbial dysbiosis. This is often achieved by metagenomics, the study of the genetic composition (genomes) of a mixed community of organisms recovered from environmental and human samples. Metagenomic studies can be performed using either high-throughput shotgun genomics (i.e., metagenomics sequencing) [107], or by the use of the polymerase chain reaction (PCR), based on 16S rRNA gene amplicon sequencing analysis, to study microbial ribosomal RNA (rRNA) [108,109]. The use of 16S rRNA amplicon sequencing allows the comprehensive phylogenetic assessment of the studied microbiome. However, microbiome researchers are currently using database-independent operational taxonomic unit (OTU)-based methods [110,111,112], which reduce the taxonomic resolution, and impair further functional analysis at the strain level.
5. Microbiota-Derived Metabolites as Potential Disease Biomarkers
Gut microbiota-derived metabolites are considered as central regulators in metabolic disorders and are important surrogates to study microbial dysbiosis [113,114,115]. For example, microbial metabolites such as bile acid derivatives, short-chain fatty acids, branched-chain amino acids, trimethylamine N-oxide, tryptophan, and indole derivatives, have been implicated in the pathogenesis of multiple metabolic disorders [115]. These metabolites are considered potential diagnostic and prognostic disease biomarkers, as well as promising targets for drug discovery and development. Both gut and serum metabolomes can be targeted to identify such metabolomics’ biomarkers. Examples on most important bacterial metabolites with biomarker potential in human disease are provided in Table 1, based on data mined from the Human Metabolome Database (HMDB) version 5.0 [116] and the Marker Database (MarkerDB) [117].

Table 1.
Important microbiota metabolites that can be explored as diagnostic biomarkers.
5.1. Short-Chain Fatty Acids (SCFAs)
These are subclasses of saturated fatty acids that contain six or fewer carbons [119]. They include acetate, propionate, butyrate, pentanoic (valeric) acid, and hexanoic (caproic) acid [120]. CSFAs are the main bacterial metabolites due to an anaerobic fermentation of indigestible dietary fiber and resistant starch by specific colonic anaerobic bacteria in the large intestine [121]. It is currently believed that SCFAs, particularly those of low molecular weights (acetate, propionate, and butyrate), play crucial role in the physiology of various systems, at both the cellular and molecular levels [122]. SCFAs play vital roles in terms of colonic health [123]. It is well established that SCFAs have anti-inflammatory, antitumorigenic, and antimicrobial activity [120]. SCFAs are now evidently involved in the pathogenesis of chronic diseases such as allergies, asthma, cancer, autoimmune and metabolic diseases, and most significantly, neurologic conditions [124]. Fecal SCFAs have the potential to be used as biomarkers for irritable bowel syndrome (IBS) [125]. Serum SCFAs have the potential to be used as biomarkers for multiple sclerosis [126,127] and colorectal cancer [128].
5.2. Branched-Chain Amino Acids (BCAAs)
These are essential amino acids whose carbon structure is marked by a branch point and which are obtained directly form sources such as meat, dairy, and legumes. They include leucine, isoleucine, and valine [129]. BCAAs supplementation is believed to have a promoting effect on anabolic pathways and may play an essential role in the protection against muscle wasting (cachexia), chronic kidney disease and liver cirrhosis, attenuating exercise-related fatigue, the promotion of wound healing, and the stimulation of insulin production [130]. BCAAs are considered potential biomarkers for insulin resistance, type 2 diabetes, the risk of cardiovascular disease, stage I and II chronic kidney disease, ischemic stroke [131,132], major depression [133], dyslipidemia [134], and chronic graft vs. host disease [135].
5.3. Tryptophan and Indole-Derivative Metabolites
Tryptophan is an essential amino acid that is necessary for normal infant growth, the production and maintenance of the body’s proteins, enzymes, and neurotransmitters [136]. It also plays an important role in regulating the sleep cycle and appetite, as it is a precursor for the synthesis of melatonin and serotonin [137]. Tryptophan is also a precursor of niacin (vitamin B3) [138]. It is found in dairy products, nutritional seeds, white meat, and fish [139,140,141,142,143]. Indole metabolites that are produced via the microbial metabolism of tryptophan include indole-3-propionic acid (IPA) and indole-3-aldehyde (IAld) [144]. These indole derivatives possess anti-inflammatory, antibiotic, antioxidant, and immunomodulatory effects [145]. In fact, the kynurenine/tryptophan ratio has been investigated as a potential blood-based biomarker in non-small cell lung cancer [146], while indole-derived metabolites have been considered as potential indicators for body mass index [147].
5.4. Trimethylamine N-Oxide (TMAO)
This is an amine oxide that is produced by the gut microbial metabolism of carnitine and choline. TMAO is evidenced to exacerbate glucose tolerance, inhibit hepatic insulin signaling, and promote inflammation; hence, it is considered as a mediating molecule to develop type-2 diabetes mellitus. Studies also suggest a crucial role of TMAO in the development of atherosclerosis and the pathophysiology of ischemic heart diseases [148]. TMAO has the potential to serve as a novel biomarker for plaque rupture in patients with ST-segment elevation myocardial infarction (STEMI) and early metabolic syndrome [149]; it is also a promising diagnostic biomarker for cardiovascular and neurological disorders [150], as well as for preeclampsia [151].
5.5. Imidazole Propionate (ImP)
This compound is identified as a novel microbial metabolite produced through the alternative metabolism of histidine in type 2 diabetes mellitus patients. ImP may be considered as a potential biomarker for elevated blood pressure in obese patients [152].
5.6. Bile Acids
These play an important role in the innate immune defense within the intestine, since they are considered as potent antimicrobials that have an essential role in the defense mechanism of the host microbiota [153]. Both host and microbiota regulate the bile acid pool. The liver bile acid–microbiome axis has been implicated in many diseases, including liver cirrhosis and hepatocarcinogenesis [154,155,156,157]. After bile acids are synthesized in the host liver, they are converted to secondary bile acids by gut microbiota. Reduced bile acid levels in the GI tract are usually associated with bacterial overgrowth and inflammation [158]. High fat diets increase the levels of bile acids in the gut, which affect the highest taxonomic levels of gut bacteria. Physiological concentrations of various intestinal bile acids play an important role in preventing the intestinal colonization by pathogens such as Clostridium difficile [159]. Increased bile acids lead to blooms of taxa, including bile acid 7α-dehydroxylating species such as Clostridium scindens and Clostridium hylemonae.
5.7. Lipopolysaccharides (LPS), Lipooligosaccharides (LOS) and Endotoxin
Lipopolysaccharides (LPS) are macromolecules consisting typically of a hydrophobic domain known as lipid A (or endotoxin), a non-repeating “core” oligosaccharide, and a distal polysaccharide (or O-antigen). They are considered important constituents of the outer membranes of Gram-negative bacteria. The term lipooligosaccharide (LOS) is used to refer to a low-molecular-weight bacterial lipopolysaccharide. Endotoxin (lipid A), the hydrophobic anchor of lipopolysaccharide (LPS), is a glucosamine-based phospholipid that is present in the outer membranes of most Gram-negative bacteria. Gut microbiota-derived endotoxin has been linked to human disease, including GI tract inflammation in Parkinson’s disease [160], nonalcoholic fatty liver disease (NAFLD) [161], and preeclampsia [151]; it is also linked to neurotoxicity [162]. Systemic exposure to bacterial endotoxin can be detected by measuring plasma LPS binding protein (LBP).
6. Microbiome Signatures as Disease Biomarkers
The microbial abundance and compositional patterns identified from metagenomic analyses can be used as disease biomarkers. However, the search for such signatures in human cohorts has been confounded by environmental factors, host factors, disease status, and the presence of other comorbidities [108]. Gut microbiome signatures are used as biomarkers for many disease conditions, including central nervous system [43], inflammatory [163], and metabolic disorders [163]. Therefore, these signatures remain as important aspects of the human microbiome regarding the identification of diagnostic biomarkers for human disease.
7. Microbiome Multi-Omics
Sequence-based methods relying on 16S ribosomal RNA (rRNA) amplicon sequencing, while very important in identifying microbiome-based biomarkers, provide very limited information on the functional relationships within microbial communities, or between the microbiota and the human host. Therefore, researchers are increasingly combining 16S rRNA analyses with the more costly shotgun metagenomics to obtain functional insight. Shotgun metagenomics allows researchers to comprehensively sample all genes in all organisms present in a given biological sample [164]. Additionally, metagenomic data can be complemented by RNA sequencing, which creates metatranscriptomic profiles for microbial communities that can be used to determine the metaproteomic and metametabolomic profiles of constituent microbial communities. This allows the validation of metagenomic findings by elucidating the mechanisms that link microbial metabolism with various diseases. Thus, metabolomics can be used to examine the crosstalk between the microbiome and the host through metabolites. The level of correlation between taxa at different taxonomic levels and metabolites has been described in the biomedical literature [112]. Additionally, certain bacterial proteins and enzymes, such as nucleases, have shown promise as diagnostic tools and treatments [165]. For example, Serratia marcescens nuclease (EC 3.1.30.2) has therapeutic value for the treatment of respiratory diseases, resulting in sputum production due to its ability to hydrolyze sputum DNA effectively [165].
8. Association Predictions of Microbiome and Other Omics Data
Multi-omics is a promising approach to predict the diagnosis, prognosis, and treatment efficiency of diseases. Genes, RNA, proteins, metabolites, microbes, and pathways, as well as pathological and medical imaging data, can all be integrated and analyzed comprehensively by means of network analysis to come up with a unified and potentially more accurate hypothesis about the disease in question [166]. Such networks enable the exploration of the relationships between biological entities to determine their function and relevance to the disease. The fusion of multimodal data for cancer diagnosis is considered a feasible research framework for radiomics and genomics [167]. Recently, some clinical trials have used diverse approaches to define characteristics of the patients who develop primary or acquired resistance to immunotherapy (e.g., NCT04243720) [168]. Such trials are aiming to develop an integrated model to predict drug resistance relying on multimodal data including radiomics, genomics, transcriptomics, epigenetics, immunophenotypic data, and fecal microbiome data [80]. There is promise that artificial intelligence models combining microbiome-based biomarkers with other omics data (e.g., radiomics) will be able to provide a more comprehensive view of the tumor microenvironment, aiding in better cancer diagnosis and allowing clinicians to non-invasively track changes in cancer phenotypes [80].
9. Diseases which Can Be Probed Using Microbiome-Based Biomarkers
Changes in the normal microbiota have been linked with different diseases such as cancer, inflammatory bowel disease, neuro-disease, cardiovascular disease, systemic infections, allergic diseases, and others. Table 2 summarizes diseases that can exploit the microbiome for diagnostic biomarkers. Important major condition groups are discussed thoroughly in the following sections. These major groups include cancer, central nervous system diseases, inflammatory bowel diseases, cardiovascular diseases, allergic diseases, and systemic infections.

Table 2.
Diseases that can exploit the human microbiome and microbiota metabolites as diagnostic biomarkers.
9.1. Cancer
Studies showed that microbiomes perform biochemical reactions affecting cancer prognosis and proliferation, as well as immunotherapy reactions [16,24]. Recurrent GI tract infections and antimicrobial drugs are linked to dysbiosis and colorectal cancer [24]. The metabolites of gut microbiota affect the intestinal lining, inducing or inhibiting carcinogenesis [33,102]. Gut microbiota contribute in colorectal cancer and hepatocellular carcinoma [33,102]. Additionally, Clostridium, Fusobacterium, and H. pylori contribute in gastric cancer [33]. Studies showed that E. coli induce lung cancer cell movement, adherence, and metastasis through Toll-like receptor 4 (TLR4) signaling via suppressing TLR4 (Eritoran), p38 mitogen-activated protein kinases (MAPK), and extracellular signal-regulated kinase (ERK1/2) phosphorylation [266]. Females with breast cancer, as opposed to healthy females, showed Staphylococcus, Enterobacteriaceae, and Bacillus in breast tissues [267]. Lactobacillus species were absent in the breast cells of breast cancer females. Moreover, Escherichia coli and Staphylococcus epidermidis were detected in cervical cancer [267]. Prostate cancer patients showed higher frequencies of Bacteroides massiliensis [61,62].
