Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer
Abstract
:1. Introduction
2. PD-L1 Expression as a Biomarker in GC
2.1. Rationale and Performance
2.2. Interpretation of PD-L1 IHC Assays in GC
2.3. Interchangeability of PD-L1 Assays in GC
2.4. Discordance between Biopsy and Resection Specimens and Inter-Observer Variation
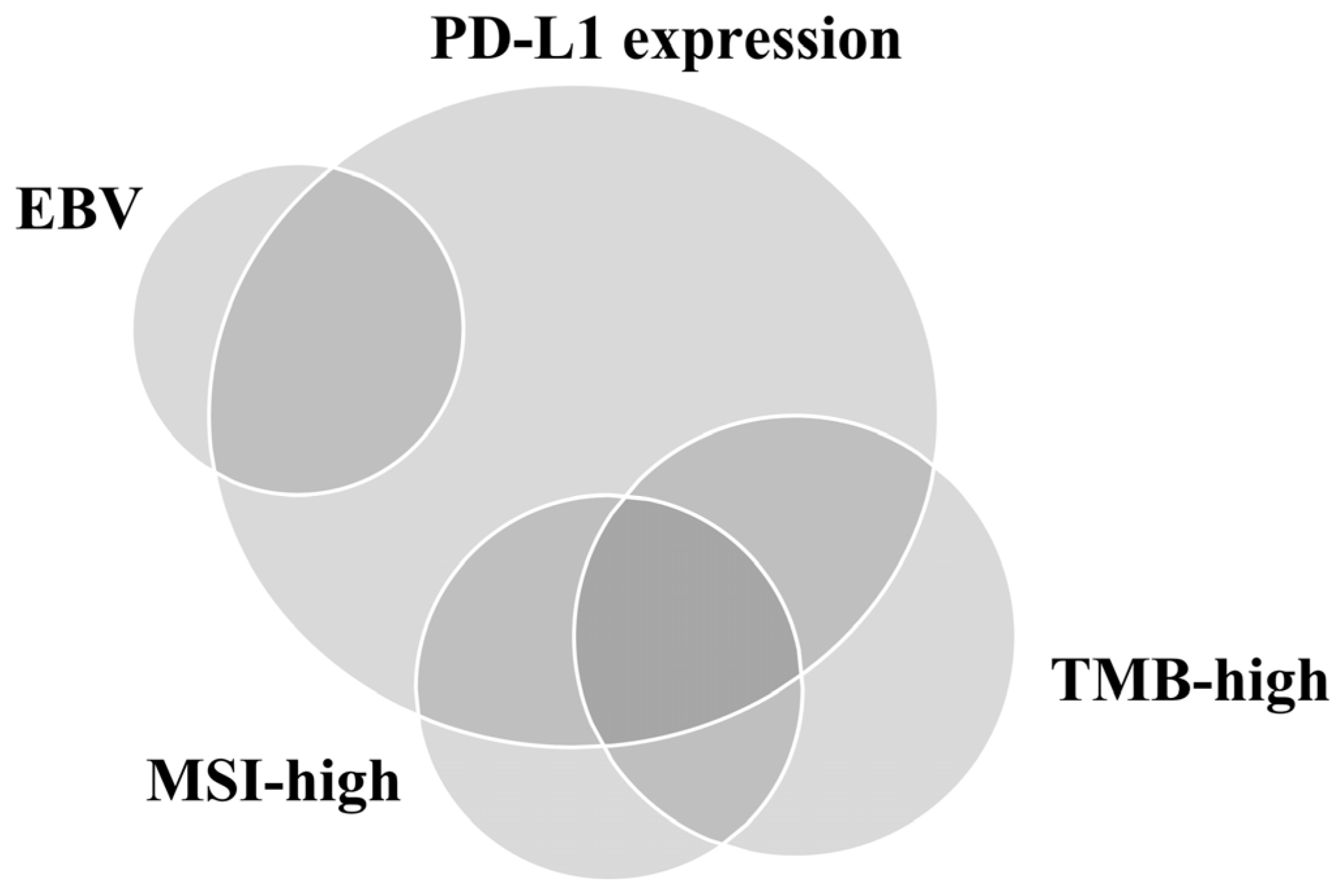
3. Other Biomarkers Associated with the Immune Microenvironment (IME) and Immunotherapy in GC
3.1. MSI/MMR-Deficiency (MMR-D)
3.2. EBV
3.3. TMB
4. Potential Predictive Biomarkers
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Na Seo, A. Molecular Pathology of Gastric Cancer. J. Gastric Cancer 2022, 22, 273–305. [Google Scholar] [CrossRef] [PubMed]
- Gullo, I.; Carneiro, F.; Oliveira, C.; Almeida, G.M. Heterogeneity in Gastric Cancer: From Pure Morphology to Molecular Classifications. Pathobiology 2018, 85, 50–63. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Doi, T.; Jang, R.W.; Muro, K.; Satoh, T.; Machado, M.; Sun, W.; Jalal, S.I.; Shah, M.A.; Metges, J.-P.; et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients with Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol. 2018, 4, e180013. [Google Scholar] [CrossRef] [PubMed]
- Shitara, K.; Özgüroğlu, M.; Bang, Y.-J.; Di Bartolomeo, M.; Mandalà, M.; Ryu, M.-H.; Fornaro, L.; Olesiński, T.; Caglevic, C.; Muro, K.; et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): A randomised, open-label, controlled, phase 3 trial. Lancet 2018, 392, 123–133. [Google Scholar] [CrossRef]
- Shitara, K.; Van Cutsem, E.; Bang, Y.-J.; Fuchs, C.; Wyrwicz, L.; Lee, K.-W.; Kudaba, I.; Garrido, M.; Chung, H.C.; Lee, J.; et al. Efficacy and Safety of Pembrolizumab or Pembrolizumab Plus Chemotherapy vs Chemotherapy Alone for Patients with First-line, Advanced Gastric Cancer: The KEYNOTE-062 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1571–1580. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Bragagnoli, A.C.; et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Kawazoe, A.; Yañez, P.; Li, N.; Lonardi, S.; Kolesnik, O.; Barajas, O.; Bai, Y.; Shen, L.; Tang, Y.; et al. The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer. Nature 2021, 600, 727–730. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef]
- Kim, H.; Chung, J.-H. PD-L1 Testing in Non-small Cell Lung Cancer: Past, Present, and Future. J. Pathol. Transl. Med. 2019, 53, 199–206. [Google Scholar] [CrossRef]
- Kwak, Y.; Na Seo, A.; Lee, H.E.; Lee, H.S. Tumor immune response and immunotherapy in gastric cancer. J. Pathol. Transl. Med. 2020, 54, 20–33. [Google Scholar] [CrossRef]
