A Novel Rolling Circle Amplification-Based Detection of SARS-CoV-2 with Multi-Region Padlock Hybridization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. RNA Isolation and Preparation
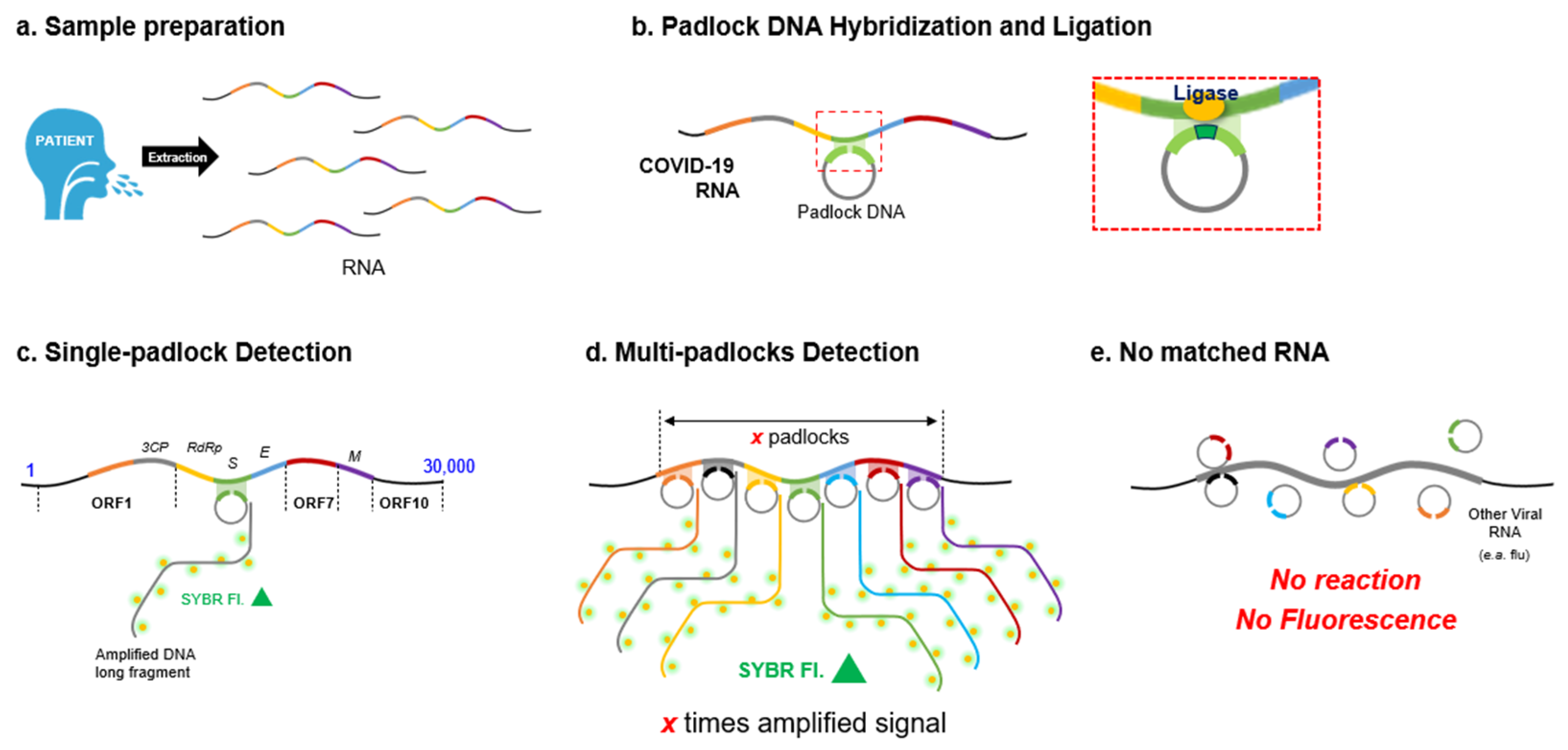
2.3. Padlock Design
2.4. MP-RCA Reaction System and Fluorescence Measurement
2.5. Circularization of Positive Control for MP-RCA Reaction
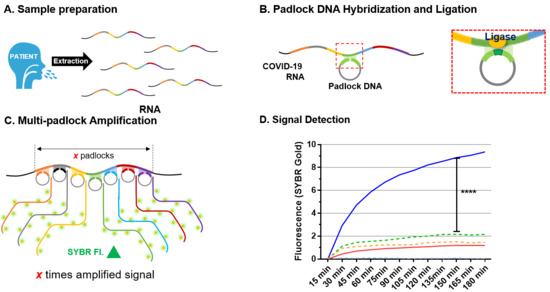
3. Results and Discussion
3.1. In Silico Comparison Analysis for the Design of the Padlocks
3.2. Recovery and Sensitivity Test of Synthesized RNA for the RCA Using SYBR Gold
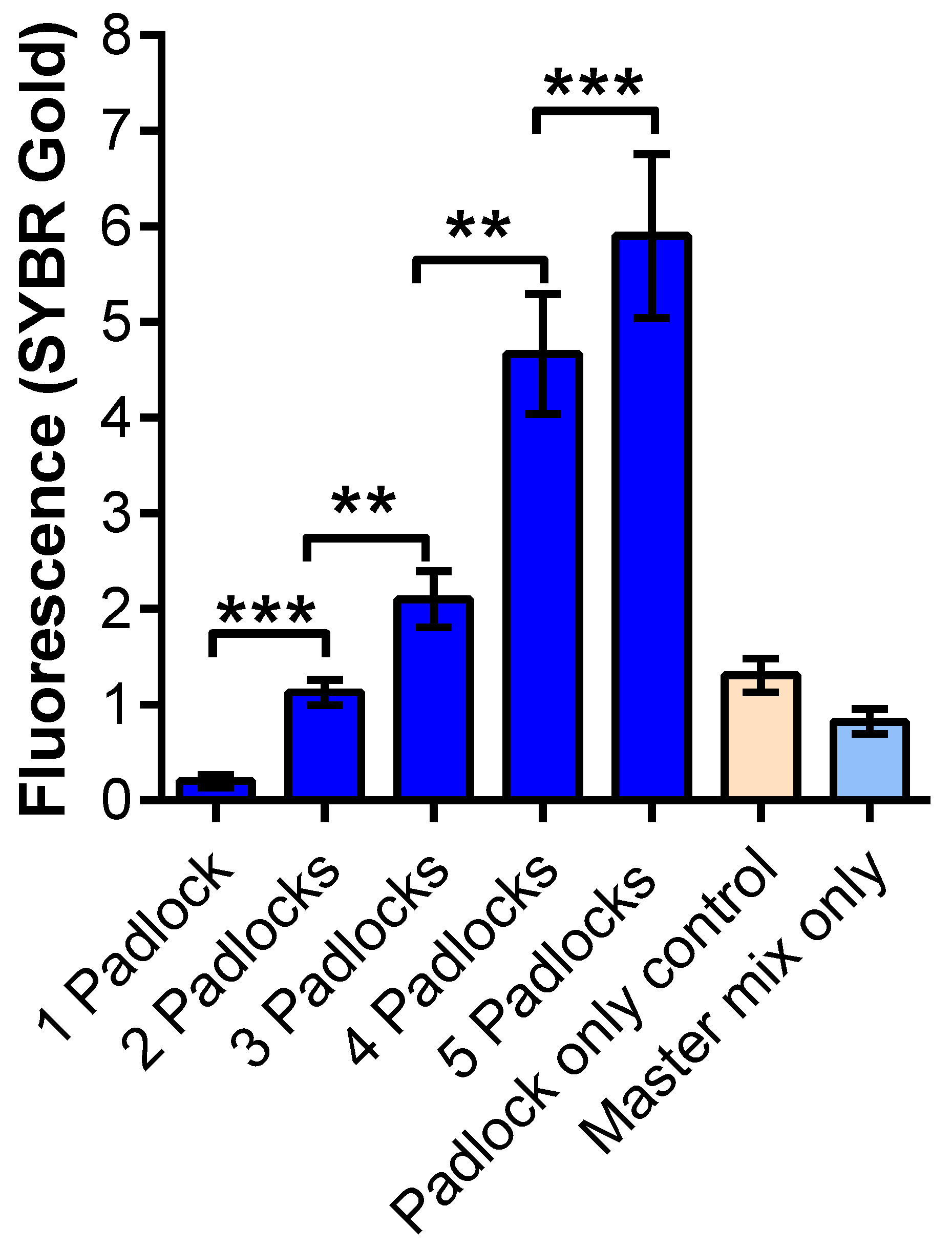
3.3. MP-RCA Optimization to the Imitated SARS-CoV-2 RNA
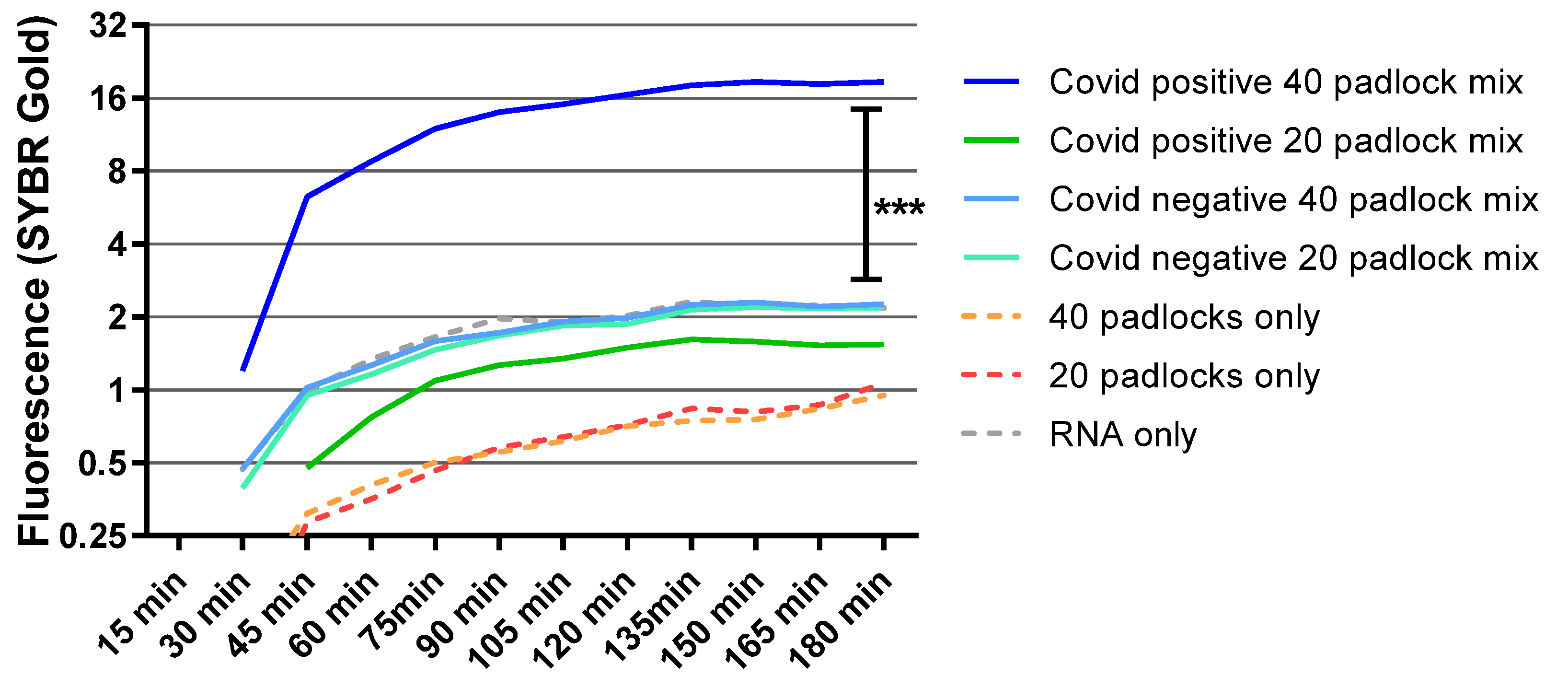
3.4. Diagnostic Test for RCA Detection of SARS-CoV-2 Patient Samples
