Early Melanoma Detection Based on a Hybrid YOLOv5 and ResNet Technique
Abstract
:1. Introduction
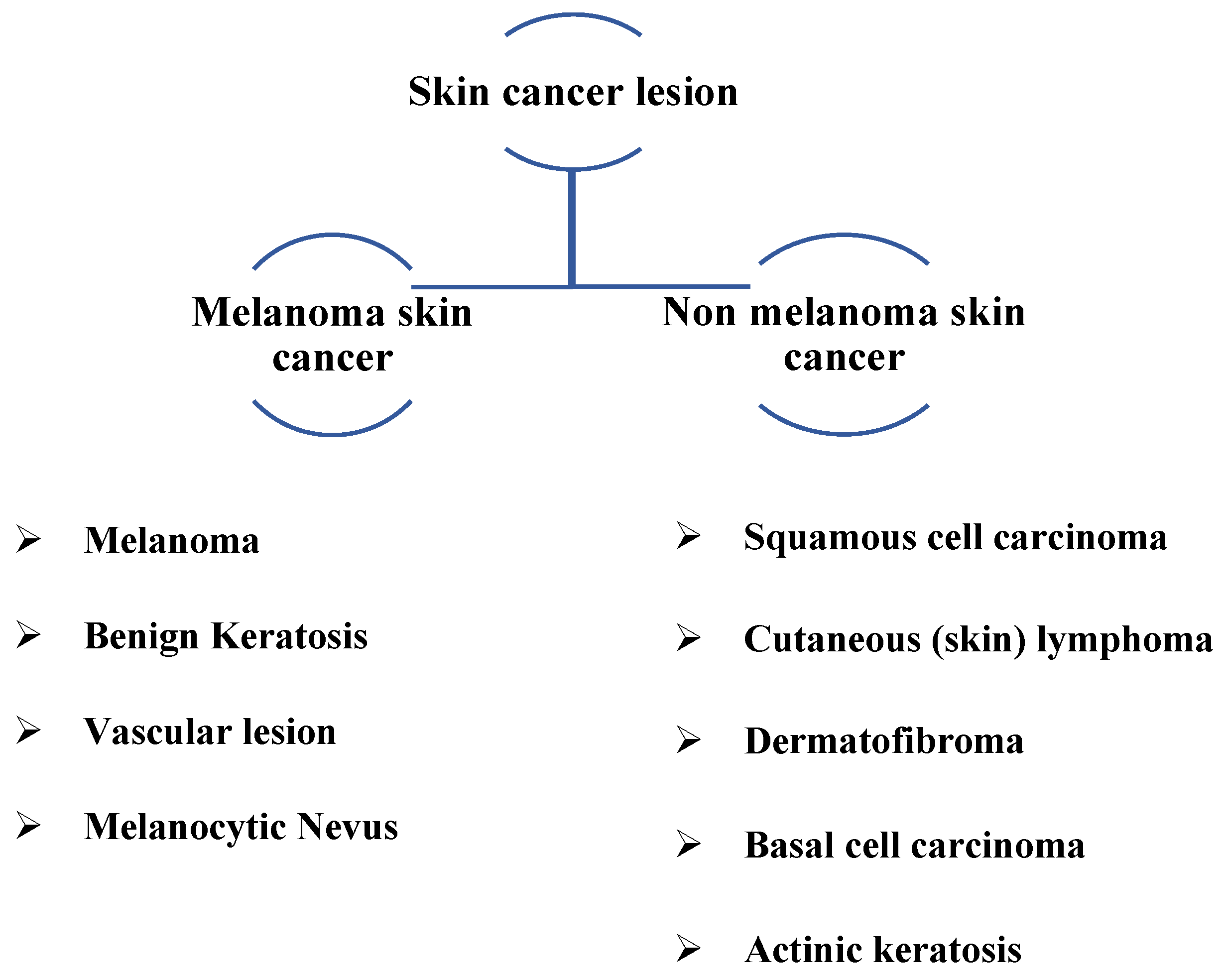
Characteristics of Lesions
- The proposed method applies to any image (dermoscopy or photographic) of pigmented skin lesions using you only look once (YOLOv5) and ResNet.
