What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine
Abstract
1. Introduction
2. Glossary
- Model architecture—describes how the data is processed, transmitted, and analysed within the machine learning algorithm, which influences its efficiency and effectiveness in solving problems.
- Data exploration—the process of analysing and summarising a large dataset to gain insight into the relationships and patterns that exist within the data.
- Binary classification—a type of classification in which the aim is to assign one of two possible classes (labels) to an object: positive or negative, true or false, etc.
- Logistic function—a sigmoidal mathematical function that transforms values from minus infinity into plus infinity to the range (0, 1), allowing not only non-linearity, but also probability, e.g., binary classification.
- Input variables—also known as independent variables, explanatory variables, predictor variables, etc., and are variables that are used to describe or explain the behaviour, trends, or decisions of the target variables.
- Target variables—also known as dependent variables, outcome variables, etc., and are variables that are studied or predicted in statistical analysis and machine learning. Target variables are dependent on and are described using explanatory variables.
- Bayes theory—used to calculate the probability of an event, having prior information about that event.
- Sentiment analysis—the process of automatically determining emotions, opinions, and moods expressed in the text. This can be in the form of product reviews, comments on online forums, tweets on Twitter, or other forms of textual communication. The purpose of sentiment analysis is to gain an automatic understanding of whether a text is positive, negative, or neutral.
- Training—the process of formatting a model to interpret the data to perform a specific task with a specific accuracy. In this case, it is the determination of the hyperplane.
- Hyperplane—a set having n − 1 dimension, relative to the n-dimensional space in which it is contained (for n = two-dimensional space it has one dimension (point); for n = three-dimensional space it has two dimensions (line)).
- Training objects—a set of objects used to determine the hyperplane with the model.
- Support vectors—at least two objects at the shortest distance from the hyperplane belonging to two classes.
- Class—a group, described on numerical ranges, to which an object can be assigned—i.e., classified.
- Cluster—a hyperplane-limited space in a data system in which the presence of an object determines the class assignment.
- Neuron—the basic element of a neural network, which connects to other neurons through transmitting data to each other.
- Weight—the characteristic that the network designer provides to the connections between the neurons to achieve the desired results.
- Recursion—referring a function to the same function using the network being trained.
- Layer—a portion of the total network architecture that is differentiated from the other parts due to having a distinctive function.
- Activation function—takes the input from one or more neurons and maps it to an output value, which is then passed onto the next layer of neurons in the network.
- Hidden layer—in an artificial neural network, this is defined as the layer between the input and output layers, where the result of their action cannot be directly observed.
- Input layer—layer where the data are collected and passed onto the next layer.
- Output layer—layer which gathers the conclusions.
- Backpropagation—sending signals in the reverse order to calculate the error associated with each individual neuron in an effort to improve the model.
- Cost function—a function that represents the errors occurring in the training process in the form of a number. It is used for subsequent optimisation.
- Receptive field—a section of the image that is individually analysed using the filter.
- Filter—a set of numbers that are used to perform computational operations on the input data on splices. It is used to extract features (e.g., the presence of edges or curvature).
- Convolution—integral of the product of the two functions after one is reflected about the y-axis and shifted.
- Pooling—reducing the amount of data representing a given area of the image.
- Matrix—a mathematical concept; a set of numbers which is used, among other things, to recalculate the data obtained from neurons.
- Skip connections—a technique used in neural networks that allows information to be passed from one layer of the network to another, while skipping intermediate layers.
3. Data and Its Relevance to Neural Networks
4. Machine Learning Models
4.1. Supervised Learning
4.2. Unsupervised Learning
5. Classical Methods of Machine Learning
5.1. k-Nearest Neighbour Algorithms
5.2. Linear Regression Algorithms
5.3. Logistic Regression Algorithms
5.4. Naive Bayes Classifier Algorithms
5.5. Support Vector Machines (SVMs)
5.6. AdaBoost

5.7. XGBoost
6. Neural Networks
- Perceptron networks: simplest neural networks with an input and output layer composed of perceptrons. Perceptrons assign a value of one or zero based on the activation threshold, dividing the set into two.
- Layered networks (feed forward): multiple layers of interconnected neurons where the outputs of the previous layer neurons serve as the inputs for the next layer. The neurons of each successive layer always have a +1 input from the previous layer. Enables the classification of non-binary sets, and are used in image, text, and speech recognition.
- Recurrent networks: neural networks with feedback loops where the output signals feed back into the input neurons. Can generate sequences of phenomena and signals until the output stabilises. Used for sentiment analysis and text generation.
- Convolutional networks, also known as braided networks, are described in the next paragraph.
