Optimized Xception Learning Model and XgBoost Classifier for Detection of Multiclass Chest Disease from X-ray Images
Abstract
1. Introduction
1.1. Limitation and Main Contributions
- Data augmentation and preprocessing steps are performed to make a dataset balanced and enhance the regions of the lungs for a better feature extraction step.
- Current solutions rely on the Inception model, which is incapable of creating space behavior and extracting feature maps from noisy areas. This is due to the significant number of false positives generated by the model, which, in the end, diminishes the model’s overall effectiveness. This is taken care of by the Xception architecture, which, in the model that has been proposed, essentially pulls features from all noisy-level portions while also enhancing the performance of the model in comparison to the one that came before it.
- Xception Net suffers from an overfitting problem due to a lack of regularization, which produces flash results on an unseen dataset and causes model performance scores to decline. To overcome this problem, regularization is used in the proposed model to improve scores and show that log functions work perfectly. The m-Xception model incorporates depth-separable convolution layers within the convolution layer, interlinked by linear residuals.
- The proposed model employs the Xception architecture as the backbone. Features are extracted from the images, and then Xception with 2D convolutional layers manages the robust features. The features pass them as input into the last layer, where the XgBoost classifier recognizes them.
- Accuracy, precision, recall, and the F1 score were used to evaluate the performance of the results. In addition, a comparison was made between the proposed work and existing methodologies to classify lung-related diseases.
1.2. Paper Organization
2. Methodological Background and Related Studies
Related Studies
3. Materials and Methods
3.1. Data Acquisition
3.2. Proposed Methodology
| Algorithm 1. Classification of X-ray chest diseases by using m-Xception and XgBoost classifier | |