Studies showed that Streptococcus and Veillonella infections in airway epithelial cells are modulated through phosphoinositide 3-kinase (PI3K) and extracellular signal-regulated kinase (ERK) signaling cascades [268]. Further studies revealed that Acidovorax, Comamonas, Klebsiella, Rhodoferax, and Polarmonas are linked to lung squamous cell carcinoma (LUSC), having tumor protein p53 (TP53) mutations [269]. Furthermore, studies reported that the pulmonary microbiome mediates lung cancer prognosis by inducing myeloid-cell-dependent interleukin (IL) (IL-1β and IL-23) and activating lung-resident T cells (Vγ6 + Vδ1 + γδ T cells) [270]. Researches showed that smoking mediates Acidimicrobiales norank, Caulobacteraceae, and Enterobacter spp. infection [271,272] in the lower respiratory tract, altering the respiratory and immunity response mechanisms, such as the dendritic cells (DCs), natural killer (NK) cells, macrophages, immunological memory (T and B) lymphocytes, CD8+, CD4+, and CD25+ Tregs. Studies showed that cigarette smoking incites pulmonary cell membrane damage, facilitating cancer proliferation and bacterial transfer to lung cancer [273]. Epidemiological studies recorded that Chlamydia pneumoniae, tuberculosis (TB), mycoplasma, and pneumococcal infection increase the risk of lung cancer [274,275,276,277,278].
Studies declared that bacterial, fungal, and viral infections are risk determinants for cancer prognosis. Particularly, 15% of cancer cases evolved by oncogenic organism infection [279], and other cases emerged by co-infection with diverse pathogens that promote the risk of cancer development [279]. Therefore, the investigation of infection-mediated cancer is necessary to impede cancer prognosis and enhance treatment protocols.
Studies showed that hepatitis B (HBV) and C viruses (HCV), (5%), human papilloma viruses (HPV) (5%), Helicobacter pylori (5%), Epstein–Barr virus (EBV) (1%), human immunodeficiency virus (HIV) (1%), human herpes virus (HSV) (1%), helminth (Schistosoma haematobium), and fungi (Aspergillus spp.) are implicated in cancer development [60,67,68,280,281]. Viruses-mediated cancer can be contracted in uterus, during adulthood, or during childhood; however, these viruses have long incubation periods prior to cancer induction. Moreover, the liability to infectious diseases is excessive in cancer patients [64].
Furthermore, cancer treatment can alter the host microbiome because of immunocompromising activity, thus enhancing infection liability and consequently, cancer prognosis [64]. Eventually, cancer microbiota has been linked to chemotherapy resistance [64]. Studies reported that the gut microbiome can affect the effectiveness of anticancer treatment, such as oxaliplatin [71].The intestinal microbiome recruits the myeloid cells for generating high concentrations of reactive oxygen species (ROS). The high levels of ROS induce oxaliplatin-accompanied DNA deterioration and consequently, promote cancer cell death [71].
Alternatively, cyclophosphamide, an alkylating chemotherapeutic agent, damages the epithelium of the small intestine and subsequently modulates anticancer activity [282]. Biological studies showed that the efficacy of 5-fluorouracil (5-FU) decreases in cells invaded by Mycoplasma hyorhinis due to bacterial thymidine phosphorylase that transforms anticancer drugs [283]. Further studies revealed that bacteria can deactivate gemcitabine due to bacterial cytidine deaminase [283]. Studies showed that the detection of Faecalibacterium spp. in gut the microbiome of melanoma patients was accompanied by the anti-programmed cell death 1 (PD-1)/PD-L1 response, whereas Anaerotruncus colihominis, Bacteroides thetaiotaomicron, and Escherichia coli were accompanied by the absence of such a response [284].
9.2. Irritable Bowel Syndrome and Inflammatory Bowel Disease (IBD) (Crohn’s Disease and Ulcerative Colitis)
Evidence suggests the existence of various pathogenic factors contributing to irritable bowel syndrome and inflammatory bowel diseases, including genetic predisposition, chronic low-grade intestinal inflammation, personality traits, and microbiome alterations [285]. Studies focusing on microbiome alternations indicated that microbiota dysbiosis invoked irregular immune reactions against body cells and tissues, resulting in autoimmune, GI tract inflammatory, and other threatening diseases [97]. A steady beneficial relationship is established between the human microbiota and the immune system. A disturbance in this relationship weakens the host’s immunity, resulting in an abnormal inflammatory response [93], such as an inflammatory bowel disease (IBD) [89]. A decrease in GI tract Firmicutes increases proinflammatory cytokines (IL12, IFN-γ) and decreases anti-inflammatory cytokine (IL-10) [89]. Reported studies demonstrated that helminth infections accompany anti-inflammatory microbes that impede IBD progress in mice models [280].
9.3. Cardiovascular Diseases
The gut microbes produce trimethylamine N-oxide (TMAO) metabolites that might contribute to cardiac disease [31,281]. Diets containing phosphatidylcholine, choline, and l-carnitine are transformed by hepatic monooxygenase to trimethylamine, which consequently metabolizes to trimethylamine N-oxide (TMAO). TMAO disrupts lipid transportation and invokes precursor production that induces atherosclerosis and artery thickening [31,281]. Clinical studies showed that disturbances in intestinal microbes are observed in cardiovascular disease patients [18,103]. Further studies showed that hypertensive patients have higher levels of Prevotella and Klebsiella in the stool. Additionally, hypertensive mice demonstrated a substantial increase in Firmicutes to Bacteriodetes ratio in the stool [18,103].
9.4. Systematic Infections
The translocation of microbes increases the probability of systemic disease in immune-deficient patients [103]. The translocated microbes generate uremic toxins, activating the inflammatory response and inducing diseases [103,286]. The misuse of antibiotics and impairment of gut epithelium induce the proliferation of anaerobic microbes and weaken the immune response [103,286]. A disturbance in GI tract microbes promotes the synthesis of nitrogenous compounds that affects the epithelial structure and in turn, facilitates the movement of microbiota and their toxins to other locations in the body [286]. Clinical studies showed that hemodialysis patients have translocated gut microbiota, implying a relationship between kidney disease and gut microbiota [31,281].
9.5. Allergic Diseases
The mucosal lining of the respiratory system is affected by gut microbiota. A disruption in the gut microbiome has an impact on the lung microbiome through microaspiration, which increases the risk of respiratory diseases [44]. Clinical studies reported that neonatal Caesarean delivery potentiates allergic diseases due to lack of maternal flora [94]. Further biological studies showed that Caesarean-delivered children have low levels of Bacteriodetes, or normal flora, in the GI tract [18]. Studies declared that a decrease in Bacteriodetes anti-inflammatory activity is accompanied by asthma and rhinitis [97]. Studies recorded that there is a substantial connection between the disruption of the microbiome and allergic antigens (IGE) [287]. Studies showed that children with lower levels of Akkermansia, Bifidobacterium, and Faecalibacterium are sensitive to numerous respiratory allergies that might lead to asthma at around 4 years of age [288]. Studies revealed that residing in a farming environment, with a variety of microbial consortium, is accompanied with a lower rate of respiratory allergies [289,290]. Earlier evidence showed that growing mice in a “farm dust” environment with a diverse bacteria community weakens the respiratory allergic response [46]. Further proof declared that exposing mice to Acinetobacter lwoffii F78 and Lactococcus lactis G121 shows a protective effect against respiratory inflammation [291].
10. The Clinical Implications of Using Microbiome-Based Biomarkers
The development of microbiome-based diagnostic biomarkers is considered one of the key aspects of precision medicine [17,292]. A large body of evidence highlighted an important role of human microbiota in modulating health and disease, through many immune and non-immune mechanisms, via changes in RNA, DNA, and metabolite networks. For example, inflammation, genotoxicity, and metabolism are fundamental mechanisms to modulate carcinogenesis by microbiota, and can therefore be exploited to develop personalized anticancer therapies [17].
However, most of the currently-available evidence linking the human microbiome to cancer and other non-inflammatory bowel diseases is considered preliminary or limited [293,294]. Therefore, there is a great need for more in vitro and in vivo confirmatory tests to prioritize reliable microbiome-based diagnostic biomarkers, drug targets, or personalized treatments [17]. The identification of validated predictive microbiome-based biomarkers could revolutionize the field of precision medicine by guiding clinical decision making about disease diagnosis and proper personalized treatments.
11. Challenges and Future Direction
The development of valid clinical biomarkers and the generation of curated microbial genetic databases are becoming essential tools for disease diagnosis and pharmacotherapy monitoring. Additionally, understanding the regulatory, microbial contamination, and safety protocols regarding microbiome bench work is important to speed up the translation of basic research into clinical interventions [295]. Many challenges in microbiome research are related to method standardization concerning biological variation [296], diet [297], complex chemical backbones [298], access to in vivo sampling locations [299], time intervals [300], collaboration of financial and human resources [301], biodiversity and clinical aspects [302], and interactions with body tissues [303]. Additionally, the determination of microbial features associated with health versus disease requires improved microbiome profiling technologies, with strain-level resolution which is still unattainable [10].
Another challenge is the difficulty in establishing the clinical importance of inter-individual differences in the gut microbiome, since some parts of the microbiome (e.g., uncultured bacteria, viruses, phage, fungi, and archaea) are poorly characterized, invoking scientists to refer to them collectively as microbial “dark matter.” Al Bataineh et al. [5] has explored the intestinal fungal dark matter, and found evidence of microbiota involvement in host metabolism and aging pathways.
Microbiome research requires novel strategies for the standardization and mechanistic validation of the identified microbial gene clusters. Integrated multi-omics methods— combined with cataloging bacterial isolates, profiling metabolites, and measuring host responses—have permitted the correlation of bacteria and bacterial metabolites with numerous diseases. However, we are now faced with new challenges concerning the causal relationships of the human microbiome in context with normal physiology and disease pathways [304,305,306]. Revolutionary research is needed to understand the causal relationships between human microbiota and human disease by understanding the underlying mechanisms through which microbes affect human health. Such understanding would advance microbiome research beyond biomarker validation to identify therapeutic drug targets. In fact, bacterial metabolites can provide colossal mechanistic insight that may accelerate the development of new therapeutic strategies for various diseases, such as the management of impaired glucose metabolism in diabetes [307]. Furthermore, the inclusion of detailed host demographical data, such as age, sex, ethnicity, geography, dietary habits, exercise, and other factors, could help in the identification of personalized diagnostic biomarkers.
12. Conclusions
The human microbiota will pave the road for a new era in biomarker research for disease diagnosis and pharmacotherapy monitoring. This will ultimately revolutionize the field of precision medicine and individualized treatments. However, more collaborative work is needed to develop robust, comprehensive, and open-source databases powered by novel methodologies that allow researchers across the world to upload, explore, visualize, and interpret their data, and also standardize their methods to be able to compare their results with those of other research groups.