- Cho, Y.A.; Lee, H.; Kim, D.G.; Kim, H.; Ha, S.Y.; Choi, Y.-L.; Jang, K.-T.; Kim, K.-M. PD-L1 Expression Is Significantly Associated with Tumor Mutation Burden and Microsatellite Instability Score. Cancers 2021, 13, 4659. [Google Scholar] [CrossRef]
- Kroeze, L.I.; de Voer, R.M.; Kamping, E.J.; von Rhein, D.; Jansen, E.A.; Hermsen, M.J.; Barberis, M.C.; Botling, J.; Garrido-Martin, E.M.; Haller, F.; et al. Evaluation of a Hybrid Capture–Based Pan-Cancer Panel for Analysis of Treatment Stratifying Oncogenic Aberrations and Processes. J. Mol. Diagn. 2020, 22, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.A.; D’amico, T.A.; Bentrem, D.J.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 167–192. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A. Tumor Immunotherapy Directed at PD-1. N. Engl. J. Med. 2012, 366, 2517–2519. [Google Scholar] [CrossRef]
- Kim, B.; Kang, S.Y.; Kim, K.-M. DNA-protein biomarkers for immunotherapy in the era of precision oncology. J. Pathol. Transl. Med. 2021, 55, 26–32. [Google Scholar] [CrossRef]
- Park, Y.S.; Kook, M.-C.; Kim, B.-H.; Lee, H.S.; Kang, D.-W.; Gu, M.-J.; Shin, O.R.; Choi, Y.; Lee, W.; Kim, H.; et al. A Standardized Pathology Report for Gastric Cancer: 2nd Edition. J. Gastric Cancer 2023, 23, 107–145. [Google Scholar] [CrossRef]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Zou, W.; Wolchok, J.D.; Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016, 8, 328rv4. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.M.; Hwu, W.-J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and Activity of Anti-PD-L1 Antibody in Patients with Advanced Cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-K.; Chen, L.-T.; Ryu, M.-H.; Oh, D.-Y.; Oh, S.C.; Chung, H.C.; Lee, K.-W.; Omori, T.; Shitara, K.; Sakuramoto, S.; et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): A randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Rashid, S.; Al-Bozom, I.A. PD–L1 immunostaining: What pathologists need to know. Diagn. Pathol. 2021, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fang, F.; Kong, Y.; ElGabry, E.A. Tumor Area Positivity (TAP) score of programmed death-ligand 1 (PD-L1): A novel visual estimation method for combined tumor cell and immune cell scoring. Diagn. Pathol. 2023, 18, 48. [Google Scholar] [CrossRef]
- Ahn, S.; Kim, K.-M. PD-L1 expression in gastric cancer: Interchangeability of 22C3 and 28-8 pharmDx assays for responses to immunotherapy. Mod. Pathol. 2021, 34, 1719–1727. [Google Scholar] [CrossRef]
- Narita, Y.; Sasaki, E.; Masuishi, T.; Taniguchi, H.; Kadowaki, S.; Ito, S.; Yatabe, Y.; Muro, K. PD-L1 immunohistochemistry comparison of 22C3 and 28-8 assays for gastric cancer. J. Gastrointest. Oncol. 2021, 12, 2696–2705. [Google Scholar] [CrossRef]