- The suggested system classifies samples and determines each class with probability.
- It interacted directly with the skin-color images that were obtained with different sizes.
- The proposed approach enhances scalability since YOLO and ResNet can detect melanoma in huge datasets. This is crucial as it allows for developing more precise and effective melanoma detection systems.
2. Related Work
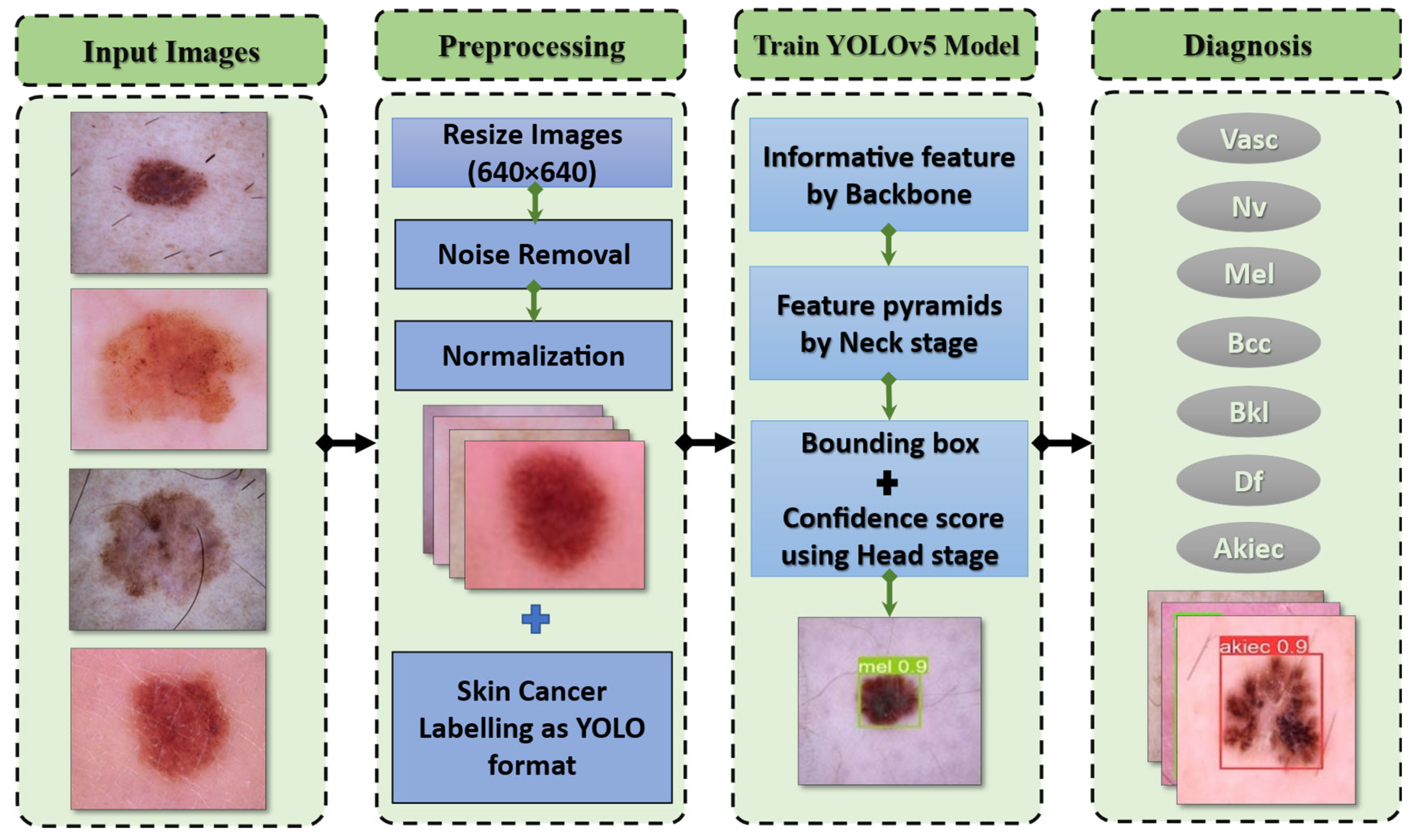
3. Proposed Melanoma Detection Technique
3.1. Dataset
3.2. Experimental Platform
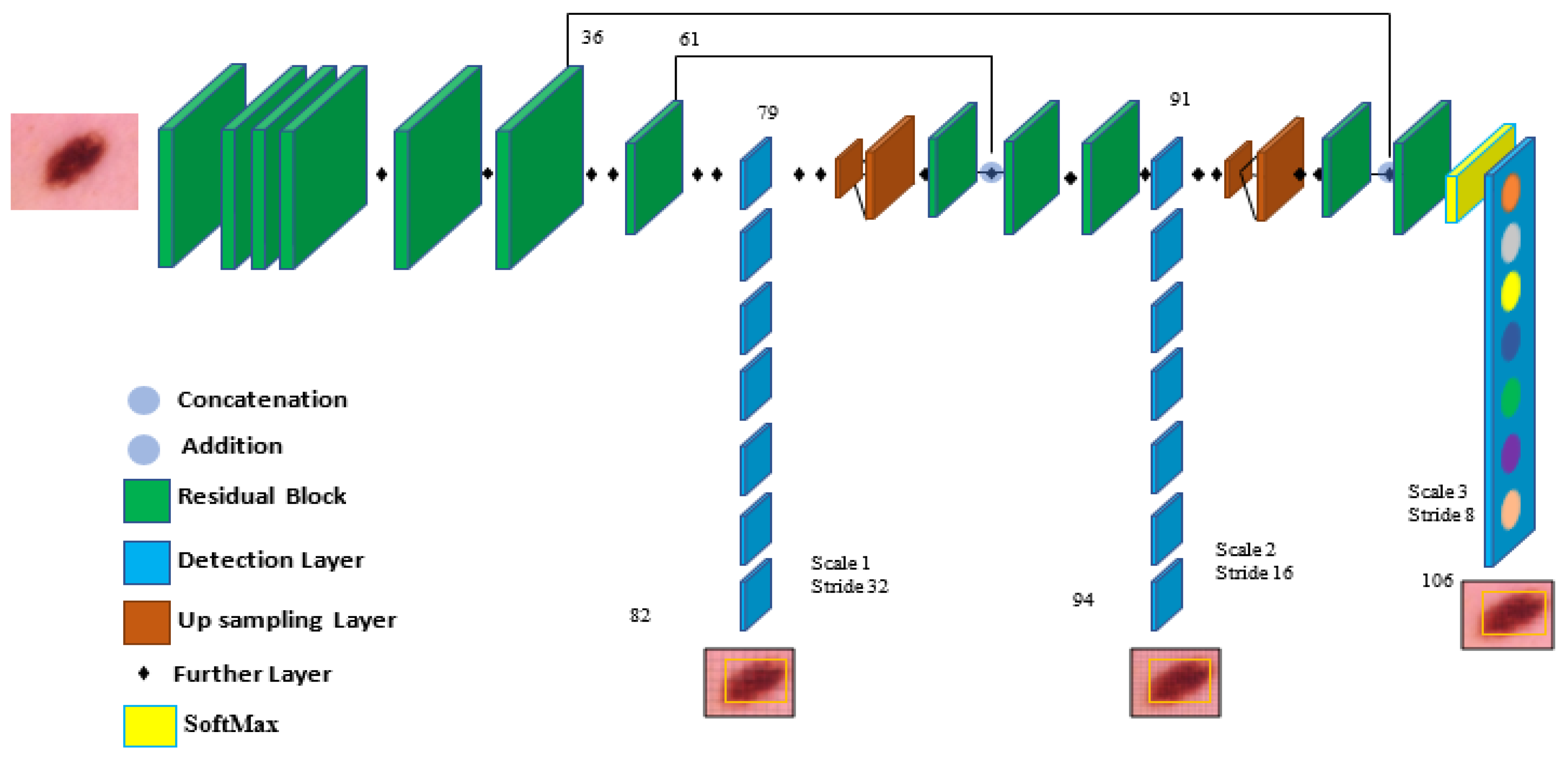
3.3. The Structure of the YOLOv5-S Model
3.4. Preprocessing
4. Experimental Results
4.1. Performance Metrics
4.2. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, S. Biochemical, Structural and Physical Changes in Aging Human Skin, and Their Relationship. Biogerontology 2022, 23, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tsui, Y.Y.; Mandal, M. Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. J. Imaging 2021, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Islami, F.; Guerra, C.E.; Minihan, A.; Yabroff, K.R.; Fedewa, S.A.; Sloan, K.; Wiedt, T.L.; Thomson, B.; Siegel, R.L.; Nargis, N. American Cancer Society’s Report on the Status of Cancer Disparities in the United States, 2021. CA A Cancer J. Clin. 2022, 72, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Saleem, S.M.; Abdullah, A.; Ameen, S.Y.; Sadeeq, M.A.M.; Zeebaree, S.R.M. Multimodal Emotion Recognition Using Deep Learning. J. Appl. Sci. Technol. Trends 2021, 2, 52–58. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Skin Cancer Classification Using Deep Learning and Transfer Learning. In Proceedings of the 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), Cairo, Egypt, 20–22 December 2018. [Google Scholar] [CrossRef]
- Premaladha, J.; Ravichandran, K. Novel Approaches for Diagnosing Melanoma Skin Lesions Through Supervised and Deep Learning Algorithms. J. Med. Syst. 2016, 40, 96. [Google Scholar] [CrossRef]
- Lee, H.; Chen, Y.-P.P. Image Based Computer Aided Diagnosis System for Cancer Detection. Expert Syst. Appl. 2015, 42, 5356–5365. [Google Scholar] [CrossRef]
- Goyal, M.; Oakley, A.; Bansal, P.; Dancey, D.; Yap, M.H. Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods. IEEE Access 2019, 8, 4171–4181. [Google Scholar] [CrossRef]
