Next-Generation Sequencing-Based Analysis of Clinical and Pathological Features of PIK3CA-Mutated Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Analysis Methodology
2.1.1. Library Preparation
2.1.2. Template Preparation and Sequencing
2.1.3. Bioinformatic Analysis
2.2. Statistical Analysis
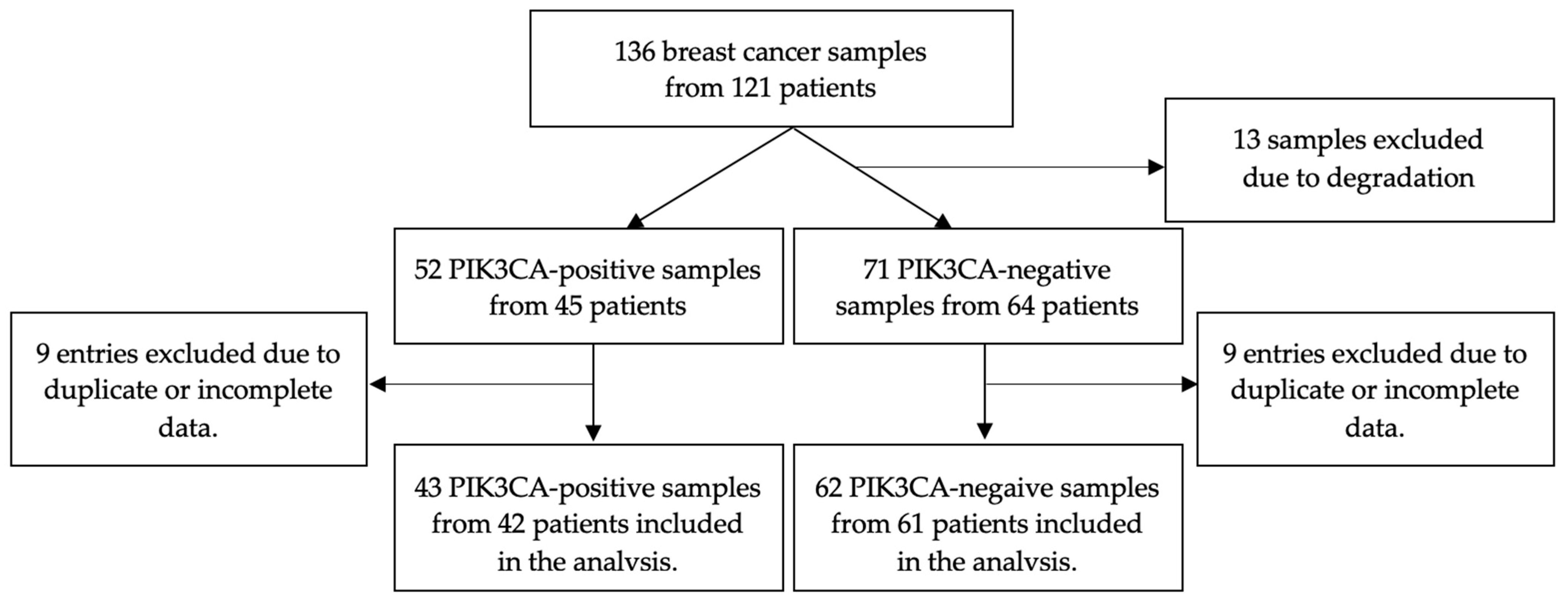
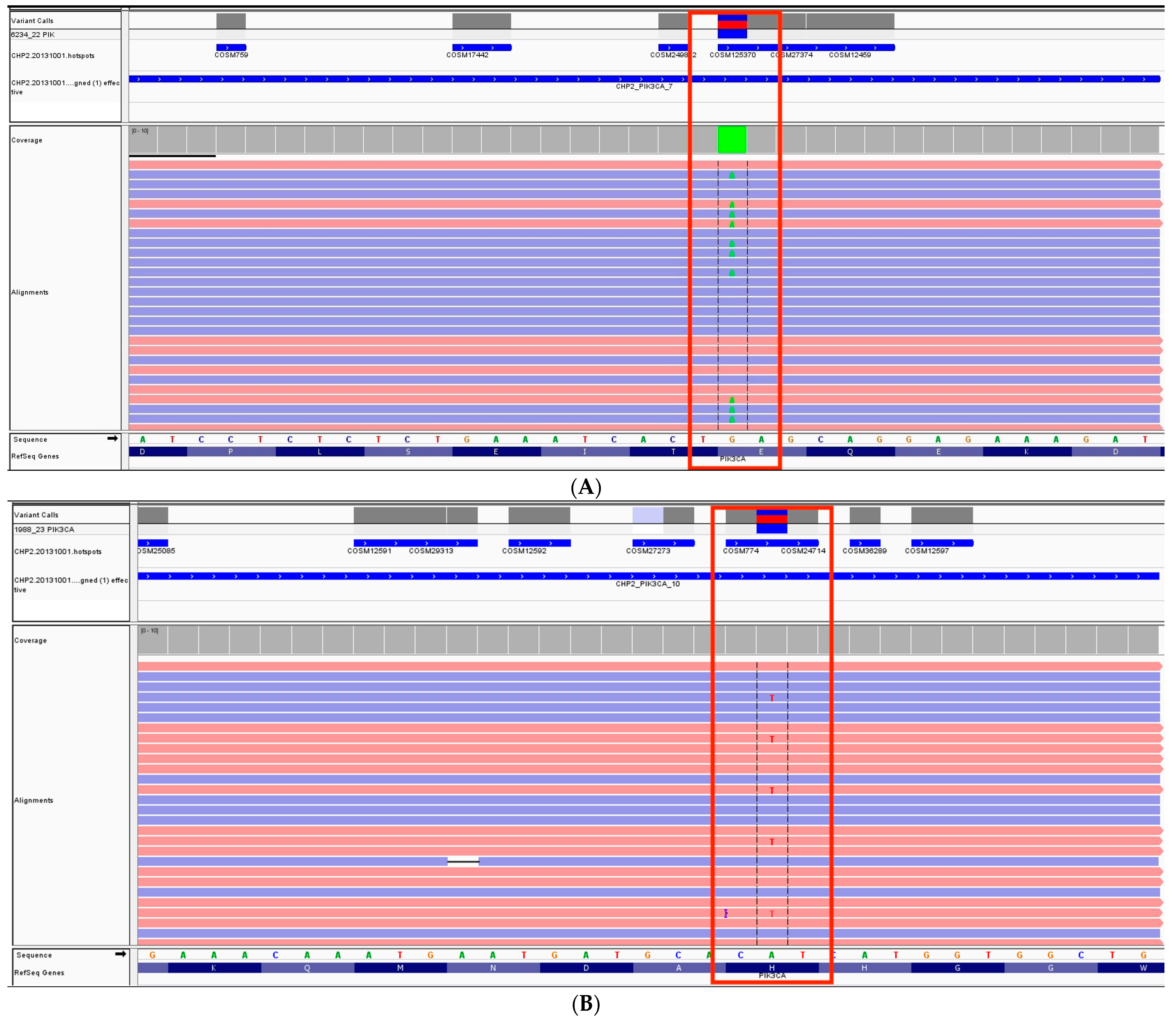
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Wojciechowska, U.; Barańska, K.; Miklewska, M.; Didkowska, J.A. Cancer incidence and mortality in Poland in 2020. Nowotw. J. Oncol. 2023, 73, 129–145. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- SEER. Cancer of the Breast (Female)—Cancer Stat Facts. Available online: http://seer.cancer.gov/statfacts/html/breast.html (accessed on 9 March 2023).