| Step 1: | Pre-process image = X and Pre-processing step is applied by using: (a) Resize Chest X-Ray image (X) to (299, 299) [Enhanced Chest X-Ray image by preprocessing steps] (a) Remove Noise using Gaussian smoothing operator, and (b) Enhance local contrast logarithmic operator |
| Step 2: | Class Balance = Augmentation (preprocessed) |
| Step 3: | [Extract Deep Feature]: (a) Feature Extraction used: Optimize the m-Inception model by first entry flow for the feature extraction |
| Step 4: | Deep-features = Deep features were extracted by the m-Xception model |
| Step 5: | Prediction = XgBoost classifier is used to classify the images into four classes: lung opacity, COVID-19, pneumonia, and normal |
| Step 6: | [End] |
3.2.1. Pre-Processing Steps
3.2.2. Data Augmentation to Control Class Imbalance
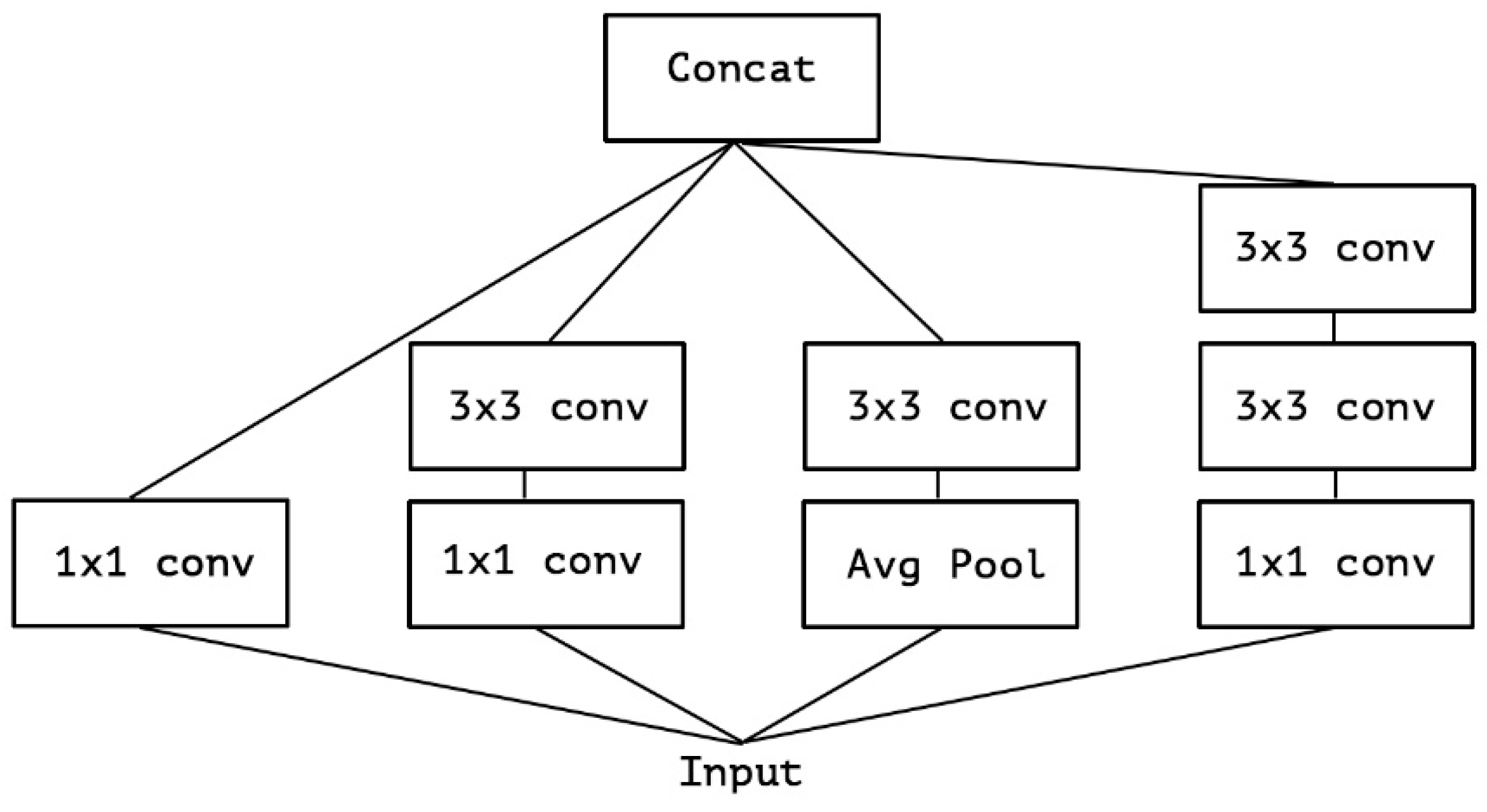
3.2.3. Proposed Model for Features Extraction
3.2.4. Formulation of the Classification Model
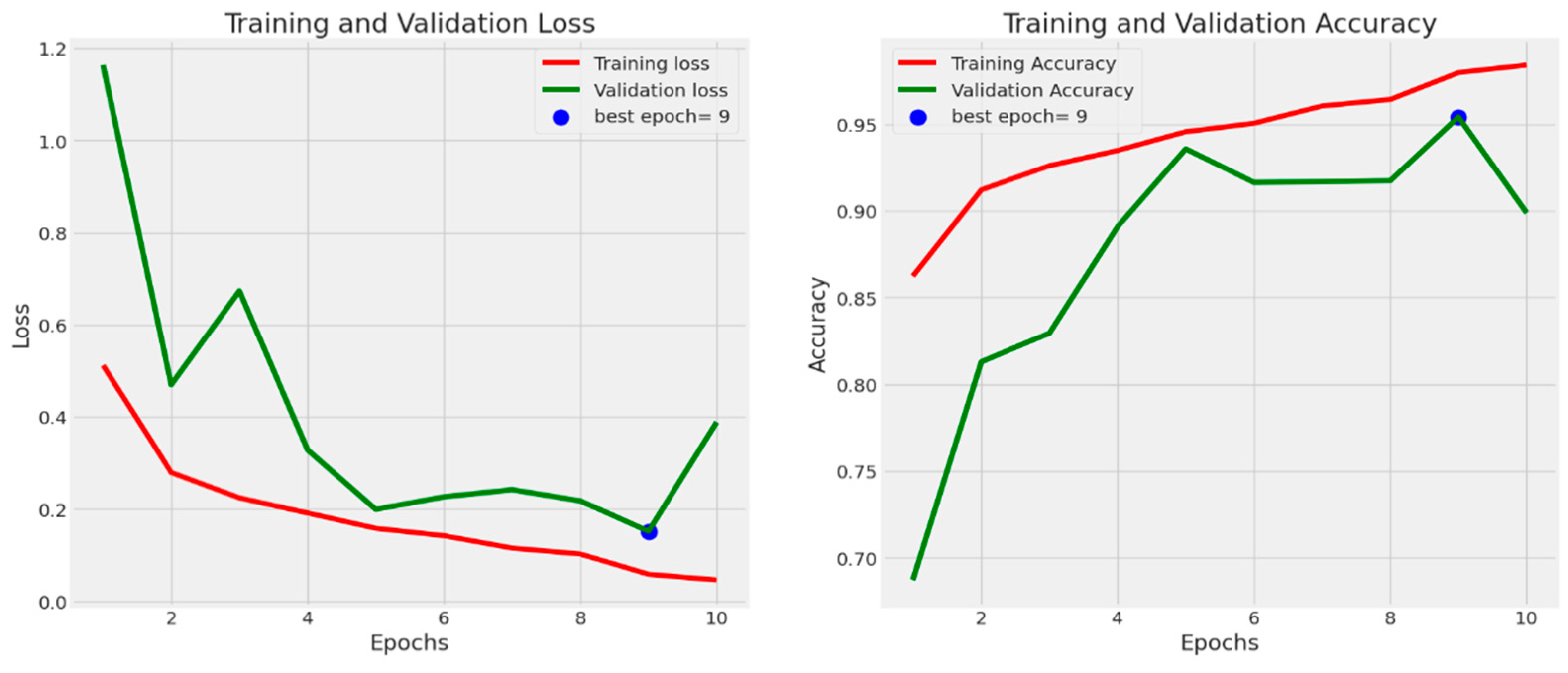
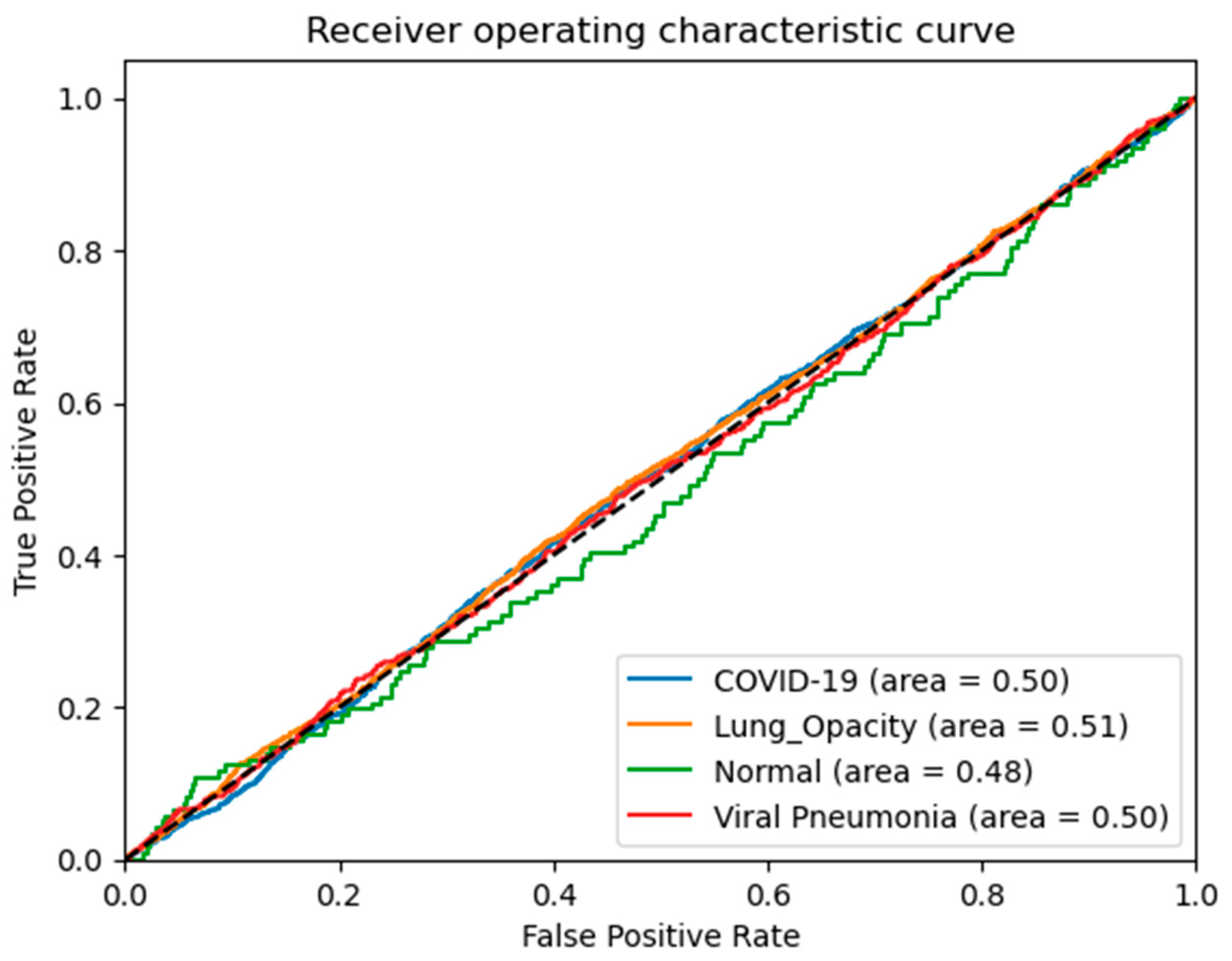
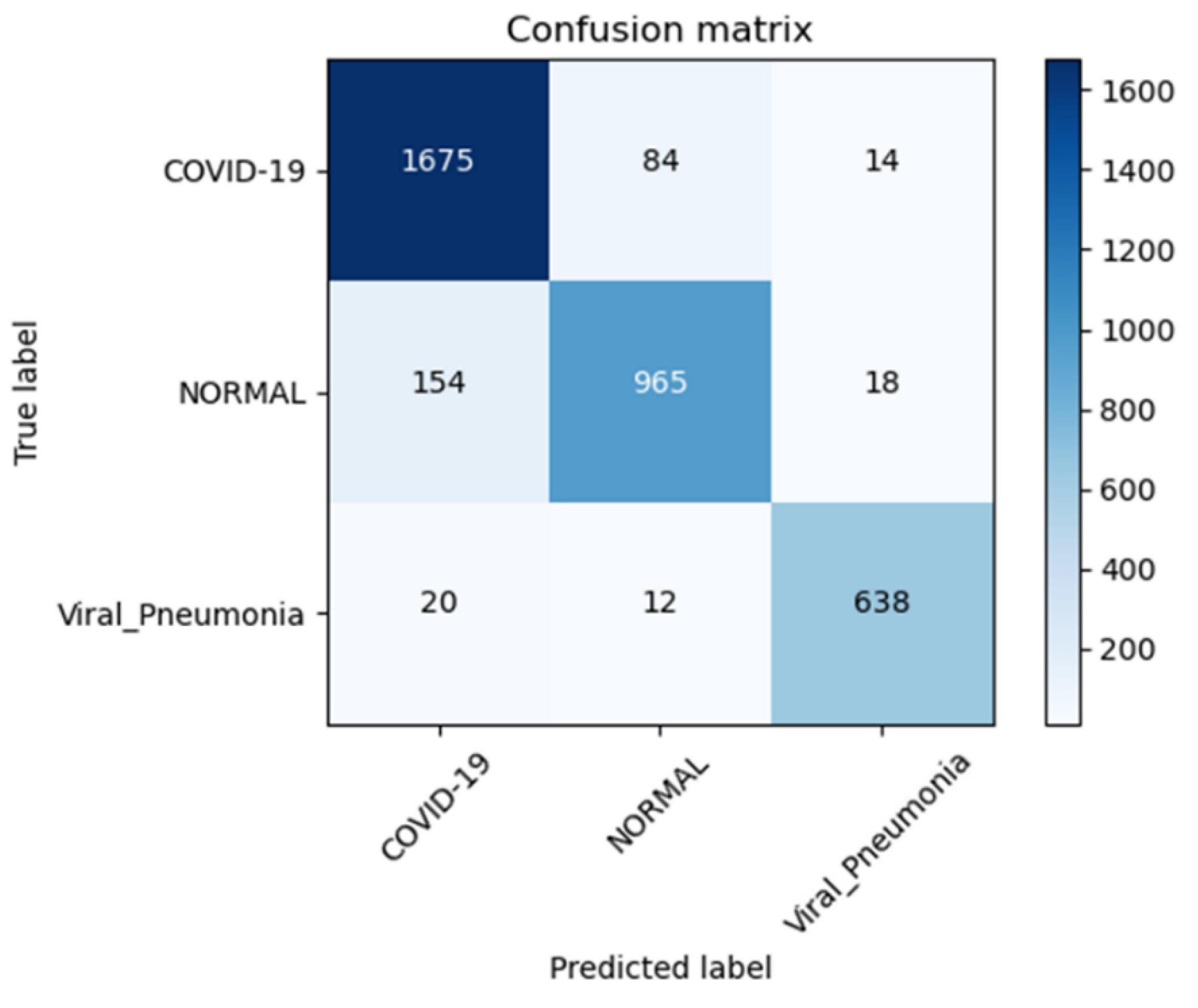
4. Experimental Results
4.1. Performance Evaluation Metrics
4.2. Results Analysis
5. Discussion
5.1. Advantages of Current Study