Author Contributions
Idea and conceptualization, R.H.; literature review, R.H., D.A.S. and A.Q.A.B.; writing—original draft preparation, R.H., D.A.S. and A.Q.A.B.; writing—review and editing, R.H. and D.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
R.H. and A.A. acknowledge support from the Deanship of Scientific Research at Al-Zaytoonah University of Jordan (Grant number 2019–2020/23/07). R.H. and D.A.S acknowledge support from the Deanship of Scientific Research at Al-Zaytoonah University of Jordan (Grant number 2020-2019/17/03).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors acknowledge funding from the Deanship of Scientific Research at Al-Zaytoonah University of Jordan.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 5-FU | 5-Fluorouracil |
| BCAAs | Branched-Chain Amino Acids |
| CD4+ | Cluster of Differentiation 4 Positive |
| CD8+ | Cluster of Differentiation 8 Positive |
| DCs | Dendritic Cells |
| CSF | Cerebrospinal Fluid |
| EBV | Epstein–Barr Virus |
| ERK | Extracellular Signal-Regulated Kinase |
| GI | Gastrointestinal |
| HBV | Hepatitis B Virus |
| HCV | Hepatitis C Virus |
| HIV | Human Immunodeficiency Virus |
| HMDB | Human Metabolome Database |
| HPV | Human Papilloma Virus |
| HSV | Herpes Simplex Virus |
| IBS | Irritable Bowel Syndrome |
| IAld | Indole-3-aldehyde |
| IGE | Immunoglobulin E |
| IL | Interleukin |
| ImP | Imidazole Propionate |
| IPA | Indole-3-propionic Acid |
| LBP | LPS Binding Protein |
| LOS | Lipooligosaccharides |
| LPS | Lipopolysaccharides |
| LUSC | Lung Squamous Cell Carcinoma |
| MAPK | Mitogen-Activated Protein Kinases |
| NAFLD | Nonalcoholic Fatty Liver Disease |
| NK | Natural Killer |
| OTU | Operational Taxonomic Unit |
| PCR | Polymerase Chain Reaction |
| PD-1 | Programmed Cell Death 1 |
| PI3K | Phosphoinositide 3-Kinase |
| RBC | Red Blood Cells |
| ROS | Reactive Oxygen Species |
| rRNA | Ribosomal RNA |
| SCFAs | Short-Chain Fatty Acids |
| TB | Tuberculosis |
| TLR | Toll-Like Receptor |
| TLR4 | Toll-Like Receptor 4 |
| TMAO | Trimethylamine N-Oxide |
| TNF | Tumor Necrosis Factor |
| TP53 | Tumor Protein p53 |
| Tregs | Regulatory T-Cells |
| WBC | White Blood Cells |
References
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Whiteside, S.A.; Razvi, H.; Dave, S.; Reid, G.; Burton, J.P. The microbiome of the urinary tract—A role beyond infection. Nat. Rev. Urol. 2015, 12, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Al Bataineh, M.T.; Alzaatreh, A.; Hajjo, R.; Banimfreg, B.H.; Dash, N.R. Compositional changes in human gut microbiota reveal a putative role of intestinal mycobiota in metabolic and biological decline during aging. Nutr. Healthy Aging 2021, 6, 269–283. [Google Scholar] [CrossRef]
- Al-Zyoud, W.; Hajjo, R.; Abu-Siniyeh, A.; Hajjaj, S. Salivary microbiome and cigarette smoking: A first of its kind investigation in Jordan. Int. J. Environ. Res. Public Health 2020, 17, 256. [Google Scholar] [CrossRef]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef]
- Tarawneh, O.; Al-Ass, A.R.; Hamed, R.; Sunoqrot, S.; Hasan, L.; Al-Sheikh, I.; Al-Qirim, R.; Alhusban, A.A.; Naser, W. Development and characterization of k-carrageenan platforms as periodontal intra-pocket films. Trop. J. Pharm. Res. 2019, 18, 1791–1798. [Google Scholar] [CrossRef]
- Tarawneh, O.; Hamadneh, I.; Huwaitat, R.; Al-Assi, A.R.; El Madani, A. Characterization of chlorhexidine-impregnated cellulose-based hydrogel films intended for the treatment of periodontitis. Biomed Res. Int. 2021, 2021, 9853977. [Google Scholar] [CrossRef]
- Shanahan, F.; Ghosh, T.S.; O’Toole, P.W. The Healthy Microbiome-What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Xiao, X.; Hu, M.; Zhang, X. Interaction between gut microbiome and cardiovascular disease. Life Sci. 2018, 214, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, T.M.; Gilbert, J.A. Introducing the Microbiome into Precision Medicine. Trends Pharmacol. Sci. 2017, 38, 81–91. [Google Scholar] [CrossRef]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef]
- Behrouzi, A.; Nafari, A.H.; Siadat, S.D. The significance of microbiome in personalized medicine. J. Transl. Med. 2019, 8, 16. [Google Scholar] [CrossRef]
- Kho, Z.Y.; Lal, S.K. The human gut microbiome–a potential controller of wellness and disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef]
- Liu, Z.; de Vries, B.; Gerritsen, J.; Smidt, H.; Zoetendal, E.G. Microbiome-based stratification to guide dietary interventions to improve human health. Nutr. Res. 2020, 82, 1–10. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Guthrie, L.; Kelly, L. Bringing microbiome-drug interaction research into the clinic. EBioMedicine 2019, 44, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Zangara, M.T.; McDonald, C. How diet and the microbiome shape health or contribute to disease: A mini-review of current models and clinical studies. Exp. Biol. Med. 2019, 244, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: A major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2016, 2, 16003. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Elmassry, M.M.; Zayed, A.; Farag, M.A. Gut homeostasis and microbiota under attack: Impact of the different types of food contaminants on gut health. Crit. Rev. Food Sci. Nutr. 2022, 62, 738–763. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Du, W.-T.; Xu, Y.-L.; Cheng, S.-Z.; Liu, Z.-J. Gut microbiome-based medical methodologies for early-stage disease prevention. Microb. Pathog. 2017, 105, 122–130. [Google Scholar] [CrossRef]
- Melli, L.C.; do Carmo-Rodrigues, M.S.; Araújo-Filho, H.B.; Solé, D.; de Morais, M.B. Intestinal microbiota and allergic diseases: A systematic review. Allergol. Et Immunopathol. 2016, 44, 177–188. [Google Scholar] [CrossRef]
- Damhorst, G.L.; Adelman, M.W.; Woodworth, M.H.; Kraft, C.S. Current Capabilities of Gut Microbiome–Based Diagnostics and the Promise of Clinical Application. J. Infect. Dis. 2021, 223, S270–S275. [Google Scholar] [CrossRef]
- Boertien, J.M.; Pereira, P.A.; Aho, V.T.; Scheperjans, F. Increasing comparability and utility of gut microbiome studies in Parkinson’s disease: A systematic review. J. Parkinsons Dis. 2019, 9, S297–S312. [Google Scholar] [CrossRef]
- Wright, E.K.; Kamm, M.A.; Teo, S.M.; Inouye, M.; Wagner, J.; Kirkwood, C.D. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: A systematic review. Inflamm. Bowel Dis. 2015, 21, 1219–1228. [Google Scholar]
- Tang, W.W.; Wang, Z.; Kennedy, D.J.; Wu, Y.; Buffa, J.A.; Agatisa-Boyle, B.; Li, X.S.; Levison, B.S.; Hazen, S.L. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Cénit, M.; Matzaraki, V.; Tigchelaar, E.; Zhernakova, A. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Hill, C. Bacteriophages of the human gut: The “known unknown” of the microbiome. Cell Host Microbe 2019, 25, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef]
- Gomez, A.; Nelson, K.E. The oral microbiome of children: Development, disease, and implications beyond oral health. Microb. Ecol. 2017, 73, 492–503. [Google Scholar] [CrossRef]
- Frame, L.A.; Costa, E.; Jackson, S.A. Current explorations of nutrition and the gut microbiome: A comprehensive evaluation of the review literature. Nutr. Rev. 2020, 78, 798–812. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef]
- Aragon, I.M.; Herrera-Imbroda, B.; Queipo-Ortuño, M.I.; Castillo, E.; Del Moral, J.S.-G.; Gomez-Millan, J.; Yucel, G.; Lara, M.F. The urinary tract microbiome in health and disease. Eur. Urol. Focus 2018, 4, 128–138. [Google Scholar] [CrossRef]
- Pascal, M.; Perez-Gordo, M.; Caballero, T.; Escribese, M.M.; Lopez Longo, M.N.; Luengo, O.; Manso, L.; Matheu, V.; Seoane, E.; Zamorano, M. Microbiome and allergic diseases. Front. immunol. 2018, 9, 1584. [Google Scholar] [CrossRef]
- Lesko, L.J.; Atkinson Jr, A. Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: Criteria, validation, strategies. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 347–366. [Google Scholar] [CrossRef]
- Vostal, J.G.; Buehler, P.W.; Gelderman, M.P.; Alayash, A.I.; Doctor, A.; Zimring, J.C.; Glynn, S.A.; Hess, J.R.; Klein, H.; Acker, J.P. Proceedings of the Food and Drug Administration’s public workshop on new red blood cell product regulatory science 2016. Transfusion 2018, 58, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Hajjo, R.; Sabbah, D.A.; Bardaweel, S.K.; Tropsha, A. Identification of Tumor-Specific MRI Biomarkers Using Machine Learning (ML). Diagnostics 2021, 11, 742. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Gong, T.; Zhang, J.; Gu, Q.; Gao, X.; Weng, X.; Liu, J.; Sun, J. Gut microbiome signatures are biomarkers for cognitive impairment in patients with ischemic stroke. Front. Aging Neurosci. 2020, 12, 297. [Google Scholar] [CrossRef] [PubMed]
- Renz, H.; Brandtzaeg, P.; Hornef, M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2011, 12, 9–23. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012, 30, 759–795. [Google Scholar] [CrossRef]
- Schuijs, M.J.; Willart, M.A.; Vergote, K.; Gras, D.; Deswarte, K.; Ege, M.J.; Madeira, F.B.; Beyaert, R.; van Loo, G.; Bracher, F.; et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 2015, 349, 1106–1110. [Google Scholar] [CrossRef]
- Hofman, P.; Vouret-Craviari, V. Microbes-induced EMT at the crossroad of inflammation and cancer. Gut Microbes 2012, 3, 176–185. [Google Scholar] [CrossRef]
- Miller, E.T.; Svanbäck, R.; Bohannan, B.J. Microbiomes as metacommunities: Understanding host-associated microbes through metacommunity ecology. Trends Ecol. Evol. 2018, 33, 926–935. [Google Scholar] [CrossRef]
- Adami, A.J.; Cervantes, J.L. The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberculosis 2015, 95, 651–658. [Google Scholar] [CrossRef]
- Bradlow, H.L. Obesity and the gut microbiome: Pathophysiological aspects. Horm. Mol. Biol. Clin. Investig. 2014, 17, 53–61. [Google Scholar] [CrossRef]
- Drago, F.; Gariazzo, L.; Cioni, M.; Trave, I.; Parodi, A. The microbiome and its relevance in complex wounds. Eur. J. Dermatol. 2019, 29, 6–13. [Google Scholar] [PubMed]
- Salaspuro, M.P. Acetaldehyde, microbes, and cancer of the digestive tract. Crit. Rev. Clin. Lab. Sci. 2003, 40, 183–208. [Google Scholar] [CrossRef]
- Khan, A.A.; Shrivastava, A.; Khurshid, M. Normal to cancer microbiome transformation and its implication in cancer diagnosis. Biochim. Biophys. Acta 2012, 1826, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef]
- Hullar, M.A.; Burnett-Hartman, A.N.; Lampe, J.W. Gut microbes, diet, and cancer. In Advances in Nutrition and Cancer; Springer: New York, NY, USA, 2014; pp. 377–399. [Google Scholar]