- Jordan, V.C. Tamoxifen: A most unlikely pioneering medicine. Nat. Rev. Drug Discov. 2003, 2, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Pietras, R.J.; Fendly, B.M.; Chazin, V.R.; Pegram, M.D.; Howell, S.B.; Slamon, D.J. Antibody to HER-2/neu receptor blocks DNA repair after cisplatin in human breast and ovarian cancer cells. Oncogene 1994, 9, 1829–1838. [Google Scholar] [PubMed]
- Goutsouliak, K.; Veeraraghavan, J.; Sethunath, V.; De Angelis, C.; Osborne, C.K.; Rimawi, M.F.; Schiff, R. Towards personalized treatment for early stage HER2-positive breast cancer. Nat. Rev. Clin. Oncol. 2020, 17, 233–250. [Google Scholar] [CrossRef]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, C.L.; Duckworth, B.C.; Auger, K.R.; Cohen, B.; Schaffhausen, B.S.; Cantley, L.C. Purification and characterization of phosphoinositide 3-kinase from rat liver. J. Biol. Chem. 1990, 265, 19704–19711. [Google Scholar] [CrossRef]
- Chang, H.W.; Aoki, M.; Fruman, D.; Auger, K.R.; Bellacosa, A.; Tsichlis, P.N.; Cantley, L.C.; Roberts, T.M.; Vogt, P.K. Transformation of Chicken Cells by the Gene Encoding the Catalytic Subunit of PI 3-Kinase. Science 1997, 276, 1848–1850. [Google Scholar] [CrossRef]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef] [PubMed]
- Cizkova, M.; Susini, A.; Vacher, S.; Cizeron-Clairac, G.; Andrieu, C.; Driouch, K.; Fourme, E.; Lidereau, R.; Bièche, I. PIK3CAmutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res. 2012, 14, R28. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Canaud, G.; Hammill, A.M.; Adams, D.; Vikkula, M.; Keppler-Noreuil, K.M. A review of mechanisms of disease across PIK3CA-related disorders with vascular manifestations. Orphanet J. Rare Dis. 2021, 16, 306. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef] [PubMed]
- Arafeh, R.; Samuels, Y. PIK3CA in cancer: The past 30 years. Semin. Cancer Biol. 2019, 59, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.; Martínez-Fernández, M.; García-Escudero, R.; Villacampa, F.; Marqués, M.; Saiz-Ladera, C.; Duarte, J.; Martínez, V.; Gómez, M.J.; Martín, M.L.; et al. PIK3CA gene alterations in bladder cancer are frequent and associate with reduced recurrence in non-muscle invasive tumors. Mol. Carcinog. 2015, 54, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Mjos, S.; Werner, H.M.J.; Birkeland, E.; Holst, F.; Berg, A.; Halle, M.K.; Tangen, I.L.; Kusonmano, K.; Mauland, K.K.; Oyan, A.M.; et al. PIK3CA exon9 mutations associate with reduced survival, and are highly concordant between matching primary tumors and metastases in endometrial cancer. Sci. Rep. 2017, 7, 10240. [Google Scholar] [CrossRef]
- Vahidnezhad, H.; Youssefian, L.; Uitto, J. Klippel-Trenaunay syndrome belongs to the PIK3CA-related overgrowth spectrum (PROS). Exp. Dermatol. 2016, 25, 17–19. [Google Scholar] [CrossRef]
- Narayan, P.; Prowell, T.M.; Gao, J.J.; Fernandes, L.L.; Li, E.; Jiang, X.; Qiu, J.; Fan, J.; Song, P.; Yu, J.; et al. FDA Approval Summary: Alpelisib Plus Fulvestrant for Patients with HR-positive, HER2-negative, PIK3CA-mutated, Advanced or Metastatic Breast Cancer. Clin. Cancer Res. 2021, 27, 1842–1849. [Google Scholar] [CrossRef]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Wilhoit, T.; Patrick, J.M.; May, M.B. Alpelisib: A Novel Therapy for Patients with PIK3CA-Mutated Metastatic Breast Cancer. J. Adv. Pract. Oncol. 2020, 11, 768–775. [Google Scholar] [PubMed]