- (1)
- To extract desirable features from enhanced chest X-ray images, we have devised a novel framework based on a convolution vision transformer and a linear residual model.
- (2)
- The XgBoost classifier was used to predict four-class lung diseases such as COVID-19, viral pneumonia, lung opacity, and normal cases using X-Ray images and a variety of train-test split techniques.
- (3)
- Performance metrics such as accuracy, precision, recall, and the F1 score were used to analyze the results. Furthermore, the planned study was compared to similar earlier work for the diagnosis of different lung disorders.
5.2. Limitations and Challenges of Current Study
- We were unable to perform the accuracy comparison on faster processing units because we lacked the processing capacity to do so. That would have allowed us to utilize hyperparameters, which would have allowed us to adjust the learning rates, processing volumes, etc. We believe that additional experiments with hyperparameters would have led to greater precision.
- However, if these parameters are computed using the CPU as opposed to the GPU, the process can take days.
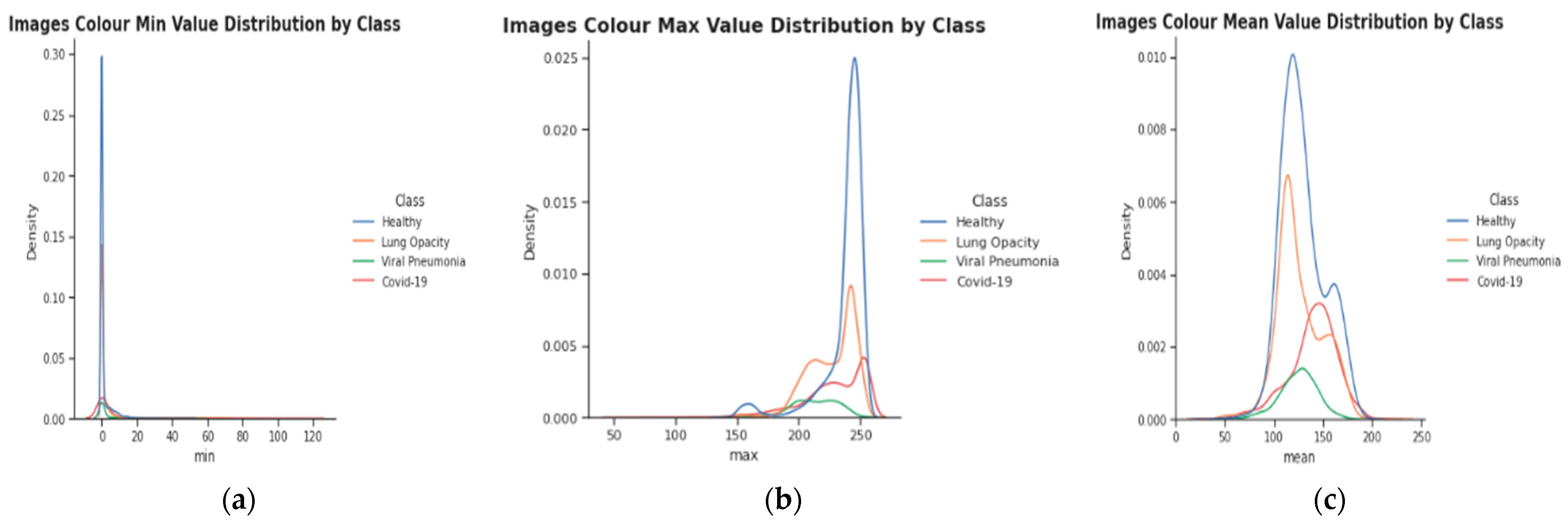
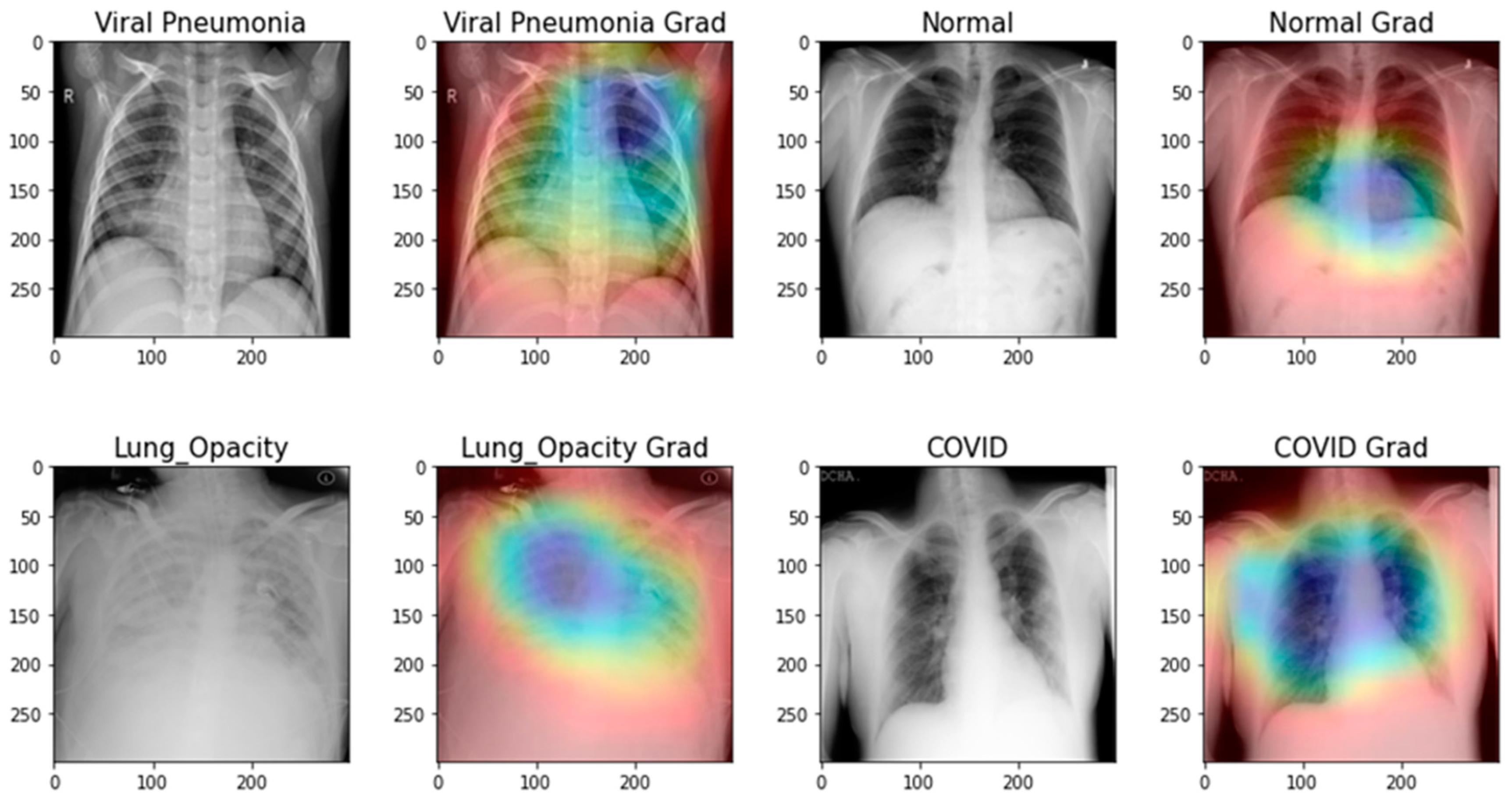
- Nearly all image enhancement methods incorrectly categorized chest X-rays in a sample case as normal, viral pneumonia, lung opacity, or normal. Gamma enhancement surpasses other enhancement methods, which is an interesting observation. As depicted in Figure 13, the Grade-Cam score indicates that the proposed TL model outperformed and clearly made a difference among lung diseases. In conclusion, this study’s detection performance for COVID-19 and other lung infections (Table 3, Table 4, Table 5 and Table 6) is consistent with that reported in recent literature. However, this research adds context that has been lacking in other recent studies. Furthermore, no article has ever reported results utilizing such massive chest X-Ray images before. Since the models in this work were trained and validated using a sizable dataset, the obtained findings are competitive with state-of-the-art methods, trustworthy, and applicable beyond the scope of the current study.