- Whisner, C.M.; Aktipis, C.A. The role of the microbiome in cancer initiation and progression: How microbes and cancer cells utilize excess energy and promote one another’s growth. Curr. Nutr. Rep. 2019, 8, 42–51. [Google Scholar] [CrossRef]
- Half, E.; Keren, N.; Reshef, L.; Dorfman, T.; Lachter, I.; Kluger, Y.; Reshef, N.; Knobler, H.; Maor, Y.; Stein, A. Fecal microbiome signatures of pancreatic cancer patients. Sci. Rep. 2019, 9, 16801. [Google Scholar] [CrossRef]
- Curty, G.; de Carvalho, P.S.; Soares, M.A. The role of the Cervicovaginal microbiome on the genesis and as a biomarker of premalignant cervical intraepithelial neoplasia and invasive cervical Cancer. Int. J. Mol. Sci. 2020, 21, 222. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef]
- Puhr, M.; De Marzo, A.; Isaacs, W.; Lucia, M.S.; Sfanos, K.; Yegnasubramanian, S.; Culig, Z. Inflammation, microbiota, and prostate cancer. Eur. Urol. Focus 2016, 2, 374–382. [Google Scholar] [CrossRef]
- Massari, F.; Mollica, V.; Di Nunno, V.; Gatto, L.; Santoni, M.; Scarpelli, M.; Cimadamore, A.; Lopez-Beltran, A.; Cheng, L.; Battelli, N. The human microbiota and prostate cancer: Friend or foe? Cancers 2019, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Celardo, I.; Melino, G.; Amelio, I. Commensal microbes and p53 in cancer progression. Biol. Direct 2020, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, M.M.; Pina-Vaz, C.; Baltazar, F. Microbes and Cancer: Friends or Faux? Int. J. Mol. Sci. 2020, 21, 3115. [Google Scholar] [CrossRef] [PubMed]
- Lérias, J.R.; Paraschoudi, G.; de Sousa, E.; Martins, J.; Condeço, C.; Figueiredo, N.; Carvalho, C.; Dodoo, E.; Castillo-Martin, M.; Beltrán, A. Microbes as master immunomodulators: Immunopathology, cancer and personalized immunotherapies. Front. Cell Dev. Biol. 2020, 7, 362. [Google Scholar] [CrossRef]
- Sawant, S.S.; Patil, S.M.; Gupta, V.; Kunda, N.K. Microbes as medicines: Harnessing the power of bacteria in advancing cancer treatment. Int. J. Mol. Sci. 2020, 21, 7575. [Google Scholar] [CrossRef]
- Guo, H.; Zhao, L.; Zhu, J.; Chen, P.; Wang, H.; Jiang, M.; Liu, X.; Sun, H.; Zhao, W.; Zheng, Z.; et al. Microbes in lung cancer initiation, treatment, and outcome: Boon or bane? Semin. Cancer Biol. 2021. [Google Scholar] [CrossRef]
- Ammer-Herrmenau, C.; Pfisterer, N.; Weingarten, M.F.; Neesse, A. The microbiome in pancreatic diseases: Recent advances and future perspectives. United Eur. Gastroenterol. J. 2020, 8, 878–885. [Google Scholar] [CrossRef]
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, e609–e616. [Google Scholar] [CrossRef]
- Temraz, S.; Nassar, F.; Nasr, R.; Charafeddine, M.; Mukherji, D.; Shamseddine, A. Gut microbiome: A promising biomarker for immunotherapy in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 4155. [Google Scholar] [CrossRef]
- Jarman, R.; Ribeiro-Milograna, S.; Kalle, W. Potential of the microbiome as a biomarker for early diagnosis and prognosis of breast cancer. J. Breast Cancer 2020, 23, 579. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Shahanavaj, K.; Gil-Bazo, I.; Castiglia, M.; Bronte, G.; Passiglia, F.; Carreca, A.P.; del Pozo, J.L.; Russo, A.; Peeters, M.; Rolfo, C. Cancer and the microbiome: Potential applications as new tumor biomarker. Expert Rev. Anticancer Ther. 2015, 15, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Totsika, M.; Morrison, M.; Punyadeera, C. Oral microbiome: A new biomarker reservoir for oral and oropharyngeal cancers. Theranostics 2017, 7, 4313. [Google Scholar] [CrossRef]
- Rüb, A.M.; Tsakmaklis, A.; Gräfe, S.K.; Simon, M.-C.; Vehreschild, M.J.G.T.; Wuethrich, I. Biomarkers of human gut microbiota diversity and dysbiosis. Biomark. Med. 2021, 15, 139–150. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Microbiome-based biomarkers for IBD. Inflamm. Bowel Dis. 2020, 26, 1463–1469. [Google Scholar] [CrossRef]
- Lin, H.; He, Q.-Y.; Shi, L.; Sleeman, M.; Baker, M.S.; Nice, E.C. Proteomics and the microbiome: Pitfalls and potential. Expert Rev. Proteomics 2019, 16, 501–511. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K. Applications of machine learning in human microbiome studies: A review on feature selection, biomarker identification, disease prediction and treatment. Front. Microbiol. 2021, 12, 313. [Google Scholar] [CrossRef]
- Kang, C.Y.; Duarte, S.E.; Kim, H.S.; Kim, E.; Park, J.; Lee, A.D.; Kim, Y.; Kim, L.; Cho, S.; Oh, Y.; et al. Artificial Intelligence-based Radiomics in the Era of Immuno-oncology. Oncologist 2022, 27, e471–e483. [Google Scholar] [CrossRef]
- Zhou, Z.; Ge, S.; Li, Y.; Ma, W.; Liu, Y.; Hu, S.; Zhang, R.; Ma, Y.; Du, K.; Syed, A.; et al. Human Gut Microbiome-Based Knowledgebase as a Biomarker Screening Tool to Improve the Predicted Probability for Colorectal Cancer. Front. Microbiol. 2020, 11, 596027. [Google Scholar] [CrossRef]
- Marech, I.; Ammendola, M.; Gadaleta, C.; Zizzo, N.; Oakley, C.; Gadaleta, C.D.; Ranieri, G. Possible biological and translational significance of mast cells density in colorectal cancer. World J. Gastroenterol. 2014, 20, 8910–8920. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Feng, Q.; Zheng, P.; Yang, L.; Zhu, D.; Chang, W.; Ji, M.; He, G.; Xu, J. Low tumor infiltrating mast cell density confers prognostic benefit and reflects immunoactivation in colorectal cancer. Int. J. Cancer 2018, 143, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, G.; Gallo, G.; Vescio, G.; Picciariello, A.; De Paola, G.; Trompetto, M.; Currò, G.; Ammendola, M. Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. J. Clin. Med. 2020, 9, 2852. [Google Scholar] [CrossRef] [PubMed]
- Groll, T.; Silva, M.; Sarker, R.S.J.; Tschurtschenthaler, M.; Schnalzger, T.; Mogler, C.; Denk, D.; Schölch, S.; Schraml, B.U.; Ruland, J.; et al. Comparative Study of the Role of Interepithelial Mucosal Mast Cells in the Context of Intestinal Adenoma-Carcinoma Progression. Cancers 2022, 14, 2248. [Google Scholar] [CrossRef]
- Pellino, G.; Gallo, G.; Pallante, P.; Capasso, R.; De Stefano, A.; Maretto, I.; Malapelle, U.; Qiu, S.; Nikolaou, S.; Barina, A.; et al. Noninvasive Biomarkers of Colorectal Cancer: Role in Diagnosis and Personalised Treatment Perspectives. Gastroenterol. Res. Pract. 2018, 2018, 2397863. [Google Scholar] [CrossRef]
- Ghareib, A.F.; Mohamed, R.H.; Abd El-Fatah, A.R.; Saadawy, S.F. Assessment of Serum MicroRNA-21 Gene Expression for Diagnosis and Prognosis of Colorectal Cancer. J. Gastrointest. Cancer 2020, 51, 818–823. [Google Scholar] [CrossRef]
- Tarallo, S.; Ferrero, G.; Gallo, G.; Francavilla, A.; Clerico, G.; Realis Luc, A.; Manghi, P.; Thomas, A.M.; Vineis, P.; Segata, N.; et al. Altered Fecal Small RNA Profiles in Colorectal Cancer Reflect Gut Microbiome Composition in Stool Samples. mSystems 2019, 4, e00289-19. [Google Scholar] [CrossRef]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K. The host microbiome regulates and maintains human health: A primer and perspective for non-microbiologists. Cancer Res. 2017, 77, 1783–1812. [Google Scholar] [CrossRef]
- Thomas-White, K.; Brady, M.; Wolfe, A.J.; Mueller, E.R. The bladder is not sterile: History and current discoveries on the urinary microbiome. Curr. Bladder Dysfunct. Rep. 2016, 11, 18–24. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, G.; Manwani, D.; Mortha, A.; Xu, C.; Faith, J.J.; Burk, R.D.; Kunisaki, Y.; Jang, J.E.; Scheiermann, C.; et al. Neutrophil ageing is regulated by the microbiome. Nature 2015, 525, 528–532. [Google Scholar] [CrossRef]
- Xu, C.; Lee, S.K.; Zhang, D.; Frenette, P.S. The Gut Microbiome Regulates Psychological-Stress-Induced Inflammation. Immunity 2020, 53, 417–428 e414. [Google Scholar] [CrossRef] [PubMed]
- Rojo, D.; Méndez-García, C.; Raczkowska, B.A.; Bargiela, R.; Moya, A.; Ferrer, M.; Barbas, C. Exploring the human microbiome from multiple perspectives: Factors altering its composition and function. FEMS Microbiol. Rev. 2017, 41, 453–478. [Google Scholar] [CrossRef] [PubMed]
- Cingi, C.; Muluk, N.B.; Scadding, G.K. Will every child have allergic rhinitis soon? Int. J. Pediatr. Otorhinolaryngol. 2019, 118, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef]
- Caussy, C.; Loomba, R. Gut microbiome, microbial metabolites and the development of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 719–720. [Google Scholar] [CrossRef]
- Ipci, K.; Altıntoprak, N.; Muluk, N.B.; Senturk, M.; Cingi, C. The possible mechanisms of the human microbiome in allergic diseases. Eur. Arch. Otorhinolaryngol. 2017, 274, 617–626. [Google Scholar] [CrossRef]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Saraswati, S.; Sitaraman, R. Aging and the human gut microbiota—from correlation to causality. Front. Microbiol. 2015, 5, 764. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Waters, J.L.; Clark, A.G.; Ley, R.E. Cross-species comparisons of host genetic associations with the microbiome. Science 2016, 352, 532–535. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef] [PubMed]
- Parfrey, L.W.; Walters, W.A.; Lauber, C.L.; Clemente, J.C.; Berg-Lyons, D.; Teiling, C.; Kodira, C.; Mohiuddin, M.; Brunelle, J.; Driscoll, M. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 2014, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Milani, C.; De Giori, G.S.; Sesma, F.; Van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Steinert, R.E.; Lee, Y.-K.; Sybesma, W. Vitamins for the gut microbiome. Trends Mol. Med. 2020, 26, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, R.D.; Adams, M.D.; White, O.; Clayton, R.A.; Kirkness, E.F.; Kerlavage, A.R.; Bult, C.J.; Tomb, J.-F.; Dougherty, B.A.; Merrick, J.M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995, 269, 496–512. [Google Scholar] [CrossRef]
- Buttó, L.F.; Haller, D. Functional relevance of microbiome signatures: The correlation era requires tools for consolidation. J. Allergy Clin. Immunol. 2017, 139, 1092–1098. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. Res. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Wakita, Y.; Shimomura, Y.; Kitada, Y.; Yamamoto, H.; Ohashi, Y.; Matsumoto, M. Taxonomic classification for microbiome analysis, which correlates well with the metabolite milieu of the gut. BMC microbiol. 2018, 18, 188. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.L.; Chilloux, J.; Sarafian, M.H.; Rahim, M.B.A.; Boulange, C.L.; Dumas, M.-E. The microbiome and its pharmacological targets: Therapeutic avenues in cardiometabolic diseases. Curr. Opin. Pharmacol. 2015, 25, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Goodarzi, M.O. Metabolites Linking the Gut Microbiome with Risk for Type 2 Diabetes. Curr. Nutr. Rep. 2020, 9, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bartok, B.; Oler, E.; Liang, K.Y.; Budinski, Z.; Berjanskii, M.; Guo, A.; Cao, X.; Wilson, M. MarkerDB: An online database of molecular biomarkers. Nucleic Acids Res. 2021, 49, D1259–D1267. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Schönfeld, P.; Wojtczak, L. Short-and medium-chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 277. [Google Scholar] [CrossRef] [PubMed]