- Yeong, J.; Lum, H.Y.J.; Teo, C.B.; Tan, B.K.J.; Chan, Y.H.; Tay, R.Y.K.; Choo, J.R.-E.; Jeyasekharan, A.D.; Miow, Q.H.; Loo, L.-H.; et al. Choice of PD-L1 immunohistochemistry assay influences clinical eligibility for gastric cancer immunotherapy. Gastric Cancer 2022, 25, 741–750. [Google Scholar] [CrossRef]
- Kim, S.-W.; Jeong, G.; Ryu, M.-H.; Park, Y.S. Comparison of PD-L1 immunohistochemical assays in advanced gastric adenocarcinomas using endoscopic biopsy and paired resected specimens. Pathology 2021, 53, 586–594. [Google Scholar] [CrossRef]
- Heo, Y.J.; Kim, B.; Kim, H.; Kim, S.; Jang, M.S.; Kim, K.-M. PD-L1 expression in paired biopsies and surgical specimens in gastric adenocarcinoma: A digital image analysis study. Pathol. Res. Pract. 2021, 218, 153338. [Google Scholar] [CrossRef]
- Yamashita, K.; Iwatsuki, M.; Harada, K.; Koga, Y.; Kiyozumi, Y.; Eto, K.; Hiyoshi, Y.; Ishimoto, T.; Iwagami, S.; Baba, Y.; et al. Can PD-L1 expression evaluated by biopsy sample accurately reflect its expression in the whole tumour in gastric cancer? Br. J. Cancer 2019, 121, 278–280. [Google Scholar] [CrossRef]
- Schoemig-Markiefka, B.; Eschbach, J.; Scheel, A.H.; Pamuk, A.; Rueschoff, J.; Zander, T.; Buettner, R.; Schroeder, W.; Bruns, C.J.; Loeser, H.; et al. Optimized PD-L1 scoring of gastric cancer. Gastric Cancer 2021, 24, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Koh, J.; Na, H.Y.; Kwak, Y.; Lee, K.-W.; Ahn, S.-H.; Park, D.J.; Kim, H.-H.; Lee, H.S. PD-L1 Testing in Gastric Cancer by the Combined Positive Score of the 22C3 PharmDx and SP263 Assay with Clinically Relevant Cut-offs. Cancer Res. Treat. 2020, 52, 661–670. [Google Scholar] [CrossRef]
- Sahin, I.H.; Akce, M.; Alese, O.; Shaib, W.; Lesinski, G.B.; El-Rayes, B.; Wu, C. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br. J. Cancer 2019, 121, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Dedeurwaerdere, F.; Claes, K.B.; Van Dorpe, J.; Rottiers, I.; Van der Meulen, J.; Breyne, J.; Swaerts, K.; Martens, G. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 2021, 11, 12880. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D.; Mitchell, M.; Sgouros, J.; Lindahl, T. Human DNA Repair Genes. Science 2001, 291, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Nicolaides, N.C.; Markowitz, S.; Willson, J.K.; Parsons, R.E.; Jen, J.; Papadopolous, N.; Peltomäki, P.; de la Chapelle, A.; Hamilton, S.R.; et al. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat. Genet. 1995, 9, 48–55. [Google Scholar] [CrossRef]