- Fuso, P.; Muratore, M.; D’angelo, T.; Paris, I.; Carbognin, L.; Tiberi, G.; Pavese, F.; Duranti, S.; Orlandi, A.; Tortora, G.; et al. PI3K Inhibitors in Advanced Breast Cancer: The Past, The Present, New Challenges and Future Perspectives. Cancers 2022, 14, 2161. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.Y.; Ma, W.L.; Lu, Y.S. Role of Alpelisib in the Treatment of PIK3CA-Mutated Breast Cancer: Patient Selection and Clinical Perspectives. TCRM 2021, 17, 193–207. [Google Scholar] [CrossRef]
- Rugo, H.S.; Raskina, K.; Schrock, A.B.; Madison, R.W.; Graf, R.P.; Sokol, E.S.; Sivakumar, S.; Lee, J.K.; Fisher, V.; Oxnard, G.R.; et al. Biology and Targetability of the Extended Spectrum of PIK3CA Mutations Detected in Breast Carcinoma. Clin. Cancer Res. 2023, 29, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Fillbrunn, M.; Signorovitch, J.; André, F.; Wang, I.; Lorenzo, I.; Ridolfi, A.; Park, J.; Dua, A.; Rugo, H.S. PIK3CA mutation status, progression and survival in advanced HR + /HER2- breast cancer: A meta-analysis of published clinical trials. BMC Cancer 2022, 22, 1002. [Google Scholar] [CrossRef]
- Cossu-Rocca, P.; Orrù, S.; Muroni, M.R.; Sanges, F.; Sotgiu, G.; Ena, S.; Pira, G.; Murgia, L.; Manca, A.; Uras, M.G.; et al. Analysis of PIK3CA Mutations and Activation Pathways in Triple Negative Breast Cancer. PLoS ONE 2015, 10, e0141763. [Google Scholar] [CrossRef]
- Arsenic, R.; Lehmann, A.; Budczies, J.; Koch, I.; Prinzler, J.; Kleine-Tebbe, A.; Schewe, C.; Loibl, S.; Dietel, M.; Denkert, C. Analysis of PIK3CA Mutations in Breast Cancer Subtypes. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 50–56. [Google Scholar] [CrossRef]
- Pogue-Geile, K.L.; Song, N.; Jeong, J.-H.; Gavin, P.G.; Kim, S.-R.; Blackmon, N.L.; Finnigan, M.; Rastogi, P.; Fehrenbacher, L.; Mamounas, E.P.; et al. Intrinsic Subtypes, PIK3CA Mutation, and the Degree of Benefit from Adjuvant Trastuzumab in the NSABP B-31 Trial. J. Clin. Oncol. 2015, 33, 1340–1347. [Google Scholar] [CrossRef]
- Padmanabhan, N.; Subramanyan, A.; Radhakrishna, S. Synchronous Bilateral Breast Cancers. J. Clin. Diagn. Res. 2015, 9, XC05-8. [Google Scholar] [CrossRef]
- Vogt, A.; Schmid, S.; Heinimann, K.; Frick, H.; Herrmann, C.; Cerny, T.; Omlin, A. Multiple primary tumours: Challenges and approaches, a review. ESMO Open 2017, 2, e000172. [Google Scholar] [CrossRef] [PubMed]
- Chaudary, M.A.; Millis, R.R.; Hoskins, E.O.L.; Halder, M.; Bulbrook, R.D.; Cuzick, J.; Hayward, J.L. Bilateral primary breast cancer: A prospective study of disease incidence. Br. J. Surg. 1984, 71, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Vogt, P.K.; Rommel, C. PI3K: From the Bench to the Clinic and Back. Curr. Top. Microbiol. Immunol. 2011, 347, 1–19. [Google Scholar]
- Carnero, A.; Paramio, J.M. The PTEN/PI3K/AKT Pathway in vivo, Cancer Mouse Models. Front. Oncol. 2014, 4, 00252. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Mollon, L.E.; Dean, J.L.; Warholak, T.L.; Aizer, A.; Platt, E.A.; Tang, D.H.; Davis, L.E. A Systematic Review of the Prevalence and Diagnostic Workup of PIK3CA Mutations in HR+/HER2– Metastatic Breast Cancer. Int. J. Breast Cancer 2020, 2020, e3759179. [Google Scholar] [CrossRef] [PubMed]
- Hamadeh, L.N.; Farhat, L.; Hilal, L.; Assi, H.; Nasr, F.; Chahine, G.; Kattan, J.; Farhat, F.; Kourie, H.; El Hachem, G.; et al. Frequency and mutational spectrum of PIK3CA gene mutations in breast cancer patients: Largest and first report from Lebanon. Gene 2023, 871, 147433. [Google Scholar] [CrossRef]
- Abramson, V.G.; Lloyd, M.C.; Ballinger, T.; Sanders, M.E.; Du, L.; Lai, D.; Su, Z.; Mayer, I.; Levy, M.; LaFrance, D.R.; et al. Characterization of breast cancers with PI3K mutations in an academic practice setting using SNaPshot profiling. Breast Cancer Res. Treat. 2014, 145, 389–399. [Google Scholar] [CrossRef]