- Dargan, S.; Kumar, M.; Ayyagari, M.R.; Kumar, G. A Survey of Deep Learning and Its Applications: A New Paradigm to Machine Learning. Arch. Comput. Methods Eng. 2019, 27, 1071–1092. [Google Scholar] [CrossRef]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images. In International Workshop on Machine Learning in Medical Imaging; Springer: Munich, Germany, 2015. [Google Scholar] [CrossRef]
- Gessert, N.; Sentker, T.; Madesta, F.; Schmitz, R.; Kniep, H.; Baltruschat, I.; Werner, R.; Schlaefer, A. Skin Lesion Diagnosis Using Ensembles, Unscaled Multi-crop Evaluation and Loss Weighting. arXiv 2018, arXiv:1808.01694. [Google Scholar]
- Waheed, Z.; Waheed, A.; Zafar, M.; Riaz, F. An Efficient Machine Learning Approach for the Detection of Melanoma Using Dermoscopic Images. In Proceedings of the 2017 International Conference on Communication, Computing and Digital Systems (C-CODE), Islamabad, Pakistan, 8–9 March 2017. [Google Scholar]
- Roy, S.; Meena, T.; Lim, S.-J. Demystifying Supervised Learning in Healthcare 4.0: A New Reality of Transforming Diagnostic Medicine. Diagnostics 2022, 12, 2549. [Google Scholar] [CrossRef]
- Srivastava, V.; Kumar, D.; Roy, S. A Median Based Quadrilateral Local Quantized Ternary Pattern Technique for the Classification of Dermatoscopic Images of Skin Cancer. Comput. Electr. Eng. 2022, 102, 108259. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 Dataset, a Large Collection of Multi-source Dermatoscopic Images of Common Pigmented Skin Lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Romero Lopez, A.; Giro Nieto, X.; Burdick, J.; Marques, O. Skin Lesion Classification from Dermoscopic Images Using Deep Learning Techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017. [Google Scholar]
- Dreiseitl, S.; Ohno-Machado, L.; Kittler, H.; Vinterbo, S.A.; Billhardt, H.; Binder, M. A Comparison of Machine Learning Methods for the Diagnosis of Pigmented Skin Lesions. J. Biomed. Inform. 2001, 34, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Ayub, H.; Khan, J.A.; Ahmad, I.S.; Salama, A.; Daradkeh, Y.I.; Javeed, D.; Ur Rehman, A.; Hamam, H. A Hybrid Deep Learning-based Approach for Brain Tumor Classification. Electronics 2022, 11, 1146. [Google Scholar] [CrossRef]