- Gated recurrent unit (GRU) and long short-term memory (LSTM) networks: perform recursive tasks with the output dependent on previous calculations. They have network memory, allowing them to remember data states across different time steps. These networks have longer training times and are applied in time series analysis (e.g., stock prices), autonomous car trajectory prediction, text-to-speech conversion, and language translation.
Vision Transformers
7. Deep Learning
7.1. Differences between the DNNs and ANNs
7.2. Deep Neural Network (DNN) Classifiers
7.3. Convolutional Neural Networks
7.4. Auto-Encoders (Unsupervised)
7.5. Segmenting Neural Networks (e.g., UNET and Lung Segmentation)
7.6. Generative Adversarial Networks
7.7. Transfer Learning
7.8. Few-Shot Learning
7.9. Deep Reinforcement Learning
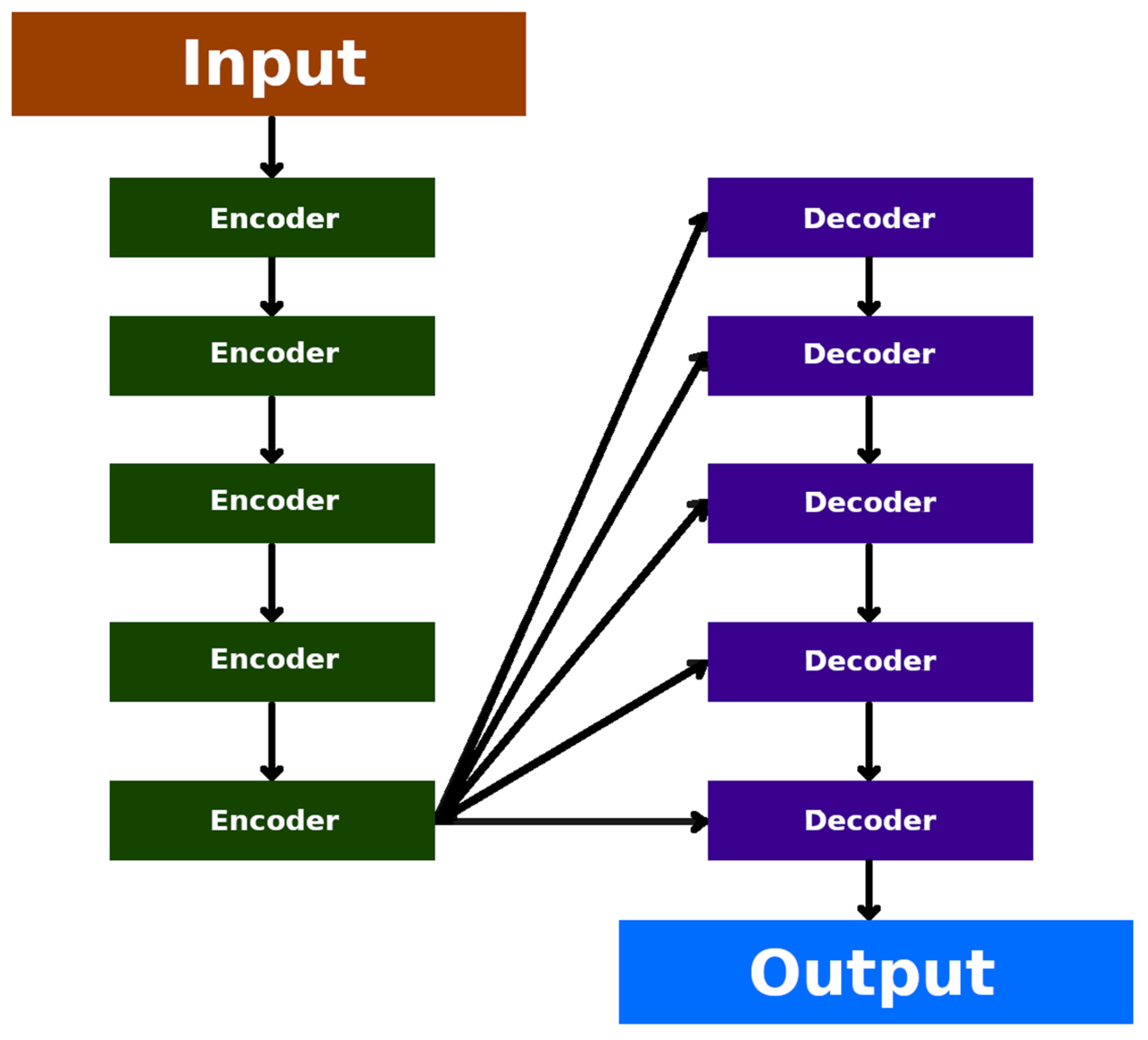
7.10. Transformer Neural Networks
7.11. Attention Mechanism
8. Examples of AI Applications in Medicine Approved by the US Food and Drug Administration (FDA)
- Apple IRNF 2.0: Apple Inc. developed the IRNF 2.0 software for the Apple Watch, utilising CNNs and machine learning algorithms. This software aims to identify cardiac rhythm disorders, particularly atrial fibrillation (AFib). The study conducted by Apple in 2021, with FDA approval, involved over 2500 participants and collected more than 3 million heart rate recordings. The algorithm successfully differentiated between AFib and non-AFib rhythms, with a sensitivity of 88.6% and a specificity of 99.3%. While effective, it is important to note that the app does not replace professional diagnosis nor target individuals that have already been diagnosed with AFib [77].