- Bae, J.M.; Yoo, S.-Y.; Kim, J.H.; Kang, G.H. Immune landscape and biomarkers for immuno-oncology in colorectal cancers. J. Pathol. Transl. Med. 2020, 54, 351–360. [Google Scholar] [CrossRef]
- Dudley, J.C.; Lin, M.-T.; Le, D.T.; Eshleman, J.R. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin. Cancer Res. 2016, 22, 813–820. [Google Scholar] [CrossRef]
- Rüschoff, J.; Baretton, G.; Bläker, H.; Dietmaier, W.; Dietel, M.; Hartmann, A.; Horn, L.-C.; Jöhrens, K.; Kirchner, T.; Knüchel, R.; et al. MSI testing. Der Pathol. 2021, 42, 110–118. [Google Scholar] [CrossRef]
- Le, D.T.; Kim, T.W.; Van Cutsem, E.; Geva, R.; Jäger, D.; Hara, H.; Burge, M.; O’neil, B.; Kavan, P.; Yoshino, T.; et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability—High/Mismatch Repair—Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2020, 38, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair—Deficient Cancer: Results from the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Fuchs, C.S.; Shitara, K.; Tabernero, J.; Muro, K.; Van Cutsem, E.; Bang, Y.-J.; De Vita, F.; Landers, G.; Yen, C.-J.; et al. Assessment of Pembrolizumab Therapy for the Treatment of Microsatellite Instability—High Gastric or Gastroesophageal Junction Cancer Among Patients in the KEYNOTE-059, KEYNOTE-061, and KEYNOTE-062 Clinical Trials. JAMA Oncol. 2021, 7, 895–902. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network; Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Setia, N.; Agoston, A.T.; Han, H.S.; Mullen, J.T.; Duda, D.G.; Clark, J.W.; Deshpande, V.; Mino-Kenudson, M.; Srivastava, A.; Lennerz, J.K.; et al. A protein and mRNA expression-based classification of gastric cancer. Mod. Pathol. 2016, 29, 772–784. [Google Scholar] [CrossRef]
- Ahn, S.; Lee, S.-J.; Kim, Y.; Kim, A.; Shin, N.; Choi, K.U.; Lee, C.-H.; Huh, G.Y.; Kim, K.-M.; Setia, N.; et al. High-throughput Protein and mRNA Expression–based Classification of Gastric Cancers Can Identify Clinically Distinct Subtypes, Concordant With Recent Molecular Classifications. Am. J. Surg. Pathol. 2017, 41, 106–115. [Google Scholar] [CrossRef]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.-M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Fukayama, M.; Abe, H.; Kunita, A.; Shinozaki-Ushiku, A.; Matsusaka, K.; Ushiku, T.; Kaneda, A. Thirty years of Epstein-Barr virus-associated gastric carcinoma. Virchows Arch. 2020, 476, 353–365. [Google Scholar] [CrossRef]
- Sakuma, K.; Chong, J.-M.; Sudo, M.; Ushiku, T.; Inoue, Y.; Shibahara, J.; Uozaki, H.; Nagai, H.; Fukayama, M. High-density methylation ofp14ARF andp16INK4A in Epstein-Barr virus-associated gastric carcinoma. Int. J. Cancer 2004, 112, 273–278. [Google Scholar] [CrossRef]