- Raza, A.; Ullah, N.; Khan, J.A.; Assam, M.; Guzzo, A.; Aljuaid, H. DeepBreastCancerNet: A Novel Deep Learning Model for Breast Cancer Detection Using Ultrasound Images. Appl. Sci. 2023, 13, 2082. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.; Utikal, J.; Enk, A.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C. Deep Learning Outperformed 11 Pathologists in the Classification of Histopathological Melanoma Images. Eur. J. Cancer 2019, 118, 91–96. [Google Scholar] [CrossRef]
- Pham, T.-C.; Luong, C.-M.; Visani, M.; Hoang, V.-D. Deep CNN and Data Augmentation for Skin Lesion Classification. In Intelligent Information and Database Systems, Proceedings of the 10th Asian Conference, ACIIDS 2018, Dong Hoi City, Vietnam, 19–21 March 2018; Springer: Dong Hoi City, Vietnam, 2018. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.-A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Li, Y.; Shen, L. Skin Lesion Analysis Towards Melanoma Detection Using Deep Learning Network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef]
- Seeja, R.D.; Suresh, A. Deep Learning Based Skin Lesion Segmentation and Classification of Melanoma Using Support Vector Machine (SVM). Asian Pac. J. Cancer Prev. 2019, 20, 1555–1561. [Google Scholar] [CrossRef]
- Nasiri, S.; Helsper, J.; Jung, M.; Fathi, M. Depict Melanoma Deep-class: A Deep Convolutional Neural Networks Approach to Classify Skin Lesion Images. Asian Pac. J. Cancer Prev. 2019, 20, 1555–1561. [Google Scholar] [CrossRef]
- Inthiyaz, S.; Altahan, B.R.; Ahammad, S.H.; Rajesh, V.; Kalangi, R.R.; Smirani, L.K.; Hossain, M.A.; Rashed, A.N.Z. Skin Disease Detection Using Deep Learning. Adv. Eng. Softw. 2023, 175, 103361. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Hsiao, Y.-P.; Mukundan, A.; Tsao, Y.-M.; Chang, W.-Y.; Wang, H.-C. Classification of Skin Cancer Using Novel Hyperspectral Imaging Engineering via YOLOv5. J. Clin. Med. 2023, 12, 1134. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Tang, H.; He, S.; Yu, Q.; Xiong, Y.; Wang, N. Performance Validation of YOLO Variants for Object Detection. In Proceedings of the 2021 International Conference on Bioinformatics and Intelligent Computing, Harbin, China, 22–24 January 2021. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Liao, H.-Y.M.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W.; Yeh, I.-H. Cspnet: A New Backbone That Can Enhance Learning Capability of CNN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- Hafiz, A.M.; Bhat, G.M. A Survey on Instance Segmentation: State of the Art. Int. J. Multimedia Inf. Retr. 2020, 9, 171–189. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. Yolov3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Gao, H.; Huang, H. Stochastic Second-order Method for Large-scale Nonconvex Sparse Learning Models. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence (IJCAI-18), Stockholm, UK, 13–19 July 2018. [Google Scholar]
- Wu, X.; Sahoo, D.; Hoi, S.C.H.; Hoi, S.C.H. Recent Advances in Deep Learning for Object Detection. Neurocomputing 2020, 396, 39–64. [Google Scholar] [CrossRef]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and Efficient Object Detection. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Thuan, D. Evolution of Yolo Algorithm and Yolov5: The State-of-the-art Object Detection Algorithm. Master’s Thesis, Oulu University of Applied Sciences, Oulu, Finland, 2021. [Google Scholar]
- Jung, H.-K.; Choi, G.-S. Improved Yolov5: Efficient Object Detection Using Drone Images Under Various Conditions. Appl. Sci. 2022, 12, 7255. [Google Scholar] [CrossRef]
- Alsaade, F.W.; Aldhyani, T.H.; Al-Adhaileh, M.H. Developing a Recognition System for Diagnosing Melanoma Skin Lesions Using Artificial Intelligence Algorithms. Comput. Math. Methods Med. 2021, 2021, 9998379. [Google Scholar] [CrossRef]
- Ali, S.; Miah, S.; Miah, S.; Haque, J.; Rahman, M.; Islam, K. An Enhanced Technique of Skin Cancer Classification Using Deep Convolutional Neural Network with Transfer Learning Models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Khaledyan, D.; Tajally, A.; Sarkhosh, A.; Shamsi, A.; Asgharnezhad, H.; Khosravi, A.; Nahavandi, S. Confidence Aware Neural Networks for Skin Cancer Detection. arXiv 2021, arXiv:2107.09118. [Google Scholar] [CrossRef]
- Chang, C.-C.; Li, Y.-Z.; Wu, H.-C.; Tseng, M.-H. Melanoma Detection Using XGB Classifier Combined with Feature Extraction and K-means SMOTE Techniques. Diagnostics 2022, 12, 1747. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, J.; Hamarneh, G. Fully Convolutional Neural Networks to Detect Clinical Dermoscopic Features. IEEE J. Biomed. Health Inform. 2019, 23, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Akram, T.; Zhang, Y.; Sharif, M. Attributes Based Skin Lesion Detection and Recognition: A Mask RCNN and Transfer Learning-based Deep Learning Framework. Pattern Recognit. Lett. 2021, 143, 58–66. [Google Scholar] [CrossRef]