- Ultromics: An AI-powered solution detects heart failure with a 90% accuracy, specifically heart failure with preserved ejection fraction (HFpEF). It analyses LV images using an AI ML-based algorithm to accurately measure LV parameters, such as volumes, left ventricular ejection fraction (LVEF), and left ventricular longitudinal strain (LVLS). This software (EchoGo Core 2.0) also classifies echocardiographic views for quality control. AI readings are more consistent than manual readings, regardless of the image quality. Additionally, AI-derived LVEF and LVLS values have been significantly associated with mortality in-hospital and at final follow-up [78].
- Aidoc: Aidoc is an AI-based platform that permits a fast and accurate analysis of X-rays and CT scans. It detects conditions, like strokes, fractures, and cancerous lesions. Their advanced AI softwares (Aidoc software from 2.0 and above) help radiologists to prioritise their critical cases and expedite patient care. Aidoc has 12 FDA-approved tools, including softwares (Aidoc software from 2.0 and above) for analysing head CT images (detecting intracranial haemorrhage), chest CT studies (identifying aortic dissection), chest X-rays (flagging a suspected pneumothorax), and abdominal CT images (indicating suspected intra-abdominal free gas) [79].
- Riverain Technologies: This AI-based platform enables an accurate and efficient analysis of lung images for detecting and monitoring lung conditions, like cancer, COPD, and bronchial disease. It includes features such as CT Detect for measuring areas of interest, ClearRead CT Compare for comparing nodules across studies, and ClearRead CT Vessel Suppress for enhancing nodule visibility. This patented technology improves the accuracy and reading performance, and seamlessly integrates processed series with the original CT series for synchronised scrolling [80].
9. Discussion and Limitations
10. Usability
11. Summary and Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ward, T.M.; Mascagni, P.; Madani, A.; Padoy, N.; Perretta, S.; Hashimoto, D.A. Surgical Data Science and Artificial Intelligence for Surgical Education. J. Surg. Oncol. 2021, 124, 221–230. [Google Scholar] [CrossRef]
- Mortani Barbosa, E.J.; Gefter, W.B.; Ghesu, F.C.; Liu, S.; Mailhe, B.; Mansoor, A.; Grbic, S.; Vogt, S. Automated Detection and Quantification of COVID-19 Airspace Disease on Chest Radiographs: A Novel Approach Achieving Expert Radiologist-Level Performance Using a Deep Convolutional Neural Network Trained on Digital Reconstructed Radiographs from Computed Tomography-Derived Ground Truth. Investig. Radiol. 2021, 56, 471–479. [Google Scholar] [CrossRef]
- Gupta, M. Introduction to Data in Machine Learning. GeeksforGeeks. Available online: https://www.geeksforgeeks.org/ml-introduction-data-machine-learning/ (accessed on 1 May 2023).
- Dorfman, E. How Much Data Is Required for Machine Learning? Postindustria. Available online: https://postindustria.com/how-much-data-is-required-for-machine-learning/ (accessed on 7 May 2023).
- Patel, H. Data-Centric Approach vs. Model-Centric Approach in Machine Learning. MLOps Blog 2023. Available online: https://neptune.ai/blog/data-centric-vs-model-centric-machine-learning (accessed on 1 May 2023).
- Brown, S. Machine Learning, Explained. MIT Sloan. Ideas Made to Matter. 2021. Available online: https://mitsloan.mit.edu/ideas-made-to-matter/machine-learning-explained (accessed on 4 May 2023).
- Christopher, A. K-Nearest Neighbor. Medium. The Startup. 2021. Available online: https://medium.com/swlh/k-nearest-neighbor-ca2593d7a3c4 (accessed on 10 May 2023).
- Hamed, A.; Sobhy, A.; Nassar, H. Accurate Classification of COVID-19 Based on Incomplete Heterogeneous Data Using a KNN Variant Algorithm. Arab J. Sci. Eng. 2021, 46, 8261–8272. [Google Scholar] [CrossRef]
- Bellino, G.; Schiaffino, L.; Battisti, M.; Guerrero, J.; Rosado-Muñoz, A. Optimization of the KNN Supervised Classification Algorithm as a Support Tool for the Implantation of Deep Brain Stimulators in Patients with Parkinson’s Disease. Entropy 2019, 21, 346. [Google Scholar] [CrossRef] [PubMed]
- What Is Linear Regression? IBM. Available online: https://www.ibm.com/topics/linear-regression (accessed on 11 May 2023).