- van der Beek, C.M.; Dejong, C.H.; Troost, F.J.; Masclee, A.A.; Lenaerts, K. Role of short-chain fatty acids in colonic inflammation, carcinogenesis, and mucosal protection and healing. Nutr. Rev. 2017, 75, 286–305. [Google Scholar] [CrossRef]
- Nagpal, R.; Shively, C.A.; Register, T.C.; Craft, S.; Yadav, H. Gut microbiome-Mediterranean diet interactions in improving host health. F1000Research 2019, 8, 699. [Google Scholar] [CrossRef] [PubMed]
- Farup, P.G.; Rudi, K.; Hestad, K. Faecal short-chain fatty acids-a diagnostic biomarker for irritable bowel syndrome? BMC Gastroenterol. 2016, 16, 51. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.; Gustavsen, S.; Nguyen, T.D.; Nyman, M.; Langkilde, A.R.; Hansen, T.H.; Sellebjerg, F.; Oturai, A.B.; Bach Søndergaard, H. Serum short-chain fatty acids and associations with inflammation in newly diagnosed patients with multiple sclerosis and healthy controls. Front. Immunol. 2021, 12, 1560. [Google Scholar] [CrossRef]
- Trend, S.; Leffler, J.; Jones, A.P.; Cha, L.; Gorman, S.; Brown, D.A.; Breit, S.N.; Kermode, A.G.; French, M.A.; Ward, N.C. Associations of serum short-chain fatty acids with circulating immune cells and serum biomarkers in patients with multiple sclerosis. Sci. Rep. 2021, 11, 5244. [Google Scholar] [CrossRef]
- Adewiah, S.; Abubakar, A.; Yusuf, F. IDDF2018-ABS-0023 Butyrate acid as a potential marker for the diversity of gut microbiota in colorectal cancer patients. Gut 2018, 67, A34. [Google Scholar] [CrossRef]
- Holeček, M. Branched-chain amino acids in health and disease: Metabolism, alterations in blood plasma, and as supplements. Nutr. Metab. 2018, 15, 33. [Google Scholar] [CrossRef]
- Holeček, M. Side effects of amino acid supplements. Physiol. Res. 2022, 71, 29. [Google Scholar] [CrossRef]
- Batch, B.C.; Hyland, K.; Svetkey, L.P. Branch chain amino acids: Biomarkers of health and disease. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 86–89. [Google Scholar] [CrossRef]
- Giesbertz, P.; Daniel, H. Branched-chain amino acids as biomarkers in diabetes. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Baranyi, A.; Amouzadeh-Ghadikolai, O.; von Lewinski, D.; Rothenhäusler, H.-B.; Theokas, S.; Robier, C.; Mangge, H.; Reicht, G.; Hlade, P.; Meinitzer, A. Branched-chain amino acids as new biomarkers of major depression-a novel neurobiology of mood disorder. PLoS ONE 2016, 11, e0160542. [Google Scholar] [CrossRef] [PubMed]
- Hamaya, R.; Mora, S.; Lawler, P.R.; Cook, N.R.; Ridker, P.M.; Buring, J.E.; Lee, I.-M.; Manson, J.E.; Tobias, D.K. Association of plasma branched-chain amino acid with biomarkers of inflammation and lipid metabolism in women. Circ. Genom. Precis. Med. 2021, 14, e003330. [Google Scholar] [CrossRef] [PubMed]
- Alborghetti, M.R.; Correa, M.E.P.; Whangbo, J.; Shi, X.; Aricetti, J.A.; Silva, A.A.d.; Miranda, E.C.M.; Sforca, M.L.; Caldana, C.; Gerszten, R.E. Clinical metabolomics identifies blood serum branched chain amino acids as potential predictive biomarkers for chronic graft vs. host disease. Front. Oncol. 2019, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Marsh, W. Tryptophan. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: New York, NY, USA, 2007; pp. 1–5. [Google Scholar] [CrossRef]
- Paredes, S.D.; Barriga, C.; Reiter, R.J.; Rodríguez, A.B. Assessment of the Potential Role of Tryptophan as the Precursor of Serotonin and Melatonin for the Aged Sleep-wake Cycle and Immune Function: Streptopelia Risoria as a Model. Int. J. Tryptophan Res. 2009, 2, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Bouma, G.; van Faassen, M.; De Vries, E.; Kema, I.; Walenkamp, A. Niacin (Vitamin B3) Suppletion in Patients with Serotonin Producing Neuroendocrine Tumors. Ann. Oncol. 2014, 25, iv402. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Milk proteins as a source of tryptophan-containing bioactive peptides. Food Funct. 2015, 6, 2115–2127. [Google Scholar] [CrossRef]
- Bertazzo, A.; Ragazzi, E.; Visioli, F. Evolution of tryptophan and its foremost metabolites’ concentrations in milk and fermented dairy products. PharmaNutrition 2016, 4, 62–67. [Google Scholar] [CrossRef]
- Comai, S.; Bertazzo, A.; Bailoni, L.; Zancato, M.; Costa, C.V.; Allegri, G. Protein and non-protein (free and protein-bound) tryptophan in legume seeds. Food Chem. 2007, 103, 657–661. [Google Scholar] [CrossRef]
- Fouad, A.M.; El-Senousey, H.K.; Ruan, D.; Wang, S.; Xia, W.; Zheng, C. Tryptophan in poultry nutrition: Impacts and mechanisms of action. J. Anim. Physiol. Anim. Nutr. 2021, 105, 1146–1153. [Google Scholar] [CrossRef]
- Hoseini, S.M.; Pérez-Jiménez, A.; Costas, B.; Azeredo, R.; Gesto, M. Physiological roles of tryptophan in teleosts: Current knowledge and perspectives for future studies. Rev. Aquac. 2019, 11, 3–24. [Google Scholar] [CrossRef]
- Konopelski, P.; Mogilnicka, I. Biological Effects of Indole-3-Propionic Acid, a Gut Microbiota-Derived Metabolite, and Its Precursor Tryptophan in Mammals’ Health and Disease. Int. J. Mol. Sci. 2022, 23, 1222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.S.; Tan, Q.W.; Guan, L.P. Antioxidant, Anti-inflammatory, Antibacterial, and Analgesic Activities and Mechanisms of Quinolines, Indoles and Related Derivatives. Mini Rev. Med. Chem. 2021, 21, 2261–2275. [Google Scholar] [CrossRef]
- Mandarano, M.; Orecchini, E.; Bellezza, G.; Vannucci, J.; Ludovini, V.; Baglivo, S.; Tofanetti, F.R.; Chiari, R.; Loreti, E.; Puma, F.; et al. Kynurenine/Tryptophan Ratio as a Potential Blood-Based Biomarker in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 4403. [Google Scholar] [CrossRef] [PubMed]
- Oluwagbemigun, K.; Anesi, A.; Ulaszewska, M.; Clarke, G.; Alexy, U.; Schmid, M.; Roden, M.; Herder, C.; Mattivi, F.; Nöthlings, U. Longitudinal relationship of amino acids and indole metabolites with long-term body mass index and cardiometabolic risk markers in young individuals. Sci. Rep. 2020, 10, 6399. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, M.T.; Ramezani, A.; Manal, A.; Raj, D.S. Trimethylamine N-oxide: The good, the bad and the unknown. Toxins 2016, 8, 326. [Google Scholar] [CrossRef]
- Barrea, L.; Annunziata, G.; Muscogiuri, G.; Di Somma, C.; Laudisio, D.; Maisto, M.; De Alteriis, G.; Tenore, G.C.; Colao, A.; Savastano, S. Trimethylamine-N-oxide (TMAO) as novel potential biomarker of early predictors of metabolic syndrome. Nutrients 2018, 10, 1971. [Google Scholar] [CrossRef]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of trimethylamine N-oxide (TMAO) in disease: Potential biomarker or new therapeutic target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef]
- Wang, J.; Gu, X.; Yang, J.; Wei, Y.; Zhao, Y. Gut Microbiota Dysbiosis and Increased Plasma LPS and TMAO Levels in Patients With Preeclampsia. Front. Cell. Infect. Microbiol. 2019, 9, 409. [Google Scholar] [CrossRef]
- van Son, J.; Serlie, M.J.; Ståhlman, M.; Bäckhed, F.; Nieuwdorp, M.; Aron-Wisnewsky, J. Plasma Imidazole Propionate Is Positively Correlated with Blood Pressure in Overweight and Obese Humans. Nutrients 2021, 13, 2706. [Google Scholar] [CrossRef]
- Sannasiddappa, T.H.; Lund, P.A.; Clarke, S.R. In vitro antibacterial activity of unconjugated and conjugated bile salts on Staphylococcus aureus. Front. Microbiol. 2017, 8, 1581. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Feng, J.; Li, J.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. The gut microbiome-bile acid axis in hepatocarcinogenesis. Biomed. Pharmacother. 2021, 133, 111036. [Google Scholar] [CrossRef]
- Delzenne, N.M.; Knudsen, C.; Beaumont, M.; Rodriguez, J.; Neyrinck, A.M.; Bindels, L.B. Contribution of the gut microbiota to the regulation of host metabolism and energy balance: A focus on the gut–liver axis. Proc. Nutr. Soc. 2019, 78, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Alves, J.M.; Hylemon, P.B.; Bajaj, J.S. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship. Gut Microbes 2013, 4, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332. [Google Scholar] [CrossRef]
- Studer, N.; Desharnais, L.; Beutler, M.; Brugiroux, S.; Terrazos, M.A.; Menin, L.; Schürch, C.M.; McCoy, K.D.; Kuehne, S.A.; Minton, N.P. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell. Infect. Microbiol. 2016, 6, 191. [Google Scholar] [CrossRef]
- Pal, G.; Shaikh, M.; Forsyth, C.; Ouyang, B.; Keshavarzian, A.; Shannon, K. Abnormal lipopolysaccharide binding protein as marker of gastrointestinal inflammation in Parkinson disease. Front. Neurosci. 2015, 9, 306. [Google Scholar] [CrossRef]
- Abu-Shanab, A.; Quigley, E.M. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 691–701. [Google Scholar] [CrossRef]
- Dempsey, J.L.; Little, M.; Cui, J.Y. Gut microbiome: An intermediary to neurotoxicity. Neurotoxicology 2019, 75, 41–69. [Google Scholar] [CrossRef]
- Metwaly, A.; Reitmeier, S.; Haller, D. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nature biotechnology 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Vafina, G.; Zainutdinova, E.; Bulatov, E.; Filimonova, M.N. Endonuclease from Gram-Negative Bacteria Serratia marcescens Is as Effective as Pulmozyme in the Hydrolysis of DNA in Sputum. Front. Pharmacol. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Lei, X.; Zhang, Y. Association predictions of genomics, proteinomics, transcriptomics, microbiome, metabolomics, pathomics, radiomics, drug, symptoms, environment factor, and disease networks: A comprehensive approach. Med. Res. Rev. 2022, 42, 441–461. [Google Scholar] [CrossRef]
- Yeh, A.C.; Li, H.; Zhu, Y.; Zhang, J.; Khramtsova, G.; Drukker, K.; Edwards, A.; McGregor, S.; Yoshimatsu, T.; Zheng, Y.; et al. Radiogenomics of breast cancer using dynamic contrast enhanced MRI and gene expression profiling. Cancer Imaging 2019, 19, 48. [Google Scholar] [CrossRef] [PubMed]
- Immune Resistance Interrogation Study (IRIS). ClinicalTrials.gov identifier: NCT04243720. Available online: https://clinicaltrials.gov/ct2/show/NCT04243720 (accessed on 5 July 2022).