- Chaturvedi, S.S.; Gupta, K.; Prasad, P.S. Skin Lesion Analyzer: An Efficient Seven-Way Multi-Class Skin Cancer Classification Using Mobilenet; Springer: Singapore, 2021. [Google Scholar] [CrossRef]



| Reference | Proposed Technique | Accuracy | Limitation |
|---|---|---|---|
| Premaladha et al. [6] | Segmentation using Otsu’s normalized algorithm and then classification | SVM (90.44), DCNN (92.89), and Hybrid AdaBoost (91.73) | Uses only three classes of skin cancer lesions |
| Codella et al. [10] | Melanoma recognition using DL, sparse coding, and SVM | 93.1% | Need to deepen features and add more cases of melanoma |
| Waheed et al. [12] | Diagnosing melanoma using the color and texture of different types of lesions | SVM (96.0%) | Need more attributes of skin lesions |
| Hekler et al. [20] | Classifying histopathologic melanoma using DCNN | 68.0% | Uses low resolution and cannot differentiate between melanoma and nevi classes |
| Pham et al. [21] | Classification using DCNN | AUC (89.2%) | Less sensitivity |
| Yu et al. [22] | Segmentation and classification using DCNN and FCRN | AUC (80.4%) | Insufficient training data |
| Li and Shen [23] | Two FCRN for melanoma segmentation and classification | AUC (91.2%) | Overfitting in AUC and low segmentation |
| Seeja and Suresh [24] | Segmenting data using form, color, and texture variables, then classification using SVM, RF, KNN, and NB | SVM (85.1%), RF (82.2%), KNN (79.2%), and NB (65.9%) | Low classification accuracy |
| Nasiri et al. [25] | Using the 19-layer model of CNN for melanoma classification | 75.0% | Need to enhance accuracy |
| VASC | NV | MEL | DF | BKL | BCC | AKIEC | |
|---|---|---|---|---|---|---|---|
| All images | 142 | 6705 | 1113 | 115 | 1099 | 514 | 327 |
| Train | 115 | 5360 | 891 | 92 | 879 | 300 | 262 |
| Test | 27 | 1345 | 222 | 23 | 220 | 214 | 65 |
| Requirement | Version | Requirement | Version |
|---|---|---|---|
| Base | matplotlib ≥ 3.2.2 | Export | coremltools ≥ 6.0 by CoreML export |
| opencv-python ≥ 4.1.1 | onnx ≥ 1.9.0 by ONNX export | ||
| Pillow ≥ 7.1.2 | onnx-simplifier ≥ 0.4.1 by ONNX simplifier | ||
| PyYAML ≥ 5.3.1 | Nvidia-pyinde by TensorRT export | ||
| requests ≥ 2.23.0 | nvidia-tensorrt by TensorRT export | ||
| scipy ≥ 1.4.1 | scikit-learn ≤ 1.1.2 by CoreML quantization | ||
| torch ≥ 1.7.0 | tensorflow ≥ 2.4 | ||