- Garcia, J.M.V.; Bahloul, M.A.; Laleg-Kirati, T.-M. A Multiple Linear Regression Model for Carotid-to-Femoral Pulse Wave Velocity Estimation Based on Schrodinger Spectrum Characterization. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 143–147. [Google Scholar]
- Co to Jest Uczenie Maszynowe? Microsoft Azure. Available online: https://azure.microsoft.com/pl-pl/resources/cloud-computing-dictionary/what-is-machine-learning-platform (accessed on 11 May 2023).
- Regresja Logistyczna. IBM. Available online: https://www.ibm.com/docs/pl/spss-statistics/28.0.0?topic=regression-logistic (accessed on 14 May 2023).
- Kleinbaum, D.G.; Klein, M. Logistic Regression. In Statistics for Biology and Health; Springer: New York, NY, USA, 2010; ISBN 978-1-4419-1741-6. [Google Scholar]
- Gruszczyński, M.; Witkowski, B.; Wiśniowski, A.; Szulc, A.; Owczarczuk, M.; Książek, M.; Bazyl, M. Mikroekonometria. Modele i Metody Analizy Danych Indywidualnych; Akademicka. Ekonomia; II; Wolters Kluwer Polska SA: Gdansk, Poland, 2012; ISBN 978-83-264-5184-3. [Google Scholar]
- Naiwny Klasyfikator Bayesa. StatSoft Internetowy Podręcznik Statystyki. Available online: https://www.statsoft.pl/textbook/stathome_stat.html?https%3A%2F%2Fwww.statsoft.pl%2Ftextbook%2Fgo_search.html%3Fq%3D%25bayersa (accessed on 2 May 2023).
- Možina, M.; Demšar, J.; Kattan, M.; Zupan, B. Nomograms for Visualization of Naive Bayesian Classifier. In Knowledge Discovery in Databases: PKDD 2004; Lecture Notes in Computer Science; Boulicaut, J.-F., Esposito, F., Giannotti, F., Pedreschi, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 3202, pp. 337–348. ISBN 978-3-540-23108-0. [Google Scholar]
- Minsky, M. Steps toward Artificial Intelligence. Proc. IRE 1961, 49, 8–30. [Google Scholar] [CrossRef]
- Zhou, S. Sparse SVM for Sufficient Data Reduction. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 5560–5571. [Google Scholar] [CrossRef] [PubMed]
- Bordes, A.; Ertekin, S.; Weston, J.; Bottou, L. Fast Kernel Classifiers with Online and Active Learning. J. Mach. Learn. 2005, 6, 1579–1619. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Noble, W.S. What Is a Support Vector Machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Winters-Hilt, S.; Merat, S. SVM Clustering. BMC Bioinform. 2007, 8, S18. [Google Scholar] [CrossRef]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, J.; Hu, X.; Chen, Q.; Tu, L.; Huang, J.; Cui, J. Diagnostic Method of Diabetes Based on Support Vector Machine and Tongue Images. BioMed Res. Int. 2017, 2017, 7961494. [Google Scholar] [CrossRef] [PubMed]
- Schapire, R.E.; Freund, Y. Boosting: Foundations and Algorithms; Adaptive Computation and Machine Learning Series; MIT Press: Cambridge, MA, USA, 2012; ISBN 978-0-262-01718-3. [Google Scholar]
- Li, S.; Zeng, Y.; Chapman, W.C.; Erfanzadeh, M.; Nandy, S.; Mutch, M.; Zhu, Q. Adaptive Boosting (AdaBoost)-based Multiwavelength Spatial Frequency Domain Imaging and Characterization for Ex Vivo Human Colorectal Tissue Assessment. J. Biophotonics 2020, 13, e201960241. [Google Scholar] [CrossRef]
- Hatwell, J.; Gaber, M.M.; Atif Azad, R.M. Ada-WHIPS: Explaining AdaBoost Classification with Applications in the Health Sciences. BMC Med. Inform. Decis. Mak. 2020, 20, 250. [Google Scholar] [CrossRef] [PubMed]
- Baniasadi, A.; Rezaeirad, S.; Zare, H.; Ghassemi, M.M. Two-Step Imputation and AdaBoost-Based Classification for Early Prediction of Sepsis on Imbalanced Clinical Data. Crit. Care Med. 2021, 49, e91–e97. [Google Scholar] [CrossRef] [PubMed]
- Takemura, A.; Shimizu, A.; Hamamoto, K. Discrimination of Breast Tumors in Ultrasonic Images Using an Ensemble Classifier Based on the AdaBoost Algorithm with Feature Selection. IEEE Trans. Med. Imaging 2010, 29, 598–609. [Google Scholar] [CrossRef]