- Dréno, B.; Dagnelie, M.A.; Khammari, A.; Corvec, S. The Skin Microbiome: A New Actor in Inflammatory Acne. Am. J. Clin. Dermatol. 2020, 21, 18–24. [Google Scholar] [CrossRef]
- Lee, Y.B.; Byun, E.J.; Kim, H.S. Potential Role of the Microbiome in Acne: A Comprehensive Review. J. Clin. Med. 2019, 8, 987. [Google Scholar] [CrossRef]
- Bowe, W.; Patel, N.B.; Logan, A.C. Acne vulgaris, probiotics and the gut-brain-skin axis: From anecdote to translational medicine. Benef. Microbes 2014, 5, 185–199. [Google Scholar] [CrossRef]
- Hall, J.B.; Cong, Z.; Imamura-Kawasawa, Y.; Kidd, B.A.; Dudley, J.T.; Thiboutot, D.M.; Nelson, A.M. Isolation and Identification of the Follicular Microbiome: Implications for Acne Research. J. Investig. Dermatol. 2018, 138, 2033–2040. [Google Scholar] [CrossRef]
- Watts, A.M.; West, N.P.; Zhang, P.; Smith, P.K.; Cripps, A.W.; Cox, A.J. The Gut Microbiome of Adults with Allergic Rhinitis Is Characterised by Reduced Diversity and an Altered Abundance of Key Microbial Taxa Compared to Controls. Int. Arch. Allergy Immunol. 2021, 182, 94–105. [Google Scholar] [CrossRef]
- Bender, M.E.; Read, T.D.; Edwards, T.S.; Hargita, M.; Cutler, A.J.; Wissel, E.F.; Wise, S.K. A Comparison of the Bacterial Nasal Microbiome in Allergic Rhinitis Patients Before and After Immunotherapy. Laryngoscope 2020, 130, E882–E888. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Green, M.; Prakash, S. The Microbiome and Alzheimer’s Disease: Potential and Limitations of Prebiotic, Synbiotic, and Probiotic Formulations. Front. Bioeng. Biotechnol. 2020, 8, 537847. [Google Scholar] [CrossRef]
- Seo, D.O.; Boros, B.D.; Holtzman, D.M. The microbiome: A target for Alzheimer disease? Cell Res. 2019, 29, 779–780. [Google Scholar] [CrossRef] [PubMed]
- Haran, J.P.; Bhattarai, S.K.; Foley, S.E.; Dutta, P.; Ward, D.V.; Bucci, V.; McCormick, B.A. Alzheimer’s Disease Microbiome Is Associated with Dysregulation of the Anti-Inflammatory P-Glycoprotein Pathway. mBio 2019, 10, e00632-19. [Google Scholar] [CrossRef]
- Boddy, S.L.; Giovannelli, I.; Sassani, M.; Cooper-Knock, J.; Snyder, M.P.; Segal, E.; Elinav, E.; Barker, L.A.; Shaw, P.J.; McDermott, C.J. The gut microbiome: A key player in the complexity of amyotrophic lateral sclerosis (ALS). BMC Med. 2021, 19, 13. [Google Scholar] [CrossRef]
- Nicholson, K.; Bjornevik, K.; Abu-Ali, G.; Chan, J.; Cortese, M.; Dedi, B.; Jeon, M.; Xavier, R.; Huttenhower, C.; Ascherio, A.; et al. The human gut microbiota in people with amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2021, 22, 186–194. [Google Scholar] [CrossRef]
- Zeng, Q.; Shen, J.; Chen, K.; Zhou, J.; Liao, Q.; Lu, K.; Yuan, J.; Bi, F. The alteration of gut microbiome and metabolism in amyotrophic lateral sclerosis patients. Sci. Rep. 2020, 10, 12998. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, X.; Wu, X.; Wu, J.; Li, X.; Wei, Q.; Zhang, X.; Fang, L.; Jin, O.; Gu, J. Adalimumab Therapy Restores the Gut Microbiota in Patients With Ankylosing Spondylitis. Front. Immunol. 2021, 12, 700570. [Google Scholar] [CrossRef]
- Cardoneanu, A.; Cozma, S.; Rezus, C.; Petrariu, F.; Burlui, A.M.; Rezus, E. Characteristics of the intestinal microbiome in ankylosing spondylitis. Exp. Ther. Med. 2021, 22, 676. [Google Scholar] [CrossRef]
- Yang, B.; Wei, J.; Ju, P.; Chen, J. Effects of regulating intestinal microbiota on anxiety symptoms: A systematic review. Gen. Psychiatry 2019, 32, e100056. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.A.; Diaz-Arteche, C.; Eliby, D.; Schwartz, O.S.; Simmons, J.G.; Cowan, C.S.M. The gut microbiota in anxiety and depression—A systematic review. Clin. Psychol. Rev. 2021, 83, 101943. [Google Scholar] [CrossRef]
- Dong, Z.; Shen, X.; Hao, Y.; Li, J.; Li, H.; Xu, H.; Yin, L.; Kuang, W. Gut Microbiome: A Potential Indicator for Differential Diagnosis of Major Depressive Disorder and General Anxiety Disorder. Front. Psychiatry 2021, 12, 651536. [Google Scholar] [CrossRef] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut⁻Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Pothmann, A.; Illing, T.; Wiegand, C.; Hartmann, A.A.; Elsner, P. The Microbiome and Atopic Dermatitis: A Review. Am. J. Clin. Dermatol. 2019, 20, 749–761. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, H.S. Microbiome of the Skin and Gut in Atopic Dermatitis (AD): Understanding the Pathophysiology and Finding Novel Management Strategies. J. Clin. Med. 2019, 8, 444. [Google Scholar] [CrossRef]
- Khadka, V.D.; Key, F.M.; Romo-González, C.; Martínez-Gayosso, A.; Campos-Cabrera, B.L.; Gerónimo-Gallegos, A.; Lynn, T.C.; Durán-McKinster, C.; Coria-Jiménez, R.; Lieberman, T.D.; et al. The Skin Microbiome of Patients With Atopic Dermatitis Normalizes Gradually During Treatment. Front. Cell. Infect. Microbiol. 2021, 11, 720674. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; Narbad, A.; Rodríguez, J.M. Autism Spectrum Disorder Associated With Gut Microbiota at Immune, Metabolomic, and Neuroactive Level. Front. Neurosci. 2020, 14, 578666. [Google Scholar] [CrossRef]
- Pulikkan, J.; Mazumder, A.; Grace, T. Role of the Gut Microbiome in Autism Spectrum Disorders. Adv. Exp. Med. Biol. 2019, 1118, 253–269. [Google Scholar] [CrossRef]
- Wan, Y.; Zuo, T.; Xu, Z.; Zhang, F.; Zhan, H.; Chan, D.; Leung, T.F.; Yeoh, Y.K.; Chan, F.K.L.; Chan, R.; et al. Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut 2022, 71, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Fouquier, J.; Moreno Huizar, N.; Donnelly, J.; Glickman, C.; Kang, D.W.; Maldonado, J.; Jones, R.A.; Johnson, K.; Adams, J.B.; Krajmalnik-Brown, R.; et al. The Gut Microbiome in Autism: Study-Site Effects and Longitudinal Analysis of Behavior Change. mSystems 2021, 6, e00848-20. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, N.; Low, L.; Wallace, G.R. Behçet’s Disease-Do Microbiomes and Genetics Collaborate in Pathogenesis? Front. Immunol. 2021, 12, 648341. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, N.; Wu, C.; Zhang, X.; Wang, Q.; Huang, X.; Du, L.; Cao, Q.; Tang, J.; Zhou, C.; et al. A metagenomic study of the gut microbiome in Behcet’s disease. Microbiome 2018, 6, 135. [Google Scholar] [CrossRef]
- Consolandi, C.; Turroni, S.; Emmi, G.; Severgnini, M.; Fiori, J.; Peano, C.; Biagi, E.; Grassi, A.; Rampelli, S.; Silvestri, E.; et al. Behçet’s syndrome patients exhibit specific microbiome signature. Autoimmun. Rev. 2015, 14, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Wei, Z.; Tian, T.; Bose, D.; Shih, N.N.C.; Feldman, M.D.; Khoury, T.; De Michele, A.; Robertson, E.S. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 2021, 12, 831. [Google Scholar] [CrossRef]
- Tzeng, A.; Sangwan, N.; Jia, M.; Liu, C.C.; Keslar, K.S.; Downs-Kelly, E.; Fairchild, R.L.; Al-Hilli, Z.; Grobmyer, S.R.; Eng, C. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 2021, 13, 60. [Google Scholar] [CrossRef]
- Liu, F.; Fan, C.; Zhang, L.; Li, Y.; Hou, H.; Ma, Y.; Fan, J.; Tan, Y.; Wu, T.; Jia, S.; et al. Alterations of Gut Microbiome in Tibetan Patients With Coronary Heart Disease. Front. Cell. Infect. Microbiol. 2020, 10, 373. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Cao, H.; Liu, X.; An, Y.; Zhou, G.; Liu, Y.; Xu, M.; Dong, W.; Wang, S.; Yan, F.; Jiang, K.; et al. Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci. Rep. 2017, 7, 10322. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Liu, W.; Alkhouri, R.; Baker, R.D.; Bard, J.E.; Quigley, E.M.; Baker, S.S. Structural changes in the gut microbiome of constipated patients. Physiol. Genom. 2014, 46, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Hoque, M.N.; Sarkar, M.M.H.; Rahman, M.S.; Akter, S.; Banu, T.A.; Goswami, B.; Jahan, I.; Hossain, M.S.; Shamsuzzaman, A.K.M.; Nafisa, T.; et al. SARS-CoV-2 infection reduces human nasopharyngeal commensal microbiome with inclusion of pathobionts. Sci. Rep. 2021, 11, 24042. [Google Scholar] [CrossRef]
- Hussain, I.; Cher, G.L.Y.; Abid, M.A.; Abid, M.B. Role of Gut Microbiome in COVID-19: An Insight Into Pathogenesis and Therapeutic Potential. Front. Immunol. 2021, 12, 765965. [Google Scholar] [CrossRef] [PubMed]
- Limbana, T.; Khan, F.; Eskander, N. Gut Microbiome and Depression: How Microbes Affect the Way We Think. Cureus 2020, 12, e9966. [Google Scholar] [CrossRef]
- Kunugi, H. Gut Microbiota and Pathophysiology of Depressive Disorder. Ann. Nutr. Metab. 2021, 77 (Suppl 2), 11–20. [Google Scholar] [CrossRef]
- Chen, Z.; Radjabzadeh, D.; Chen, L.; Kurilshikov, A.; Kavousi, M.; Ahmadizar, F.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Association of Insulin Resistance and Type 2 Diabetes With Gut Microbial Diversity: A Microbiome-Wide Analysis From Population Studies. JAMA Netw. Open 2021, 4, e2118811. [Google Scholar] [CrossRef]
- Li, W.Z.; Stirling, K.; Yang, J.J.; Zhang, L. Gut microbiota and diabetes: From correlation to causality and mechanism. World J. Diabetes 2020, 11, 293–308. [Google Scholar] [CrossRef]
- Li, Y.; Xia, S.; Jiang, X.; Feng, C.; Gong, S.; Ma, J.; Fang, Z.; Yin, J.; Yin, Y. Gut Microbiota and Diarrhea: An Updated Review. Front. Cell. Infect. Microbiol. 2021, 11, 625210. [Google Scholar] [CrossRef]
- Rouhani, S.; Griffin, N.W.; Yori, P.P.; Gehrig, J.L.; Olortegui, M.P.; Salas, M.S.; Trigoso, D.R.; Moulton, L.H.; Houpt, E.R.; Barratt, M.J.; et al. Diarrhea as a Potential Cause and Consequence of Reduced Gut Microbial Diversity Among Undernourished Children in Peru. Clin. Infect. Dis. 2020, 71, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Relman, D.A. Unrest at home: Diarrheal disease and microbiota disturbance. Genome Biol. 2014, 15, 120. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lum, G.R.; Olson, C.A.; Hsiao, E.Y. Emerging roles for the intestinal microbiome in epilepsy. Neurobiol. Dis. 2020, 135, 104576. [Google Scholar] [CrossRef]