3.5. Detection of S-Gene Target Failure Variants with Multi-Padlocks
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jindal, H.; Jain, S.; Suvvari, T.K.; Kutikuppala, L.; Rackimuthu, S.; Rocha, I.C.N.; Goyal, S. False-Negative RT-PCR Findings and Double Mutant Variant as Factors of an Overwhelming Second Wave of COVID-19 in India: An Emerging Global Health Disaster. SN Compr. Clin. Med. 2021, 3, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, T.C. Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. J. Vis. Exp. 2012, 63, e3998. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (accessed on 10 March 2022).
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Siddle, K.J.; Krasilnikova, L.A.; Moreno, G.K.; Schaffner, S.F.; Vostok, J.; Fitzgerald, N.A.; Lemieux, J.E.; Barkas, N.; Loreth, C.; Specht, I.; et al. Transmission from vaccinated individuals in a large SARS-CoV-2 Delta variant outbreak. Cell 2022, 185, 485–492.e10. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, J.E.; Li, J.Z. Uncovering Ways That Emerging Severe Acute Respiratory Syndrome Coronavirus 2 Lineages May Increase Transmissibility. J. Infect. Dis. 2021, 223, 1663–1665. [Google Scholar] [CrossRef]
- Parums, V. Editorial: Revised World Health Organization (WHO) Terminology for Variants of Concern and Variants of Interest of SARS-CoV-2. Med. Sci. Monit. 2021, 27, e933622. [Google Scholar] [CrossRef] [PubMed]
- Association, A.M. What Is the BA.2 or “Stealth” Omicron Subvariant? 2022. Available online: https://www.ama-assn.org/delivering-care/public-health/what-ba2-or-stealth-omicron-subvariant (accessed on 10 March 2022).