- Salcedo-Sanz, S.; Pérez-Aracil, J.; Ascenso, G.; Del Ser, J.; Casillas-Pérez, D.; Kadow, C.; Fister, D.; Barriopedro, D.; García-Herrera, R.; Restelli, M.; et al. Analysis, Characterization, Prediction and Attribution of Extreme Atmospheric Events with Machine Learning: A Review. arXiv 2022, arXiv:2207.07580. [Google Scholar] [CrossRef]
- Moore, A.; Bell, M. XGBoost, A Novel Explainable AI Technique, in the Prediction of Myocardial Infarction: A UK Biobank Cohort Study. Clin. Med. Insights Cardiol. 2022, 16, 117954682211336. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, T.; Xia, M.; Liu, Y.; Wang, Y.; Wang, X.; Zhuang, L.; Zhong, D.; Zhu, J.; He, H.; et al. Predicting the Prognosis of Patients in the Coronary Care Unit: A Novel Multi-Category Machine Learning Model Using XGBoost. Front. Cardiovasc. Med. 2022, 9, 764629. [Google Scholar] [CrossRef]
- Subha Ramakrishnan, M.; Ganapathy, N. Extreme Gradient Boosting Based Improved Classification of Blood-Brain-Barrier Drugs. In Studies in Health Technology and Informatics; Séroussi, B., Weber, P., Dhombres, F., Grouin, C., Liebe, J.-D., Pelayo, S., Pinna, A., Rance, B., Sacchi, L., Ugon, A., et al., Eds.; IOS Press: Amsterdam, The Netherlands, 2022; ISBN 978-1-64368-284-6. [Google Scholar]
- Inoue, T.; Ichikawa, D.; Ueno, T.; Cheong, M.; Inoue, T.; Whetstone, W.D.; Endo, T.; Nizuma, K.; Tominaga, T. XGBoost, a Machine Learning Method, Predicts Neurological Recovery in Patients with Cervical Spinal Cord Injury. Neurotrauma Rep. 2020, 1, 8–16. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the NIPS 2017, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar] [CrossRef]
- Shao, R.; Shi, Z.; Yi, J.; Chen, P.-Y.; Hsieh, C.-J. On the Adversarial Robustness of Vision Transformers. arXiv 2021, arXiv:2103.15670. [Google Scholar] [CrossRef]
- Qureshi, J. What Is the Difference between Neural Networks and Deep Neural Networks? Quora 2018. Available online: https://www.quora.com/What-is-the-difference-between-neural-networks-and-deep-neural-networks (accessed on 3 May 2023).
- Jeffrey, C. Explainer: What Is Machine Learning? TechSpot 2020. Available online: https://www.techspot.com/article/2048-machine-learning-explained/ (accessed on 3 May 2023).
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Tridandapani, S.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.-P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. In Deep Learning in Medical Image Analysis; Advances in Experimental Medicine and Biology; Lee, G., Fujita, H., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 1213, pp. 3–21. ISBN 978-3-030-33127-6. [Google Scholar]
- Kriegeskorte, N.; Golan, T. Neural Network Models and Deep Learning. Curr. Biol. 2019, 29, R231–R236. [Google Scholar] [CrossRef] [PubMed]
- Bajić, F.; Orel, O.; Habijan, M. A Multi-Purpose Shallow Convolutional Neural Network for Chart Images. Sensors 2022, 22, 7695. [Google Scholar] [CrossRef]
- Han, R.; Yang, Y.; Li, X.; Ouyang, D. Predicting Oral Disintegrating Tablet Formulations by Neural Network Techniques. Asian J. Pharm. Sci. 2018, 13, 336–342. [Google Scholar] [CrossRef]
- Egger, J.; Gsaxner, C.; Pepe, A.; Pomykala, K.L.; Jonske, F.; Kurz, M.; Li, J.; Kleesiek, J. Medical Deep Learning—A Systematic Meta-Review. Comput. Methods Programs Biomed. 2022, 221, 106874. [Google Scholar] [CrossRef]
- Jafari, R.; Spincemaille, P.; Zhang, J.; Nguyen, T.D.; Luo, X.; Cho, J.; Margolis, D.; Prince, M.R.; Wang, Y. Deep Neural Network for Water/Fat Separation: Supervised Training, Unsupervised Training, and No Training. Magn. Reson. Med. 2021, 85, 2263–2277. [Google Scholar] [CrossRef]
- Hou, J.-J.; Tian, H.-L.; Lu, B. A Deep Neural Network-Based Model for Quantitative Evaluation of the Effects of Swimming Training. Comput. Intell. Neurosci. 2022, 2022, 5508365. [Google Scholar] [CrossRef]