- Gong, X.; Liu, X.; Chen, C.; Lin, J.; Li, A.; Guo, K.; An, D.; Zhou, D.; Hong, Z. Alteration of Gut Microbiota in Patients With Epilepsy and the Potential Index as a Biomarker. Front. Microbiol. 2020, 11, 517797. [Google Scholar] [CrossRef] [PubMed]
- Erdrich, S.; Hawrelak, J.A.; Myers, S.P.; Harnett, J.E. Determining the association between fibromyalgia, the gut microbiome and its biomarkers: A systematic review. BMC Musculoskelet. Disord. 2020, 21, 181. [Google Scholar] [CrossRef] [PubMed]
- Minerbi, A.; Gonzalez, E.; Brereton, N.J.B.; Anjarkouchian, A.; Dewar, K.; Fitzcharles, M.A.; Chevalier, S.; Shir, Y. Altered microbiome composition in individuals with fibromyalgia. Pain 2019, 160, 2589–2602. [Google Scholar] [CrossRef]
- Walke, J.B.; Belden, L.K. Harnessing the Microbiome to Prevent Fungal Infections: Lessons from Amphibians. PLoS Pathog. 2016, 12, e1005796. [Google Scholar] [CrossRef]
- Arzani, M.; Jahromi, S.R.; Ghorbani, Z.; Vahabizad, F.; Martelletti, P.; Ghaemi, A.; Sacco, S.; Togha, M. Gut-brain Axis and migraine headache: A comprehensive review. J. Headache Pain 2020, 21, 15. [Google Scholar] [CrossRef]
- Koay, W.L.A.; Siems, L.V.; Persaud, D. The microbiome and HIV persistence: Implications for viral remission and cure. Curr. Opin. HIV AIDS 2018, 13, 61–68. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Cui, P.; Luo, L.; Chen, H.; Liang, B.; Jiang, J.; Ning, C.; Tian, L.; Zhong, X.; et al. Gut Microbiome Changes Associated With HIV Infection and Sexual Orientation. Front. Cell. Infect. Microbiol. 2020, 10, 434. [Google Scholar] [CrossRef]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Andrews, C.N.; Sidani, S.; Marshall, J.K. Clinical Management of the Microbiome in Irritable Bowel Syndrome. J. Can. Assoc. Gastroenterol. 2021, 4, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, M.; Lembo, A. Microbiome and Its Role in Irritable Bowel Syndrome. Dig. Dis. Sci. 2020, 65, 829–839. [Google Scholar] [CrossRef]
- Ramírez-Labrada, A.G.; Isla, D.; Artal, A.; Arias, M.; Rezusta, A.; Pardo, J.; Gálvez, E.M. The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy. Trends Cancer 2020, 6, 86–97. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Zang, D.; Zhang, M.; Li, X.; Liu, D.; Gao, B.; Zhou, H.; Sun, J.; Han, X.; et al. The Role of Gut Microbiota in Lung Cancer: From Carcinogenesis to Immunotherapy. Front. Oncol. 2021, 11, 720842. [Google Scholar] [CrossRef]
- Lee, K.A.; Luong, M.K.; Shaw, H.; Nathan, P.; Bataille, V.; Spector, T.D. The gut microbiome: What the oncologist ought to know. Br. J. Cancer 2021, 125, 1197–1209. [Google Scholar] [CrossRef]
- Mrázek, J.; Mekadim, C.; Kučerová, P.; Švejstil, R.; Salmonová, H.; Vlasáková, J.; Tarasová, R.; Čížková, J.; Červinková, M. Melanoma-related changes in skin microbiome. Folia Microbiol. 2019, 64, 435–442. [Google Scholar] [CrossRef]
- Dabke, K.; Hendrick, G.; Devkota, S. The gut microbiome and metabolic syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef]
- Wutthi-In, M.; Cheevadhanarak, S.; Yasom, S.; Kerdphoo, S.; Thiennimitr, P.; Phrommintikul, A.; Chattipakorn, N.; Kittichotirat, W.; Chattipakorn, S. Gut microbiota profiles of treated metabolic syndrome patients and their relationship with metabolic health. Sci. Rep. 2020, 10, 10085. [Google Scholar] [CrossRef]
- Huang, Y.S.; Lai, L.C.; Chen, Y.A.; Lin, K.Y.; Chou, Y.H.; Chen, H.C.; Wang, S.S.; Wang, J.T.; Chang, S.C. Colonization With Multidrug-Resistant Organisms Among Healthy Adults in the Community Setting: Prevalence, Risk Factors, and Composition of Gut Microbiome. Front. Microbiol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Gargiullo, L.; Del Chierico, F.; D’Argenio, P.; Putignani, L. Gut Microbiota Modulation for Multidrug-Resistant Organism Decolonization: Present and Future Perspectives. Front. Microbiol. 2019, 10, 1704. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Repáraz, J.; Kirby, T.O.; Kasper, L.H. The Gut Microbiome and Multiple Sclerosis. Cold Spring Harb. Perspect. Med. 2018, 8, a029017. [Google Scholar] [CrossRef] [PubMed]
- Boziki, M.K.; Kesidou, E.; Theotokis, P.; Mentis, A.A.; Karafoulidou, E.; Melnikov, M.; Sviridova, A.; Rogovski, V.; Boyko, A.; Grigoriadis, N. Microbiome in Multiple Sclerosis; Where Are We, What We Know and Do Not Know. Brain Sci. 2020, 10, 234. [Google Scholar] [CrossRef]
- Proal, A.; Marshall, T. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome in the Era of the Human Microbiome: Persistent Pathogens Drive Chronic Symptoms by Interfering With Host Metabolism, Gene Expression, and Immunity. Front. Pediatrics 2018, 6, 373. [Google Scholar] [CrossRef]
- Lupo, G.F.D.; Rocchetti, G.; Lucini, L.; Lorusso, L.; Manara, E.; Bertelli, M.; Puglisi, E.; Capelli, E. Potential role of microbiome in Chronic Fatigue Syndrome/Myalgic Encephalomyelits (CFS/ME). Sci. Rep. 2021, 11, 7043. [Google Scholar] [CrossRef]
- Mehrian-Shai, R.; Reichardt, J.K.V.; Harris, C.C.; Toren, A. The Gut-Brain Axis, Paving the Way to Brain Cancer. Trends Cancer 2019, 5, 200–207. [Google Scholar] [CrossRef]
- Yang, J.; Moon, H.E.; Park, H.W.; McDowell, A.; Shin, T.S.; Jee, Y.K.; Kym, S.; Paek, S.H.; Kim, Y.K. Brain tumor diagnostic model and dietary effect based on extracellular vesicle microbiome data in serum. Exp. Mol. Med. 2020, 52, 1602–1613. [Google Scholar] [CrossRef]
- Campo, L.; Eiseler, S.; Apfel, T.; Pyrsopoulos, N. Fatty Liver Disease and Gut Microbiota: A Comprehensive Update. J. Clin. Transl. Hepatol. 2019, 7, 56–60. [Google Scholar] [CrossRef]
- He, L.H.; Yao, D.H.; Wang, L.Y.; Zhang, L.; Bai, X.L. Gut Microbiome-Mediated Alteration of Immunity, Inflammation, and Metabolism Involved in the Regulation of Non-alcoholic Fatty Liver Disease. Front. Microbiol. 2021, 12, 761836. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Cantone, E.; Cassarano, S.; Tuccinardi, D.; Barrea, L.; Savastano, S.; Colao, A. Gut microbiota: A new path to treat obesity. Int. J. Obes. Suppl. 2019, 9, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Aoun, A.; Darwish, F.; Hamod, N. The Influence of the Gut Microbiome on Obesity in Adults and the Role of Probiotics, Prebiotics, and Synbiotics for Weight Loss. Prev. Nutr. Food Sci. 2020, 25, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, L.H.; Xing, C.; Liu, T. Pain regulation by gut microbiota: Molecular mechanisms and therapeutic potential. Br. J. Anaesth. 2019, 123, 637–654. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Wang, Y.; Zhang, P.; Yuan, Y.; Zhang, Y.; Chen, G. Gut microbiota regulates neuropathic pain: Potential mechanisms and therapeutic strategy. J. Headache Pain 2020, 21, 103. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jin, M.; Liu, Y.; Jin, L. Gut Microbiota: Its Potential Roles in Pancreatic Cancer. Front. Cell. Infect. Microbiol. 2020, 10, 572492. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, Y.; Guo, S.; Mei, Z.; Liao, H.; Dong, H.; Wu, K.; Ye, H.; Zhu, Y.; Lang, J.; et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun. Biol. 2021, 4, 1019. [Google Scholar] [CrossRef]
- Keshavarzian, A.; Engen, P.; Bonvegna, S.; Cilia, R. The gut microbiome in Parkinson’s disease: A culprit or a bystander? Prog. Brain Res. 2020, 252, 357–450. [Google Scholar] [CrossRef]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 62. [Google Scholar] [CrossRef]
- Pinheiro de Oliveira, F.; Mendes, R.H.; Dobbler, P.T.; Mai, V.; Pylro, V.S.; Waugh, S.G.; Vairo, F.; Refosco, L.F.; Roesch, L.F.; Schwartz, I.V. Phenylketonuria and Gut Microbiota: A Controlled Study Based on Next-Generation Sequencing. PLoS ONE 2016, 11, e0157513. [Google Scholar] [CrossRef]
- Mancilla, V.J.; Mann, A.E.; Zhang, Y.; Allen, M.S. The Adult Phenylketonuria (PKU) Gut Microbiome. Microorganisms 2021, 9, 530. [Google Scholar] [CrossRef]
- Dei-Cas, I.; Giliberto, F.; Luce, L.; Dopazo, H.; Penas-Steinhardt, A. Metagenomic analysis of gut microbiota in non-treated plaque psoriasis patients stratified by disease severity: Development of a new Psoriasis-Microbiome Index. Sci. Rep. 2020, 10, 12754. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef]
- Bodkhe, R.; Balakrishnan, B.; Taneja, V. The role of microbiome in rheumatoid arthritis treatment. Ther. Adv. Musculoskelet. Dis. 2019, 11, 1759720X19844632. [Google Scholar] [CrossRef]
- Gupta, V.K.; Cunningham, K.Y.; Hur, B.; Bakshi, U.; Huang, H.; Warrington, K.J.; Taneja, V.; Myasoedova, E.; Davis, J.M., 3rd; Sung, J. Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis. Genome Med. 2021, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Tutka, K.; Żychowska, M.; Reich, A. Diversity and Composition of the Skin, Blood and Gut Microbiome in Rosacea-A Systematic Review of the Literature. Microorganisms 2020, 8, 1756. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Lee, W.H.; Ho, H.J.; Tseng, C.H.; Wu, C.Y. An altered fecal microbial profiling in rosacea patients compared to matched controls. J. Formos. Med. Assoc. 2021, 120, 256–264. [Google Scholar] [CrossRef]
- Sepulveda, M.; Pirozzolo, I.; Alegre, M.L. Impact of the microbiota on solid organ transplant rejection. Curr. Opin. Organ Transplant. 2019, 24, 679–686. [Google Scholar] [CrossRef]
- Tabibian, J.H.; Kenderian, S.S. The Microbiome and Immune Regulation After Transplantation. Transplantation 2017, 101, 56–62. [Google Scholar] [CrossRef]