| tqdm ≥ 4.64.0 | tensorflowjs ≥ 3.9.0 by TF.js export | ||
| protobuf ≤ 3.20.1 | openvino-dev by OpenVINO export | ||
| Plotting | pandas ≥ 1.1.4 | Extras | ipython by interactive notebook |
| seaborn ≥ 0.11.0 | psutil by system utilization | ||
| Logging | tensorboard ≥ 2.4. clearml ≥ 1.2.0 | thop ≥ 0.1.1 by FLOPs computation | |
| by albumentations ≥ 1.0.3 | |||
| by pycocotools ≥ 2.0 |
| Parameter | First Run | Second Run | Definition |
|---|---|---|---|
| Epoch | 300 | 100 | The frequency with which the learning algorithm operates |
| Batch_size | 16 | 32 | How many training instances are used in a single iteration |
| lr0 | 0.001 | 0.001 | Initial learning rate (SGD = 1 × 10−2; Adam = 1 × 10−3) |
| Lrf | 0.2 | 0.2 | Final OneCycleLR learning rate (lr0 × lrf) |
| Momentum | 0.937 | 0.937 | SGD momentum/Adam beta1 |
| warmup_epochs | 3.0 | 3.0 | Warmup epochs (fractions ok) |
| weight_decay | 0.0005 | 0.0005 | Optimizer weight decay, 5 × 10−4 |
| warmup_momentum | 0.8 | 0.8 | Warmup initial momentum |
| warmup_bias_lr | 0.1 | 0.1 | Warmup initial bias learning rate |
| Box | 0.05 | 0.05 | Box loss gain |
| Cls | 0.5 | 0.5 | Class loss gain |
| cls_pw | 1.0 | 1.0 | Cls BCELoss positive_weight |
| Obj | 1.0 | 1.0 | Obj loss gain (scale with pixels) |
| obj_pw | 1.0 | 1.0 | Obj BCELoss positive_weight |
| anchor_t | 4.0 | 4.0 | Anchor-multiple threshold |
| iou_t | 0.20 | 0.20 | IOU training threshold |
| Scale | 0.5 | 0.5 | Image scale (+/−gain) |
| Shear | 0.0 | 0.0 | Image shear (+/−deg) |
| Perspective | 0.0 | 0.0 | Image perspective (+/−fraction), range 0–0.001 |
| Precision (%) | Recall (%) | DSC (%) | MAP 0.0:0.5 (%) | MAP 0.5:0.95 (%) | Accuracy (%) | |
|---|---|---|---|---|---|---|
| AKIEC | 99.1 | 94.9 | 96.9 | 99.7 | 95.2 | 95.2 |
| BKL | 95.3 | 96.8 | 96.0 | 95.3 | 94.5 | 96.1 |
| VASC | 97.0 | 95.6 | 96.2 | 98.7 | 95.5 | 97.2 |
| BCC | 97.1 | 97.6 | 97.3 | 97.5 | 96.4 | 97.3 |
| DF | 98.7 | 99.5 | 99.0 | 94.3 | 94.8 | 98.8 |
| NV | 100.0 | 98.6 | 99.2 | 96.4 | 99.5 | 98.1 |
| MEL | 98.8 | 100.0 | 99.3 | 98.2 | 98.6 | 100.0 |
| Average | 98.1 | 97.5 | 97.7 | 97.1 | 96.3 | 97.5 |
| Precision (%) | Recall (%) | DSC (%) | MAP 0.0:0.5 (%) | MAP 0.5:0.95 (%) | Accuracy (%) | |
|---|---|---|---|---|---|---|
| AKIEC | 100.0 | 96.7 | 98.3 | 98.9 | 99.7 | 98.8 |
| BKL | 98.2 | 98.2 | 98.2 | 97.6 | 94.9 | 98.9 |
| VASC | 98.8 | 99.6 | 99.1 | 97.9 | 97.9 | 99.4 |