- Singh, A.; Ardakani, A.A.; Loh, H.W.; Anamika, P.V.; Acharya, U.R.; Kamath, S.; Bhat, A.K. Automated Detection of Scaphoid Fractures Using Deep Neural Networks in Radiographs. Eng. Appl. Artif. Intell. 2023, 122, 106165. [Google Scholar] [CrossRef]
- Gülmez, B. A Novel Deep Neural Network Model Based Xception and Genetic Algorithm for Detection of COVID-19 from X-Ray Images. Ann. Oper. Res. 2022. [CrossRef]
- Tsai, K.-J.; Chou, M.-C.; Li, H.-M.; Liu, S.-T.; Hsu, J.-H.; Yeh, W.-C.; Hung, C.-M.; Yeh, C.-Y.; Hwang, S.-H. A High-Performance Deep Neural Network Model for BI-RADS Classification of Screening Mammography. Sensors 2022, 22, 1160. [Google Scholar] [CrossRef]
- Sharrock, M.F.; Mould, W.A.; Ali, H.; Hildreth, M.; Awad, I.A.; Hanley, D.F.; Muschelli, J. 3D Deep Neural Network Segmentation of Intracerebral Hemorrhage: Development and Validation for Clinical Trials. Neuroinform 2021, 19, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Yuan, J.; Sodimu, O.M.; Qiang, Y.; Ding, Y. Deep Neural Network-Aided Histopathological Analysis of Myocardial Injury. Front. Cardiovasc. Med. 2022, 8, 724183. [Google Scholar] [CrossRef]
- Rajput, J.S.; Sharma, M.; Kumar, T.S.; Acharya, U.R. Automated Detection of Hypertension Using Continuous Wavelet Transform and a Deep Neural Network with Ballistocardiography Signals. IJERPH 2022, 19, 4014. [Google Scholar] [CrossRef] [PubMed]
- Voigt, I.; Boeckmann, M.; Bruder, O.; Wolf, A.; Schmitz, T.; Wieneke, H. A Deep Neural Network Using Audio Files for Detection of Aortic Stenosis. Clin. Cardiol. 2022, 45, 657–663. [Google Scholar] [CrossRef]
- Ma, L.; Yang, T. Construction and Evaluation of Intelligent Medical Diagnosis Model Based on Integrated Deep Neural Network. Comput. Intell. Neurosci. 2021, 2021, 7171816. [Google Scholar] [CrossRef]
- Ragab, M.; AL-Ghamdi, A.S.A.-M.; Fakieh, B.; Choudhry, H.; Mansour, R.F.; Koundal, D. Prediction of Diabetes through Retinal Images Using Deep Neural Network. Comput. Intell. Neurosci. 2022, 2022, 7887908. [Google Scholar] [CrossRef]
- Min, J.K.; Yang, H.-J.; Kwak, M.S.; Cho, C.W.; Kim, S.; Ahn, K.-S.; Park, S.-K.; Cha, J.M.; Park, D.I. Deep Neural Network-Based Prediction of the Risk of Advanced Colorectal Neoplasia. Gut Liver 2021, 15, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis Using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef]
- Mohamed, E.A.; Gaber, T.; Karam, O.; Rashed, E.A. A Novel CNN Pooling Layer for Breast Cancer Segmentation and Classification from Thermograms. PLoS ONE 2022, 17, e0276523. [Google Scholar] [CrossRef]
- Chamberlin, J.; Kocher, M.R.; Waltz, J.; Snoddy, M.; Stringer, N.F.C.; Stephenson, J.; Sahbaee, P.; Sharma, P.; Rapaka, S.; Schoepf, U.J.; et al. Automated Detection of Lung Nodules and Coronary Artery Calcium Using Artificial Intelligence on Low-Dose CT Scans for Lung Cancer Screening: Accuracy and Prognostic Value. BMC Med. 2021, 19, 55. [Google Scholar] [CrossRef] [PubMed]
- Alajanbi, M.; Malerba, D.; Liu, H. Distributed Reduced Convolution Neural Networks. Mesopotamian J. Big Data 2021, 2021, 26–29. [Google Scholar] [CrossRef]
- Le, Q.V. A Tutorial on Deep Learning Part 2: Autoencoders, Convolutional Neural Networks and Recurrent Neural Networks. Google Brain 2015, 20, 1–20. [Google Scholar]
- Ren, L.; Sun, Y.; Cui, J.; Zhang, L. Bearing Remaining Useful Life Prediction Based on Deep Autoencoder and Deep Neural Networks. J. Manuf. Syst. 2018, 48, 71–77. [Google Scholar] [CrossRef]
- Dertat, A. Applied Deep Learning-Part 3: Autoencoders. Medium. Towards Data Science. 2017. Available online: https://towardsdatascience.com/applied-deep-learning-part-3-autoencoders-1c083af4d798 (accessed on 3 May 2023).