- An FPGA-based implementation [43] of the described model can provide performance boosts in terms of faster inference times, hardware acceleration, reduced power consumption, and optimized resource usage. However, it requires expertise in FPGA programming and careful consideration of cost and resource constraints. The potential advantages of FPGA-based implementations are particularly attractive for applications with real-time processing needs or resource-constrained environments. However, this is not primarily concerned with this research. This point of view will be addressed in future applications of this proposed model.
- This paper used the Xception TL model as the backbone of the architecture. However, the Xception model is an improvement over InceptionV3 in several aspects. Both models are based on the concept of “Inception” modules, which use multiple filters of different sizes to capture features at various scales. However, Xception improves upon InceptionV3 by introducing depthwise separable convolutions, resulting in better efficiency, improved representation learning, and a smaller model size, while maintaining or even surpassing the performance of InceptionV3. Therefore, the implementation of the m-Xception model should be tested on an application in a resource-constrained environment.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, H.; Stratton, C.W.; Tang, Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020, 92, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Shaheed, K.; Szczuko, P.; Abbas, Q.; Hussain, A.; Albathan, M. Computer-Aided Diagnosis of COVID-19 from Chest X-ray Images Using Hybrid-Features and Random Forest Classifier. Healthcare 2023, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Turkoglu, M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl. Intell. 2021, 51, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Gaur, L.; Bhatia, U.; Jhanjhi, N.Z.; Muhammad, G.; Masud, M. Medical image-based detection of COVID-19 using deep convolution neural networks. Multimed. Syst. 2023, 29, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Gou, B.; Khishe, M.; Mohammadi, M.; Rashidi, S.; Moradpour, R.; Mirjalili, S. Improved deep convolutional neural networks using chimp optimization algorithm for Covid19 diagnosis from the X-ray images. Expert Syst. Appl. 2023, 213, 119206. [Google Scholar] [CrossRef]
- Kathamuthu, N.D.; Subramaniam, S.; Le, Q.H.; Muthusamy, S.; Panchal, H.; Sundararajan, S.C.M.; Alrubaieg, A.J.; Zahra, M.M.A. A deep transfer learning-based convolution neural network model for COVID-19 detection using computed tomography scan images for medical applications. Adv. Eng. Softw. 2023, 175, 1–20. [Google Scholar] [CrossRef]
- Qureshi, I.; Yan, J.; Abbas, Q.; Shaheed, K.; Riaz, A.B.; Wahid, A.; Khan, M.W.; Szczuko, P. Medical image segmentation using deep semantic-based methods: A review of techniques, applications and emerging trends. Inf. Fusion 2022, 90, 316–352. [Google Scholar]
- Karnati, M.; Seal, A.; Sahu, G.; Yazidi, A.; Krejcar, O. A novel multi-scale based deep convolutional neural network for detecting COVID-19 from X-rays. Appl. Soft Comput. 2022, 125, 109109. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, S.; Hao, Y.; Fang, Y.; Fang, Z.; Wu, W.; Liu, Z.; Li, S. Auxiliary diagnosis for COVID-19 with deep transfer learning. J. Digit. Imaging 2021, 34, 231–241. [Google Scholar] [CrossRef]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Wei, X.-S.; Xie, C.-W.; Wu, J.; Shen, C. Mask-CNN: Localizing parts and selecting descriptors for fine-grained bird species categorization. Pattern Recognit. 2018, 76, 704–714. [Google Scholar] [CrossRef]
- Himeur, Y.; Al-Maadeed, S.; Varlamis, I.; Al-Maadeed, N.; Abualsaud, K.; Mohamed, A. Face mask detection in smart cities using deep and transfer learning: Lessons learned from the COVID-19 pandemic. Systems 2023, 11, 107. [Google Scholar]
- George, G.S.; Mishra, P.R.; Sinha, P.; Prusty, M.R. COVID-19 detection on chest X-ray images using Homomorphic Transformation and VGG inspired deep convolutional neural network. Biocybern. Biomed. Eng. 2023, 43, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ismael, A.M.; Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.H.; Geng, H.; Chiu, T.L.; Yu, S.K.; Cho, D.C.; Heo, J.; Choi, M.S.; Choi, I.H.; Cung Van, C.; Nhung, N.V. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020, 7, 427. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Gupta, M.; Gupta, D.; Tiwari, S. Novel deep transfer learning model for COVID-19 patient detection using X-ray chest images. J. Ambient Intell. Humaniz. Comput. 2023, 14, 469–478. [Google Scholar] [CrossRef]