- Böger, C.; Krüger, S.; Behrens, H.M.; Bock, S.; Haag, J.; Kalthoff, H.; Röcken, C. Epstein–Barr virus-associated gastric cancer reveals intratumoral heterogeneity of PIK3CA mutations. Ann. Oncol. 2017, 28, 1005–1014. [Google Scholar] [CrossRef]
- Kim, H.-N.; Ahn, S.; Kim, K.-M. Gastric cancer with Epstein-Barr virus heterogeneity: Evaluation of the frequency, clinicopathologic features, and genomic profiles. Pathol. Res. Pract. 2022, 238, 154108. [Google Scholar] [CrossRef]
- Fancello, L.; Gandini, S.; Pelicci, P.G.; Mazzarella, L. Tumor mutational burden quantification from targeted gene panels: Major advancements and challenges. J. Immunother. Cancer 2019, 7, 183. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Wagle, N.; Stojanov, P.; Perrin, D.L.; Cibulskis, K.; Marlow, S.; Jane-Valbuena, J.; Friedrich, D.C.; Kryukov, G.; Carter, S.L.; et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014, 20, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Meléndez, B.; Van Campenhout, C.; Rorive, S.; Remmelink, M.; Salmon, I.; D’haene, N. Methods of measurement for tumor mutational burden in tumor tissue. Transl. Lung Cancer Res. 2018, 7, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.M.; McShane, L.M.; Fabrizio, D.; Funari, V.; Chen, S.J.; White, J.R.; Wenz, P.; Baden, J.; Barrett, J.C.; Chaudhary, R.; et al. Establishing guidelines to harmonize tumor mutational burden (TMB): In silico assessment of variation in TMB quantification across diagnostic platforms: Phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer 2020, 8, e000147. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, A.; Sholl, L.; Reardon, B.; Taylor-Weiner, A.; Amin-Mansour, A.; Miao, D.; Liu, D.; Oliver, N.; MacConaill, L.; Ducar, M.; et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016, 8, 79. [Google Scholar] [CrossRef]
- Kim, S.; Park, C.; Ji, Y.; Kim, D.G.; Bae, H.; van Vrancken, M.; Kim, D.-H.; Kim, K.-M. Deamination Effects in Formalin-Fixed, Paraffin-Embedded Tissue Samples in the Era of Precision Medicine. J. Mol. Diagn. 2017, 19, 137–146. [Google Scholar] [CrossRef]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Lee, K.-W.; Van Cutsem, E.; Bang, Y.-J.; Fuchs, C.S.; Kudaba, I.; Garrido, M.; Chung, H.C.; Lee, J.; Castro, H.R.; Chao, J.; et al. Association of Tumor Mutational Burden with Efficacy of Pembrolizumab ± Chemotherapy as First-Line Therapy for Gastric Cancer in the Phase III KEYNOTE-062 Study. Clin. Cancer Res. 2022, 28, 3489–3498. [Google Scholar] [CrossRef] [PubMed]