- Baur, C.; Denner, S.; Wiestler, B.; Navab, N.; Albarqouni, S. Autoencoders for Unsupervised Anomaly Segmentation in Brain MR Images: A Comparative Study. Med. Image Anal. 2021, 69, 101952. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv 2022, arXiv:2105.05537. [Google Scholar] [CrossRef]
- Shamim, S.; Awan, M.J.; Mohd Zain, A.; Naseem, U.; Mohammed, M.A.; Garcia-Zapirain, B. Automatic COVID-19 Lung Infection Segmentation through Modified Unet Model. J. Healthc. Eng. 2022, 2022, 6566982. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, H.; Li, S.; Cui, Y.; Liu, Z.; Yang, G.; Hu, J. Transfer Learning with Deep Recurrent Neural Networks for Remaining Useful Life Estimation. Appl. Sci. 2018, 8, 2416. [Google Scholar] [CrossRef]
- Rios, A.; Kavuluru, R. Neural Transfer Learning for Assigning Diagnosis Codes to EMRs. Artif. Intell. Med. 2019, 96, 116–122. [Google Scholar] [CrossRef]
- Snell; Swersky, K.; Zemel, R.S. Prototypical Networks for Few-Shot Learning. In Proceedings of the NIPS 2017, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.-M.; Li, Q.; Gu, J.; Xiang, W.; Zhang, L.; Li, V.O.K. Large Margin Few-Shot Learning. arXiv 2018, arXiv:1807.02872. [Google Scholar]
- Berger, M.; Yang, Q.; Maier, A. X-ray Imaging. In Medical Imaging Systems; Lecture Notes in Computer Science; Maier, A., Steidl, S., Christlein, V., Hornegger, J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11111, pp. 119–145. ISBN 978-3-319-96519-2. [Google Scholar]
- Nie, M.; Chen, D.; Wang, D. Reinforcement Learning on Graphs: A Survey. arXiv 2022, arXiv:2204.06127. [Google Scholar] [CrossRef]
- Giacaglia, G. How Transformers Work. The Neural Network Used by Open AI and DeepMind. Towards Data Science 2019. Available online: https://towardsdatascience.com/transformers-141e32e69591 (accessed on 1 May 2023).
- Luo, X.; Gandhi, P.; Zhang, Z.; Shao, W.; Han, Z.; Chandrasekaran, V.; Turzhitsky, V.; Bali, V.; Roberts, A.R.; Metzger, M.; et al. Applying Interpretable Deep Learning Models to Identify Chronic Cough Patients Using EHR Data. Comput. Methods Programs Biomed. 2021, 210, 106395. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, J.; Hou, L.; Zhai, Y.; Shi, J.; Cui, F. An Attention-Based Deep Learning Model for Clinical Named Entity Recognition of Chinese Electronic Medical Records. BMC Med. Inform. Decis. Mak. 2019, 19, 235. [Google Scholar] [CrossRef] [PubMed]
- Available online. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf21/K212516.pdf (accessed on 5 May 2023).
- Asch, F.M.; Descamps, T.; Sarwar, R.; Karagodin, I.; Singulane, C.C.; Xie, M.; Tucay, E.S.; Tude Rodrigues, A.C.; Vasquez-Ortiz, Z.Y.; Monaghan, M.J.; et al. Human versus Artificial Intelligence–Based Echocardiographic Analysis as a Predictor of Outcomes: An Analysis from the World Alliance Societies of Echocardiography COVID Study. J. Am. Soc. Echocardiogr. 2022, 35, 1226–1237.e7. [Google Scholar] [CrossRef]
- Aidoc. Available online: https://www.aidoc.com/solutions/radiology/ (accessed on 5 May 2023).
- Riverain Technologies. Available online: https://www.riveraintech.com/clearread-ai-solutions/clearread-ct/ (accessed on 5 May 2023).