- Poola, R.G.; Pl, L. COVID-19 diagnosis: A comprehensive review of pre-trained deep learning models based on feature extraction algorithm. Results Eng. 2023, 18, 101020. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Slima, M.B.; BenHamida, A. Deep efficient-nets with transfer learning assisted detection of COVID-19 using chest X-ray radiology imaging. Multimed. Tools Appl. 2023. [Google Scholar] [CrossRef]
- Sahin, M.E.; Ulutas, H.; Yuce, E.; Erkoc, M.F. Detection and classification of COVID-19 by using faster R-CNN and mask R-CNN on CT images. Neural Comput. Appl. 2023, 35, 13597–13611. [Google Scholar] [CrossRef]
- Tang, S.; Wang, C.; Nie, J.; Kumar, N.; Zhang, Y.; Xiong, Z.; Barnawi, A. EDL-COVID: Ensemble deep learning for COVID-19 case detection from chest x-ray images. IEEE Trans. Ind. Inform. 2021, 17, 6539–6549. [Google Scholar] [CrossRef]
- Sahlol, A.T.; Yousri, D.; Ewees, A.A.; Al-qaness, M.A.A.; Damasevicius, R.; Elaziz, M.A. COVID-19 image classification using deep features and fractional-order marine predators algorithm. Sci. Rep. 2020, 10, 15364. [Google Scholar] [CrossRef]
- Sharma, A.; Rani, S.; Gupta, D. Artificial intelligence-based classification of chest X-ray images into COVID-19 and other infectious diseases. Int. J. Biomed. Imaging 2020, 2020, 8889023. [Google Scholar] [CrossRef] [PubMed]
- Bougourzi, F.; Dornaika, F.; Mokrani, K.; Taleb-Ahmed, A.; Ruichek, Y. Fusion Transformed Deep and Shallow features (FTDS) for Image-Based Facial Expression Recognition. Expert Syst. Appl. 2020, 156, 113459. [Google Scholar] [CrossRef]
- Bougourzi, F.; Mokrani, K.; Ruichek, Y.; Dornaika, F.; Ouafi, A.; Taleb-Ahmed, A. Fusion of transformed shallow features for facial expression recognition. IET Image Process. 2019, 13, 1479–1489. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Sahin, M.E. Deep learning-based approach for detecting COVID-19 in chest X-rays. Biomed. Signal Process. Control 2022, 78, 103977. [Google Scholar] [CrossRef]
- Mangal, A.; Kalia, S.; Rajgopal, H.; Rangarajan, K.; Namboodiri, V.; Banerjee, S.; Arora, C. CovidAID: COVID-19 Detection Using Chest X-ray. arXiv 2020, arXiv:2004.09803. [Google Scholar]
- Al-Waisy, A.S.; Al-Fahdawi, S.; Mohammed, M.A.; Abdulkareem, K.H.; Mostafa, S.A.; Maashi, M.S.; Arif, M.; Garcia-Zapirain, B. COVID-CheXNet: Hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2023, 27, 2657–2672. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Vantaggiato, E.; Paladini, E.; Bougourzi, F.; Distante, C.; Hadid, A.; Taleb-Ahmed, A. COVID-19 recognition using ensemble-cnns in two new chest x-ray databases. Sensors 2021, 21, 1742. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Gupta, K.; Bajaj, V. Deep learning models-based CT-scan image classification for automated screening of COVID-19. Biomed. Signal Process. Control. 2023, 80, 104268. [Google Scholar] [CrossRef] [PubMed]
- Ur Rehman, T. COVID-19 Radiography Database. Kaggle. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 20 July 2023).
- Xiyuan, P.; Jinxiang, Y.; Bowen, Y.; Liansheng, L.; Yu, P. A Review of FPGA-Based Custom Computing Architecture for Convolutional Neural Network Inference. Chin. J. Electron. 2021, 30, 1–17. [Google Scholar] [CrossRef]







| Approach | Models | Class | * Output | Criterion | Constraints |
|---|---|---|---|---|---|
| Hemdan [28], 2020 | VGG-19 ResNet-v2 DenseNet-201 | COVID-19 and Normal | ACC = 88%, F1-score = 84%, SEN = 96%, SP = 81% | 4.1 million trainable | Complex image preprocessing and classification stages render the method computationally challenging. Two divisions are recognized. |
| Apostolopoulos [27], 2020 | VGG-19 MobileNet-v2 Inception Xception Inception-ResNet v2 | COVID-19, pneumonia (bacterial, viral, and normal) | ACC = 98%, F1-score = 96%, SEN = 95%, SP = 95% | 5.8 million trainable | A small number of COVID-19 samples used but five-classes of lung diseases were identified. |
| Edoardo [38], 2021 | ResNetXt-50, Inception-v3, DenseNet-161 | COVID-19, viral pneumonia, bacterial pneumonia, lung opacity, normal | ACC = 81% | 12.00 million trainable | Only focused on lung infections without preprocessing and classify COVID-19 patients. Ensemble approach to obtaining multi-class label infection labels. |