| BCC | 97.1 | 96.9 | 96.9 | 99.5 | 99.1 | 99.7 |
| DF | 99.6 | 98.9 | 99.2 | 98.6 | 96.2 | 100.0 |
| MV | 100.0 | 100.0 | 100.0 | 96.2 | 100.0 | 99.8 |
| MEL | 99.9 | 100.0 | 99.9 | 99.8 | 98.9 | 100.0 |
| Average | 99.0 | 98.6 | 98.8 | 98.3 | 98.7 | 99.5 |
| Reference | Year | Method | Precision (%) | Recall (%) | DSC (%) | Accuracy (%) | Dataset |
|---|---|---|---|---|---|---|---|
| Nasiri et al. [25] | 2020 | KNN | 73.0 | 55.0 | 79.0 | 67.0 | ISIC dataset |
| SVM | 58.0 | 47.0 | 66.0 | 62.0 | |||
| CNN | 77.0 | 73.0 | 78.0 | 75.0 | |||
| Alsaade et al. [38] | 2021 | CNN | 81.2 | 92.9 | 87.5 | 97.5 | PH2 |
| Ali et al. [39] | 2021 | CNN | 96.5 | 93.6 | 95.0 | 91.9 | HAM10000 |
| Khaledyan et al. [40] | 2021 | Ensemble Bayesian Networks | 88.6 | 73.4 | 90.7 | 83.6 | HAM10000 |
| Chang et al. [41] | 2022 | XGB classifier | 97.4 | 87.8 | 90.5 | 94.1 | ISIC |
| Kawahara et al. [42] | 2019 | FCNN | 97.6 | 81.3 | 93.0 | 98.0 | ISIC |
| Khan et al. [43] | 2021 | Mask RCNN | 88.5% | 88.5% | 88.6% | 93.6 | ISIC |
| Chaturvedi et al. [44] | 2020 | Mobile Net | 83.0% | 83.0% | 89.0% | 83.1 | HAM10000 |
| Proposed model | 2023 | YOLOv5 + ResNet | 99.0 | 98.6 | 98.8 | 99.5 | HAM10000 |
| Database | Description |
|---|---|
| PH2 |
|
| ISIC |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshahawy, M.; Elnemr, A.; Oproescu, M.; Schiopu, A.-G.; Elgarayhi, A.; Elmogy, M.M.; Sallah, M. Early Melanoma Detection Based on a Hybrid YOLOv5 and ResNet Technique. Diagnostics 2023, 13, 2804. https://doi.org/10.3390/diagnostics13172804
Elshahawy M, Elnemr A, Oproescu M, Schiopu A-G, Elgarayhi A, Elmogy MM, Sallah M. Early Melanoma Detection Based on a Hybrid YOLOv5 and ResNet Technique. Diagnostics. 2023; 13(17):2804. https://doi.org/10.3390/diagnostics13172804
Chicago/Turabian StyleElshahawy, Manar, Ahmed Elnemr, Mihai Oproescu, Adriana-Gabriela Schiopu, Ahmed Elgarayhi, Mohammed M. Elmogy, and Mohammed Sallah. 2023. "Early Melanoma Detection Based on a Hybrid YOLOv5 and ResNet Technique" Diagnostics 13, no. 17: 2804. https://doi.org/10.3390/diagnostics13172804
APA StyleElshahawy, M., Elnemr, A., Oproescu, M., Schiopu, A.-G., Elgarayhi, A., Elmogy, M. M., & Sallah, M. (2023). Early Melanoma Detection Based on a Hybrid YOLOv5 and ResNet Technique. Diagnostics, 13(17), 2804. https://doi.org/10.3390/diagnostics13172804