- Alzubaidi, L.; Bai, J.; Al-Sabaawi, A.; Santamaría, J.; Albahri, A.S.; Al-dabbagh, B.S.N.; Fadhel, M.A.; Manoufali, M.; Zhang, J.; Al-Timemy, A.H.; et al. A Survey on Deep Learning Tools Dealing with Data Scarcity: Definitions, Challenges, Solutions, Tips, and Applications. J. Big Data 2023, 10, 46. [Google Scholar] [CrossRef]
- Albahri, A.S.; Duhaim, A.M.; Fadhel, M.A.; Alnoor, A.; Baqer, N.S.; Alzubaidi, L.; Albahri, O.S.; Alamoodi, A.H.; Bai, J.; Salhi, A.; et al. A Systematic Review of Trustworthy and Explainable Artificial Intelligence in Healthcare: Assessment of Quality, Bias Risk, and Data Fusion. Inf. Fusion 2023, 96, 156–191. [Google Scholar] [CrossRef]
- Hephzipah, J.J.; Vallem, R.R.; Sheela, M.S.; Dhanalakshmi, G. An Efficient Cyber Security System Based on Flow-Based Anomaly Detection Using Artificial Neural Network. Mesopotamian J. Cybersecur. 2023, 2023, 48–56. [Google Scholar] [CrossRef]
- Oliver, M.; Renou, A.; Allou, N.; Moscatelli, L.; Ferdynus, C.; Allyn, J. Image Augmentation and Automated Measurement of Endotracheal-Tube-to-Carina Distance on Chest Radiographs in Intensive Care Unit Using a Deep Learning Model with External Validation. Crit. Care 2023, 27, 40. [Google Scholar] [CrossRef]
- Moon, J.-H.; Hwang, H.-W.; Yu, Y.; Kim, M.-G.; Donatelli, R.E.; Lee, S.-J. How Much Deep Learning Is Enough for Automatic Identification to Be Reliable? Angle Orthod. 2020, 90, 823–830. [Google Scholar] [CrossRef]
- Albrecht, T.; Slabaugh, G.; Alonso, E.; Al-Arif, S.M.R. Deep Learning for Single-Molecule Science. Nanotechnology 2017, 28, 423001. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, A.; Sudjianto, A. Enhancing Explainability of Neural Networks Through Architecture Constraints. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Stock, P.; Cisse, M. ConvNets and ImageNet Beyond Accuracy: Understanding Mistakes and Uncovering Biases. arXiv 2017, arXiv:1711.11443. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, L.; Zeng, B.; Li, J.; Guo, K.; Liang, Y.; Liao, G. Artificial Intelligence Performance in Detecting Tumor Metastasis from Medical Radiology Imaging: A Systematic Review and Meta-Analysis. EClinicalMedicine 2021, 31, 100669. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Rahim, S.; Zubair, M.; Abdul-Ghafar, J. Artificial Intelligence (AI) in Medicine, Current Applications and Future Role with Special Emphasis on Its Potential and Promise in Pathology: Present and Future Impact, Obstacles Including Costs and Acceptance among Pathologists, Practical and Philosophical Considerations. A Comprehensive Review. Diagn. Pathol. 2021, 16, 24. [Google Scholar] [CrossRef]
- Syed, A.; Zoga, A. Artificial Intelligence in Radiology: Current Technology and Future Directions. Semin. Musculoskelet. Radiol. 2018, 22, 540–545. [Google Scholar] [CrossRef]
- McDougall, R.J. Computer Knows Best? The Need for Value-Flexibility in Medical AI. J. Med. Ethics 2019, 45, 156–160. [Google Scholar] [CrossRef]
- Dave, M.; Patel, N. Artificial Intelligence in Healthcare and Education. Br. Dent. J. 2023, 234, 761–764. [Google Scholar] [CrossRef]













Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kufel, J.; Bargieł-Łączek, K.; Kocot, S.; Koźlik, M.; Bartnikowska, W.; Janik, M.; Czogalik, Ł.; Dudek, P.; Magiera, M.; Lis, A.; et al. What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine. Diagnostics 2023, 13, 2582. https://doi.org/10.3390/diagnostics13152582
Kufel J, Bargieł-Łączek K, Kocot S, Koźlik M, Bartnikowska W, Janik M, Czogalik Ł, Dudek P, Magiera M, Lis A, et al. What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine. Diagnostics. 2023; 13(15):2582. https://doi.org/10.3390/diagnostics13152582
Chicago/Turabian StyleKufel, Jakub, Katarzyna Bargieł-Łączek, Szymon Kocot, Maciej Koźlik, Wiktoria Bartnikowska, Michał Janik, Łukasz Czogalik, Piotr Dudek, Mikołaj Magiera, Anna Lis, and et al. 2023. "What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine" Diagnostics 13, no. 15: 2582. https://doi.org/10.3390/diagnostics13152582
APA StyleKufel, J., Bargieł-Łączek, K., Kocot, S., Koźlik, M., Bartnikowska, W., Janik, M., Czogalik, Ł., Dudek, P., Magiera, M., Lis, A., Paszkiewicz, I., Nawrat, Z., Cebula, M., & Gruszczyńska, K. (2023). What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine. Diagnostics, 13(15), 2582. https://doi.org/10.3390/diagnostics13152582