- Mazaika, E.; Homsy, J. Digital Droplet PCR: CNV Analysis and Other Applications. Curr. Protoc. Hum. Genet. 2014, 82, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zhong, W.; Tan, Y.; Wang, G.A.; Li, F.; Liu, Y. DNA Strand Displacement Reaction: A Powerful Tool for Discriminating Single Nucleotide Variants. Top. Curr. Chem. 2020, 378, 10. [Google Scholar] [CrossRef]
- Kashir, J.; Yaqinuddin, A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef] [PubMed]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mannan, K.S.; Chowdhury, A.; Mazumdar, R.M.; Hossain, M.N.; Islam, S.; Chowdhury, M.A. Nucleic acid amplification: Alternative methods of polymerase chain reaction. J. Pharm. Bioallied Sci. 2013, 5, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Konry, T.; Lim, J.W. Colorimetric and Multiplexed Isothermal RNA-and Antibody-Based Assay for SARS-CoV-2 and Other Viral Diagnostics and Cell Analysis. WO2021202815A1, 7 October 2021. [Google Scholar]
- Liu, D.; Daubendiek, S.L.; Zillman, M.A.; Ryan, K.; Kool, E.T. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J. Am. Chem. Soc. 1996, 118, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yu, Q.; He, C.; Duan, N. Colorimetric aptasensor for the detection of mercury based on signal intensification by rolling circle amplification. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 224, 117387. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Wang, S.; Wang, J. Rolling circle amplification based colorimetric determination of Staphylococcus aureus. Mikrochim. Acta 2020, 187, 119. [Google Scholar] [CrossRef]
- Hamidi, S.V.; Perreault, J. Simple rolling circle amplification colorimetric assay based on pH for target DNA detection. Talanta 2019, 201, 419–425. [Google Scholar] [CrossRef]
- Hao, L.; Wang, W.; Shen, X.; Wang, S.; Li, Q.; An, F.; Wu, S. A Fluorescent DNA Hydrogel Aptasensor Based on the Self-Assembly of Rolling Circle Amplification Products for Sensitive Detection of Ochratoxin A. J. Agric. Food Chem. 2020, 68, 369–375. [Google Scholar] [CrossRef]
- Ge, J.; Zhang, L.L.; Liu, S.J.; Yu, R.Q.; Chu, X. A highly sensitive target-primed rolling circle amplification (TPRCA) method for fluorescent in situ hybridization detection of microRNA in tumor cells. Anal. Chem. 2014, 86, 1808–1815. [Google Scholar] [CrossRef]
- Goryunova, M.S.; Arzhanik, V.K.; Zavriev, S.K.; Ryazantsev, D.Y. Rolling circle amplification with fluorescently labeled dUTP-balancing the yield and degree of labeling. Anal. Bioanal. Chem. 2021, 413, 3737–3748. [Google Scholar] [CrossRef]
- Konry, T.; Lerner, A.; Yarmush, M.L.; Smolina, I.V. Target DNA detection and quantitation on a single cell with single base resolution. Technology (Singap. World Sci.) 2013, 1, 88. [Google Scholar] [CrossRef]
- Konry, T.; Hayman, R.B.; Walt, D.R. Microsphere-based rolling circle amplification microarray for the detection of DNA and proteins in a single assay. Anal. Chem. 2009, 81, 5777–5782. [Google Scholar] [CrossRef]
- Gabanella, F.; Barbato, C.; Corbi, N.; Fiore, M.; Petrella, C.; de Vincentiis, M.; Greco, A.; Ferraguti, G.; Corsi, A.; Ralli, M.; et al. Exploring Mitochondrial Localization of SARS-CoV-2 RNA by Padlock Assay: A Pilot Study in Human Placenta. Int. J. Mol. Sci. 2022, 23, 2100. [Google Scholar] [CrossRef]
- Hamidi, S.V.; Ghourchian, H.; Tavoosidana, G. Real-time detection of H5N1 influenza virus through hyperbranched rolling circle amplification. Analyst 2015, 140, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef]
- Ning, B.; Youngquist, B.M.; Li, D.D.; Lyon, C.J.; Zelazny, A.; Maness, N.J.; Tian, D.; Hu, T.Y. Rapid detection of multiple SARS-CoV-2 variants of concern by PAM-targeting mutations. Cell Rep. Methods 2022, 2, 100173. [Google Scholar] [CrossRef]