- Wood, M.R.; Yu, E.A.; Mehta, S. The Human Microbiome in the Fight Against Tuberculosis. Am. J. Trop. Med. Hyg. 2017, 96, 1274–1284. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Wu, C. Microbiota and Tuberculosis: A Potential Role of Probiotics, and Postbiotics. Front. Nutr. 2021, 8, 626254. [Google Scholar] [CrossRef]
- Verbanic, S.; Shen, Y.; Lee, J.; Deacon, J.M.; Chen, I.A. Microbial predictors of healing and short-term effect of debridement on the microbiome of chronic wounds. NPJ Biofilms Microbiomes 2020, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.R.; Gómez, B.I.; McIntyre, M.K.; Dubick, M.A.; Christy, R.J.; Nicholson, S.E.; Burmeister, D.M. The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. Int. J. Mol. Sci. 2018, 19, 2699. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Fang, Z.; Xue, Y.; Zhang, J.; Zhu, J.; Gao, R.; Yao, S.; Ye, Y.; Wang, S.; Lin, C. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 2020, 11, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Tsay, J.-C.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.-A.; Lhakhang, T.; Olsen, E.; et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef]
- Greathouse, K.L.; White, J.R.; Vargas, A.J.; Bliskovsky, V.V.; Beck, J.A.; von Muhlinen, N.; Polley, E.C.; Bowman, E.D.; Khan, M.A.; Robles, A.I.; et al. Author Correction: Interaction between the microbiome and TP53 in human lung cancer. Genome biology 2020, 21, 41. [Google Scholar] [CrossRef]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S. Commensal microbiota promote lung cancer development via γδ T cells. Cell 2019, 176, 998–1013.e1016. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, L.; Cao, L.; Li, K.J.; Huang, Y.; Luan, X.Q.; Li, G. Effects of smoking on the lower respiratory tract microbiome in mice. Respir. Res. 2018, 19, 253. [Google Scholar] [CrossRef]
- Reinhold, L.; Möllering, A.; Wallis, S.; Palade, E.; Schäfer, K.; Drömann, D.; Rupp, J.; Graspeuntner, S.; Dalhoff, K. Dissimilarity of Airway and Lung Tissue Microbiota in Smokers Undergoing Surgery for Lung Cancer. Microorganisms 2020, 8, 794. [Google Scholar] [CrossRef]
- Jungnickel, C.; Wonnenberg, B.; Karabiber, O.; Wolf, A.; Voss, M.; Wolf, L.; Honecker, A.; Kamyschnikow, A.; Herr, C.; Bals, R.; et al. Cigarette smoke-induced disruption of pulmonary barrier and bacterial translocation drive tumor-associated inflammation and growth. Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 309, L605–L613. [Google Scholar] [CrossRef][Green Version]
- Hua-Feng, X.; Yue-Ming, W.; Hong, L.; Junyi, D. A meta-analysis of the association between Chlamydia pneumoniae infection and lung cancer risk. Indian J. Cancer 2015, 52 (Suppl 2), e112–e115. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Huang, W.-Y.; Lin, J.-C.; Lin, C.-L.; Sung, F.-C.; Kao, C.-H.; Yeh, J.-J. Increased Lung Cancer Risk Among Patients with Pneumococcal Pneumonia: A Nationwide Population-Based Cohort Study. Lung 2014, 192, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-H.; Liao, C.-C.; Hsu, W.-H.; Chen, H.-J.; Liao, W.-C.; Muo, C.-H.; Sung, F.-C.; Chen, C.-Y. Increased Lung Cancer Risk among Patients with Pulmonary Tuberculosis: A Population Cohort Study. J. Thorac. Oncol. 2011, 6, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.M.; Roh, Y.H.; Lim, D.; Kong, H.J.; Cho, H.; Hwangbo, B.; Won, Y.J.; Jung, K.W.; Oh, K. Pulmonary Tuberculosis is Associated with Elevated Risk of Lung cancer in Korea: The Nationwide Cohort Study. J. Cancer 2020, 11, 1899–1906. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, S.; Langenfeld, J.; Lo, S.-C.; Rogers, M.B. Mycoplasma infection transforms normal lung cells and induces bone morphogenetic protein 2 expression by post-transcriptional mechanisms. J. Cell. Biochem. 2008, 104, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, J.-I.; Matsuoka, M. Oncogenic spiral by infectious pathogens: Cooperation of multiple factors in cancer development. Cancer Sci. 2018, 109, 24–32. [Google Scholar] [CrossRef]
- Kaiser, J. Gut microbes shape response to cancer immunotherapy. Science 2017, 358, 573. [Google Scholar] [CrossRef]
- Reiner, M.F.; Müller, D.; Gobbato, S.; Stalder, O.; Limacher, A.; Bonetti, N.R.; Pasterk, L.; Méan, M.; Rodondi, N.; Aujesky, D. Gut microbiota-dependent trimethylamine-N-oxide (TMAO) shows a U-shaped association with mortality but not with recurrent venous thromboembolism. Thromb. Res. 2019, 174, 40–47. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Bronckaers, A.; Balzarini, J.; Liekens, S. The cytostatic activity of pyrimidine nucleosides is strongly modulated by Mycoplasma hyorhinis infection: Implications for cancer therapy. Biochem. Pharmacol. 2008, 76, 188–197. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Farmer, A.D.; Wood, E.; Ruffle, J.K. An approach to the care of patients with irritable bowel syndrome. CMAJ 2020, 192, E275–E282. [Google Scholar] [CrossRef] [PubMed]
- Otani, T.; Furuse, M. Tight junction structure and function revisited. Trends Cell Biol. 2020, 30, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-Y.; Chan, Y.-L.; Tsai, M.-H.; Wang, C.-J.; Chiang, M.-H.; Chiu, C.-C. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies. World Allergy Organ. J. 2019, 12, 100021. [Google Scholar] [CrossRef] [PubMed]
- Wise, S.K.; Lin, S.Y.; Toskala, E. International consensus statement on allergy and rhinology: Allergic rhinitis—executive summary. Int. Forum Allergy Rhinol. 2018, 8, 85–107. [Google Scholar] [CrossRef]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.; Muraro, A.; Fleisher, T.A. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017, 139, 1099–1110. [Google Scholar] [CrossRef]
- Ege, M.J.; Mayer, M.; Normand, A.C.; Genuneit, J.; Cookson, W.O.; Braun-Fahrländer, C.; Heederik, D.; Piarroux, R.; von Mutius, E. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 2011, 364, 701–709. [Google Scholar] [CrossRef]
- Debarry, J.; Garn, H.; Hanuszkiewicz, A.; Dickgreber, N.; Blümer, N.; von Mutius, E.; Bufe, A.; Gatermann, S.; Renz, H.; Holst, O.; et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J. Allergy Clin. Immunol. 2007, 119, 1514–1521. [Google Scholar] [CrossRef]
- Stafford, P.; Cichacz, Z.; Woodbury, N.W.; Johnston, S.A. Immunosignature system for diagnosis of cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E3072–E3080. [Google Scholar] [CrossRef]
- Zackular, J.P.; Rogers, M.A.; Ruffin, M.T.t.; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef]
- Wynendaele, E.; Verbeke, F.; D’Hondt, M.; Hendrix, A.; Van De Wiele, C.; Burvenich, C.; Peremans, K.; De Wever, O.; Bracke, M.; De Spiegeleer, B. Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides 2015, 64, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Mimee, M.; Citorik, R.J.; Lu, T.K. Microbiome therapeutics — Advances and challenges. Adv. Drug Deliv. Rev. 2016, 105, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, U.P.; Nielsen, H.B.; Hildebrand, F.; Raes, J.; Sicheritz-Ponten, T.; Kouskoumvekaki, I.; Panagiotou, G. The chemical interactome space between the human host and the genetically defined gut metabotypes. ISME J. 2013, 7, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis: Supplementary issue: Bioinformatics methods and applications for big metagenomics data. Evol. Bioinform. 2016, 12, EBO. S36436. [Google Scholar] [CrossRef]
- Levy, M.; Blacher, E.; Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 2017, 35, 8–15. [Google Scholar] [CrossRef]
- McHardy, I.H.; Goudarzi, M.; Tong, M.; Ruegger, P.M.; Schwager, E.; Weger, J.R.; Graeber, T.G.; Sonnenburg, J.L.; Horvath, S.; Huttenhower, C.; et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 2013, 1, 17. [Google Scholar] [CrossRef]
- Uhr, G.T.; Dohnalová, L.; Thaiss, C.A. The Dimension of Time in Host-Microbiome Interactions. mSystems 2019, 4, e00216–e00218. [Google Scholar] [CrossRef]
- Kaput, J.; van Ommen, B.; Kremer, B.; Priami, C.; Monteiro, J.P.; Morine, M.; Pepping, F.; Diaz, Z.; Fenech, M.; He, Y.; et al. Consensus statement understanding health and malnutrition through a systems approach: The ENOUGH program for early life. Genes Nutr. 2013, 9, 378. [Google Scholar] [CrossRef][Green Version]
- Young, V.B.; Kahn, S.A.; Schmidt, T.M.; Chang, E.B. Studying the Enteric Microbiome in Inflammatory Bowel Diseases: Getting through the Growing Pains and Moving Forward. Front. Microbiol. 2011, 2, 144. [Google Scholar] [CrossRef]
- Shoaie, S.; Nielsen, J. Elucidating the interactions between the human gut microbiota and its host through metabolic modeling. Front. Genet. 2014, 5, 86. [Google Scholar] [CrossRef]
- Issa Isaac, N.; Philippe, D.; Nicholas, A.; Raoult, D.; Eric, C. Metaproteomics of the human gut microbiota: Challenges and contributions to other OMICS. Clin. Mass Spectrom. 2019, 14, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.B.-M.; Ofosu, F.K.; Chelliah, R.; Lee, B.H.; Oh, D.-H. Challenges and Perspective in Integrated Multi-Omics in Gut Microbiota Studies. Biomolecules 2021, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Sarwal, M.M. Recent advances in biomarker discovery in solid organ transplant by proteomics. Expert Rev. Proteom. 2011, 8, 705–715. [Google Scholar] [CrossRef]
- Molinaro, A.; Bel Lassen, P.; Henricsson, M.; Wu, H.; Adriouch, S.; Belda, E.; Chakaroun, R.; Nielsen, T.; Bergh, P.-O.; Rouault, C.; et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun. 2020, 11, 5881. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).