- Sundar, R.; Huang, K.; Qamra, A.; Kim, K.-M.; Kim, S.; Kang, W.; Tan, A.; Lee, J.; Tan, P. Epigenomic promoter alterations predict for benefit from immune checkpoint inhibition in metastatic gastric cancer. Ann. Oncol. 2019, 30, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Qamra, A.; Xing, M.; Padmanabhan, N.; Kwok, J.J.T.; Zhang, S.; Xu, C.; Leong, Y.S.; Lim, A.P.L.; Tang, Q.; Ooi, W.F.; et al. Epigenomic Promoter Alterations Amplify Gene Isoform and Immunogenic Diversity in Gastric Adenocarcinoma. Cancer Discov. 2017, 7, 630–651. [Google Scholar] [CrossRef]
- Kim, J.; Kim, B.; Kang, S.Y.; Heo, Y.J.; Park, S.H.; Kim, S.T.; Kang, W.K.; Lee, J.; Kim, K.-M. Tumor Mutational Burden Determined by Panel Sequencing Predicts Survival After Immunotherapy in Patients with Advanced Gastric Cancer. Front. Oncol. 2020, 10, 314. [Google Scholar] [CrossRef] [PubMed]
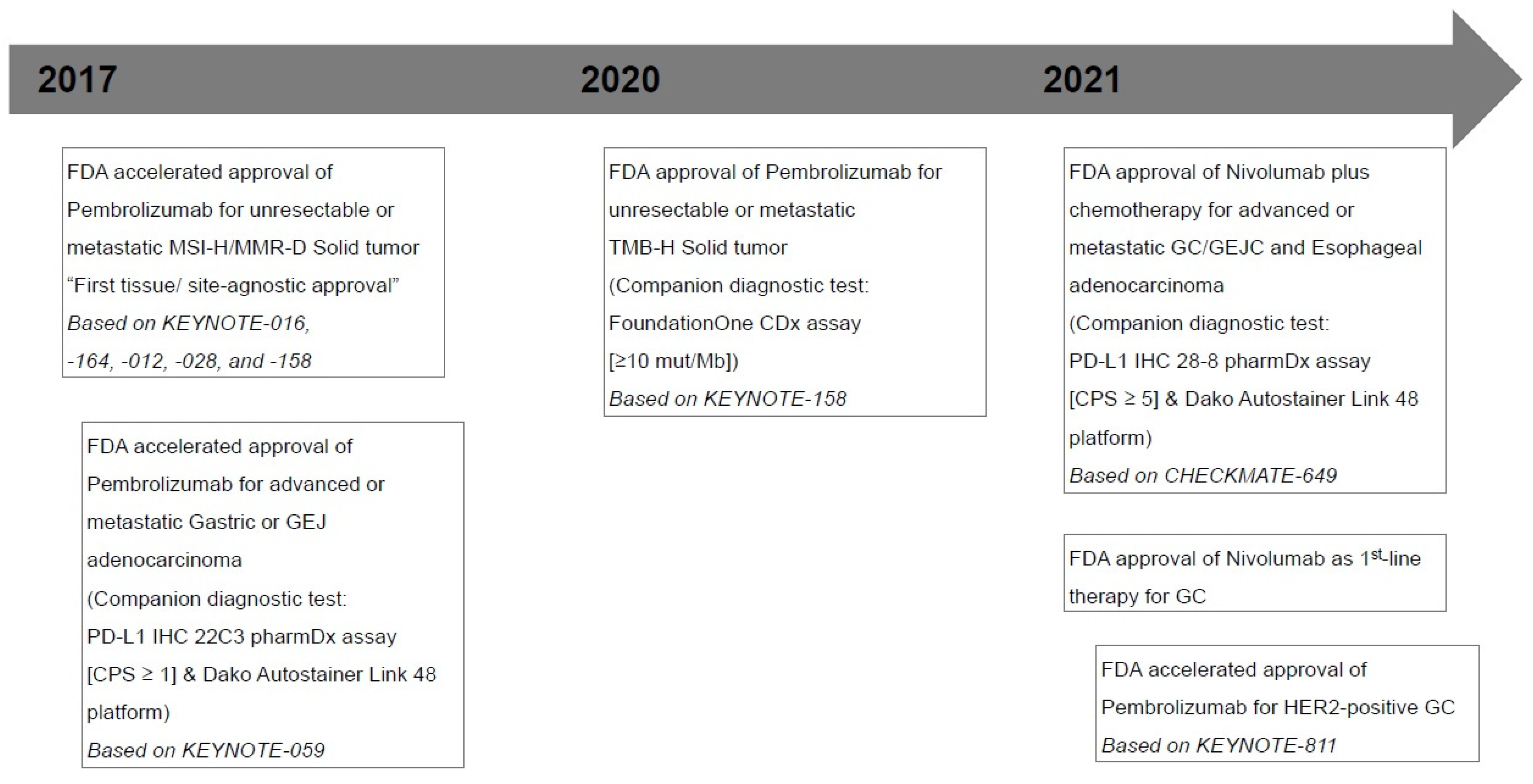
| Trials | Clone | Study Design | Number of Patients | Efficacy Findings |
|---|---|---|---|---|
| Pembrolizumab | ||||
| KEYNOTE-059 | phase 2, global, open-label, single-arm, multicohort | 259 (148 with CPS ≥ 1) | Objective response rate (ORR) (CR + PR) 11.6% | |
| [4] | Recurrent or metastatic G/GEJ cancer | Median response duration 8.4 (1.6+ to 17.3+) | ||
| pembrolizumab, 200 mg, 3rd-line | Median PFS 2.0 months | |||
| Median OS 5.6 months | ||||
| KEYNOTE-061 | phase 3, global, randomized, open-label, multicohort | 592 (395 with CPS ≥ 1) | Pembrolizumab vs. Paclitaxel | |
| [5] | PD-L1 IHC 22C3 | Advanced/unresectable or metastatic G/GEJ cancer | Median PFS 1.5 vs. 4.1 months | |
| pharmDx assay | pembrolizumab, 200 mg, 2nd-line | Median OS 9.1 vs. 8.3 months | ||
| KEYNOTE-062 | CPS ≥ 1 | phase 3, global, randomized, controlled, partially blind | 763 with CPS ≥ 1 | Pembrolizumab vs. Chemotherapy (CTx) |
| [6] | Advanced/unresectable or metastatic G/GEJ cancer | (281 with CPS ≥ 10) | Median OS 10.6 vs. 11.1 months | |
| pembrolizumab, 200 mg, 1st-line | Median PFS 2.0 vs. 6.4 months | |||
| Pembrolizumab + CTx vs. CTx | ||||
| Median OS 12.5 vs. 11.1 months | ||||
| Median PFS 6.9 vs. 6.4 months | ||||
| KEYNOTE-811 | phase 3, global, randomized, placebo-controlled, double-blind | 692 (582 with CPS ≥ 1) | Pembrolizumab vs. Placebo | |
| [8] | HER2-positive unresectable or metastatic G/GEJ cancer | ORR 74% vs. 52% (p < 0.0001) | ||
| pembrolizumab, 200 mg, 1st-line | Median duration of response 10.6 vs. 9.5 months | |||
| or placebo (normal saline or dextrose) | ||||
| Nivolumab | ||||
| CheckMate-649 | PD-L1 IHC 28-8 | phase 3, randomized, open-label, multicenter | 1581 (955 with CPS ≥ 5) | Nivolumab + CTx vs. CTx (patients with CPS ≥ 5) |
| [7] | pharmDx assay | Untreated, unresectable, HER2-negative | Median OS 14.4 vs. 11.1 months (p < 0.0001) | |