- Ciftci, S.; Neumann, F.; Hernandez-Neuta, I.; Hakhverdyan, M.; Balint, A.; Herthnek, D.; Madaboosi, N.; Nilsson, M. A novel mutation tolerant padlock probe design for multiplexed detection of hypervariable RNA viruses. Sci. Rep. 2019, 9, 2872. [Google Scholar] [CrossRef]
- Jackson, B.R. The dangers of false-positive and false-negative test results: False-positive results as a function of pretest probability. Clin. Lab. Med. 2008, 28, 305–319. [Google Scholar] [CrossRef]
- Mouliou, D.S.; Gourgoulianis, K.I. False-positive and false-negative COVID-19 cases: Respiratory prevention and management strategies, vaccination, and further perspectives. Expert Rev. Respir. Med. 2021, 15, 993–1002. [Google Scholar] [CrossRef]
- Stougaard, M.; Juul, S.; Andersen, F.F.; Knudsen, B.R. Strategies for highly sensitive biomarker detection by Rolling Circle Amplification of signals from nucleic acid composed sensors. Integr. Biol. 2011, 3, 982–992. [Google Scholar] [CrossRef]
- Xu, L.; Duan, J.; Chen, J.; Ding, S.; Cheng, W. Recent advances in rolling circle amplification-based biosensing strategies-A review. Anal. Chim. Acta 2021, 1148, 238187. [Google Scholar] [CrossRef]
- Linck, L.; Resch-Genger, U. Identification of efficient fluorophores for the direct labeling of DNA via rolling circle amplification (RCA) polymerase phi29. Eur. J. Med. Chem. 2010, 45, 5561–5566. [Google Scholar] [CrossRef]
- Wang, F.; Lu, C.H.; Liu, X.; Freage, L.; Willner, I. Amplified and multiplexed detection of DNA using the dendritic rolling circle amplified synthesis of DNAzyme reporter units. Anal. Chem. 2014, 86, 1614–1621. [Google Scholar] [CrossRef]
- Liu, S.; Fang, H.; Sun, C.; Wang, N.; Li, J. Highly sensitive and multiplexed miRNA analysis based on digitally encoded silica microparticles coupled with RCA-based cascade amplification. Analyst 2018, 143, 5137–5144. [Google Scholar] [CrossRef] [PubMed]
- Heo, H.Y.; Chung, S.; Kim, Y.T.; Kim, D.H.; Seo, T.S. A valveless rotary microfluidic device for multiplex point mutation identification based on ligation-rolling circle amplification. Biosens. Bioelectron. 2016, 78, 140–146. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, R.; Lim, J.W.; Sullivan, M.R.; Malampy, R.; Baush, C.; Smolina, I.; Robin, H.; Demidov, V.V.; Ugolini, G.S.; Auclair, J.R.; et al. A Novel Rolling Circle Amplification-Based Detection of SARS-CoV-2 with Multi-Region Padlock Hybridization. Diagnostics 2022, 12, 2252. https://doi.org/10.3390/diagnostics12092252
Kumari R, Lim JW, Sullivan MR, Malampy R, Baush C, Smolina I, Robin H, Demidov VV, Ugolini GS, Auclair JR, et al. A Novel Rolling Circle Amplification-Based Detection of SARS-CoV-2 with Multi-Region Padlock Hybridization. Diagnostics. 2022; 12(9):2252. https://doi.org/10.3390/diagnostics12092252
Chicago/Turabian StyleKumari, Rajesh, Ji Won Lim, Matthew Ryan Sullivan, Rachel Malampy, Connor Baush, Irina Smolina, Howard Robin, Vadim V. Demidov, Giovanni Stefano Ugolini, Jared R. Auclair, and et al. 2022. "A Novel Rolling Circle Amplification-Based Detection of SARS-CoV-2 with Multi-Region Padlock Hybridization" Diagnostics 12, no. 9: 2252. https://doi.org/10.3390/diagnostics12092252
APA StyleKumari, R., Lim, J. W., Sullivan, M. R., Malampy, R., Baush, C., Smolina, I., Robin, H., Demidov, V. V., Ugolini, G. S., Auclair, J. R., & Konry, T. (2022). A Novel Rolling Circle Amplification-Based Detection of SARS-CoV-2 with Multi-Region Padlock Hybridization. Diagnostics, 12(9), 2252. https://doi.org/10.3390/diagnostics12092252