| Mangal [35], 2020 | COVIDAID | COVID-19, pneumonia and normal | ACC = 93% | 11.78 million | The method has a degraded performance and is based on three classes only and no generalized solution. |
| Yoo et al. [5], 2020 | AXIR 1 to 4 | Normal, abnormal, TB, non-TB and COVID-19 | ACC = 90% | NS | Training complexity several deep learning models was used for training. |
| Turkoglu [3], 2020 | COVIDetectioNet | COVID-19, pneumonia and Normal | ACC = 99%. SEN = 100%, SP = 98% | NS | Non-effective evaluation of deep learning model because only one train-test split strategy is used. |
| Rehman et.al [19] | AlexNet, ResNet18, DenseNet201, and SqueezeNet | CT Images | ACC = 93%, | 23 million | Limited in terms of data used in the work. No class imbalance and no preprocessing used. |
| Nasiri et al. [20] | ReseNet-50 | Pneumonia, uninfected, and infected with COVID-19 | ACC = 98%. | NS | Limited to three classes only, no preprocessing and data augmentation. |
| Category | Images | Data Augmentation |
|---|---|---|
| Normal | 375 | 12,000 |
| Pneumonia | 345 | 12,000 |
| COVID-19 | 375 | 12,000 |
| Lung Opacity | 400 | 12,000 |
| Total | 1495 | 48,000 |
| Pathology | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| COVID-19 | 97.56% | 95.33% | 95.34% | 97.16% |
| Lung Opacity | 94.82% | 89.33% | 94.64% | 94.38% |
| Normal | 96.22% | 96.67% | 90.23% | 95.80% |
| Viral Pneumonia | 94.13% | 95.33% | 96.33% | 97.33% |
| Pathology | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| COVID-19 | 97.56% | 95.33% | 95.34% | 97.16% |
| Lung Opacity | 94.82% | 89.33% | 94.64% | 94.38% |
| Normal | 96.22% | 96.67% | 90.23% | 95.80% |
| Viral Pneumonia | 94.13% | 95.33% | 96.33% | 97.33% |
| Pathology | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| COVID-19 | 97.23% | 95.00% | 95.00% | 97.26% |
| Lung Opacity | 94.20% | 89.11% | 94.20% | 94.08% |
| Normal | 96.20% | 96.20% | 90.10% | 95.10% |
| Viral Pneumonia | 94.10% | 95.13% | 96.00% | 97.11% |
| Models/Methods | 10 Epochs | 30 Epochs | ||||||
|---|---|---|---|---|---|---|---|---|
| Training Acc. % | Validation Acc. % | Training Losses | Validation Losses | Training Acc. % | Validation Acc. % | Training Losses | Validation Losses | |
| Hemdan [28] | 75.6 | 75.4 | 88.14 | 88.41 | 73 | 71 | 83 | 82 |
| Apostolopoulos [27] | 82 | 82 | 76.44 | 73 | 76 | 75 | 85 | 81 |
| Edoardo [38] | 83.24 | 84 | 73.1 | 71 | 81 | 79 | 81 | 75 |
| Mangal [35] | 87.1 | 87 | 74 | 77 | 80 | 72 | 73 | 80 |
| Yoo [15] | 88.54 | 88 | 89 | 82 | 81 | 78 | 77 | 75 |
| Turkoglu [3] | 94.11 | 95.32 | 71 | 73 | 82 | 81 | 79 | 72 |
| m-Xception | 96.78 | 95 | 62.1 | 55 | 92 | 91 | 61 | 56 |
| Pre-Trained Models | 10 Epochs | 30 Epochs | ||||||
|---|---|---|---|---|---|---|---|---|
| Training Acc. % | Validation Acc. % | Training Losses | Validation Losses | Training Acc. % | Validation Acc. % | Training Losses | Validation Losses | |
| VGG-19 [15] | 75 | 74 | 55 | 52 | 67 | 70.5 | 70.3 | 72.5 |
| DenseNet-121 [16] | 77 | 75 | 52 | 53 | 74 | 78.1 | 70 | 70.4 |
| Inception-V3 [17] | 78 | 82 | 50 | 52 | 81 | 83.9 | 65.2 | 62.5 |
| ResNet-V2 [18] | 80 | 81 | 51 | 55 | 84 | 85.2 | 61.9 | 60.3 |
| InceptionResNet-V2 [19] | 83 | 84 | 50 | 50 | 89 | 88.9 | 59.4 | 59 |
| Xception [20] | 84 | 87 | 45 | 50 | 90.6 | 90 | 58 | 53.1 |
| m-Xception | 96 | 95 | 32 | 45 | 96.82 | 97.4 | 53.1 | 52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaheed, K.; Abbas, Q.; Hussain, A.; Qureshi, I. Optimized Xception Learning Model and XgBoost Classifier for Detection of Multiclass Chest Disease from X-ray Images. Diagnostics 2023, 13, 2583. https://doi.org/10.3390/diagnostics13152583
Shaheed K, Abbas Q, Hussain A, Qureshi I. Optimized Xception Learning Model and XgBoost Classifier for Detection of Multiclass Chest Disease from X-ray Images. Diagnostics. 2023; 13(15):2583. https://doi.org/10.3390/diagnostics13152583
Chicago/Turabian StyleShaheed, Kashif, Qaisar Abbas, Ayyaz Hussain, and Imran Qureshi. 2023. "Optimized Xception Learning Model and XgBoost Classifier for Detection of Multiclass Chest Disease from X-ray Images" Diagnostics 13, no. 15: 2583. https://doi.org/10.3390/diagnostics13152583
APA StyleShaheed, K., Abbas, Q., Hussain, A., & Qureshi, I. (2023). Optimized Xception Learning Model and XgBoost Classifier for Detection of Multiclass Chest Disease from X-ray Images. Diagnostics, 13(15), 2583. https://doi.org/10.3390/diagnostics13152583