| CPS ≥ 5 | G/GEJ or esophageal adenocarcinoma | Nivolumab + CTx vs. CTx (All randomized patients) | ||
| Nivolumab, 360 mg or 240 mg, 1st-line | Median OS 13.8 vs. 11.6 months (p = 0.0002) | |||
| ATTRACTION-4 | PD-L1 IHC 28-8 | phase 2-3, randomized, double-blind, placebo-controlled, | 724 (114 with TPS ≥ 1) | Nivolumab + CTx vs. CTx |
| [22] | pharmDx assay | multicenter across Japan, South Korea, and Taiwan | Median PFS 10.45 versus 8.34 months (p = 0.0007) | |
| TPS ≥ 1 | HER2-negative, unresectable, advanced or recurrent | Median OS 17.45 vs. 17.15 months (p = 0.26) | ||
| G/GEJ cancer | ||||
| Nivolumab, 360 mg, 1st-line |
| Predictive Markers | Comments |
|---|---|
| PD-L1 testing | |
| Immunohistochemistry (Clone) | Platform: Autostainer Link 48 |
| 22C3 | Pembrolizumab (Companion diagnostic) |
| 28-8 | Nivolumab (Companion diagnostic) |
| MSI testing | |
| Immunohistochemistry | Four MMR proteins: MLH1, MSH2, PMS2, and MSH6 |
| MMR-D is determined in the absence of nuclear expression of at least one MMR protein. | |
| Polymerase chain reaction | Bethesda/National Cancer Institute panel: |
| Two mononucleotide (BAT-25 and BAT-26) & Three dinucleotide (D5S346, D2S123, and D17S250) | |
| Five poly-A mononucleotide panel: | |
| NR-21, NR-24, NR-27 [or Mono-27], BAT-25, and BAT-26 | |
| The five poly-A panel shows higher sensitivity and specificity. | |
| MSI-H is determined as instability of two or more of five microsatellite loci. | |
| Next-generation sequencing (NGS) | The major advantage of NGS is that MSI analysis and TMB determination can be performed simultaneously. |
| EBV testing | |
| In situ hybridization | It is the most suitable and widely used method to identify EBV in FFPE specimens. |
| TMB testing | |
| Whole exome sequencing | Optimal for evaluating TMB measurements |
| Targeted sequencing | Lower sequencing cost, shorter turn-around time, and lower DNA input amounts |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Jeong, J.Y.; Seo, A.N. Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer. Diagnostics 2023, 13, 2782. https://doi.org/10.3390/diagnostics13172782
Kim M, Jeong JY, Seo AN. Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer. Diagnostics. 2023; 13(17):2782. https://doi.org/10.3390/diagnostics13172782
Chicago/Turabian StyleKim, Moonsik, Ji Yun Jeong, and An Na Seo. 2023. "Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer" Diagnostics 13, no. 17: 2782. https://doi.org/10.3390/diagnostics13172782
APA StyleKim, M., Jeong, J. Y., & Seo, A. N. (2023). Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer. Diagnostics, 13(17), 2782. https://doi.org/10.3390/diagnostics13172782