- Arthur, L.M.; Turnbull, A.K.; Renshaw, L.; Keys, J.; Thomas, J.S.; Wilson, T.R.; Lackner, M.R.; Sims, A.H.; Dixon, J.M. Changes in PIK3CA mutation status are not associated with recurrence, metastatic disease or progression in endocrine-treated breast cancer. Breast Cancer Res. Treat. 2014, 147, 211–219. [Google Scholar] [CrossRef]
- Kalinsky, K.; Jacks, L.M.; Heguy, A.; Patil, S.; Drobnjak, M.; Bhanot, U.K.; Hedvat, C.V.; Traina, T.A.; Solit, D.; Gerald, W.; et al. PIK3CA Mutation Associates with Improved Outcome in Breast Cancer. Clin. Cancer Res. 2009, 15, 5049–5059. [Google Scholar] [CrossRef]
- Ogenyi, S.I.; Onu, J.A.; Ibeh, N.C.; Madukwe, J.U.; Onu, O.A.; Menkiti, F.E. PIK3CA, KI67, Estrogen (ER) and Progesterone Receptors (PR) Expression Pattern of in HER2 Positive Breast Cancers. Asian Pac. J. Cancer Biol. 2021, 6, 281–287. [Google Scholar] [CrossRef]
- Pang, B.; Cheng, S.; Sun, S.-P.; An, C.; Liu, Z.-Y.; Feng, X.; Liu, G.-J. Prognostic role of PIK3CA mutations and their association with hormone receptor expression in breast cancer: A meta-analysis. Sci. Rep. 2014, 4, 6255. [Google Scholar] [CrossRef] [PubMed]
- Saal, L.H.; Holm, K.; Maurer, M.; Memeo, L.; Su, T.; Wang, X.; Yu, J.S.; Malmström, P.O.; Mansukhani, M.; Enoksson, J.; et al. PIK3CA Mutations Correlate with Hormone Receptors, Node Metastasis, and ERBB2, and Are Mutually Exclusive with PTEN Loss in Human Breast Carcinoma. Cancer Res. 2005, 65, 2554–2559. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tenorio, G.; Alkhori, L.; Olsson, B.; Waltersson, M.A.; Nordenskjöld, B.; Rutqvist, L.E.; Skoog, L.; Stål, O. PIK3CA Mutations and PTEN Loss Correlate with Similar Prognostic Factors and Are Not Mutually Exclusive in Breast Cancer. Clin. Cancer Res. 2007, 13, 3577–3584. [Google Scholar] [CrossRef] [PubMed]
- Esteva, F.J.; Guo, H.; Zhang, S.; Santa-Maria, C.; Stone, S.; Lanchbury, J.S.; Sahin, A.A.; Hortobagyi, G.N.; Yu, D. PTEN, PIK3CA, p-AKT, and p-p70S6K Status: Association with Trastuzumab Response and Survival in Patients with HER2-Positive Metastatic Breast Cancer. Am. J. Pathol. 2010, 177, 1647–1656. [Google Scholar] [CrossRef]
- Reinhardt, K.; Stückrath, K.; Hartung, C.; Kaufhold, S.; Uleer, C.; Hanf, V.; Lantzsch, T.; Peschel, S.; John, J.; Pöhler, M.; et al. PIK3CA-mutations in breast cancer. Breast Cancer Res. Treat. 2022, 196, 483–493. [Google Scholar] [CrossRef]
- Kandula, M.; Chennaboina, K.K.; Raju, Y.A.; Raju, S. Phosphatidylinositol 3-kinase (PI3KCA) Oncogene Mutation Analysis and Gene Expression Profiling in Primary Breast Cancer Patients. Asian Pac. J. Cancer Prev. 2013, 14, 5067–5072. [Google Scholar] [CrossRef]
- Kumar, S.; Bal, A.; Das, A.; Loriya, I.; Khare, S.; Bhattacharya, S.; Singh, G. Spectrum of PIK3CA/AKT mutations across molecular subtypes of triple-negative breast cancer. Breast Cancer Res. Treat. 2021, 187, 625–633. [Google Scholar] [CrossRef]
- Pascual, J.; Turner, N.C. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann. Oncol. 2019, 30, 1051–1060. [Google Scholar] [CrossRef]
- Zagami, P.; Carey, L.A. Triple negative breast cancer: Pitfalls and progress. NPJ Breast Cancer 2022, 8, 95. [Google Scholar] [CrossRef]
- López-Knowles, E.; Segal, C.V.; Gao, Q.; Garcia-Murillas, I.; Turner, N.C.; Smith, I.; Martin, L.-A.; Dowsett, M. Relationship of PIK3CA mutation and pathway activity with antiproliferative response to aromatase inhibition. Breast Cancer Res. 2014, 16, R68. [Google Scholar] [CrossRef]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Dien, A.T.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.-P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.M.; Juric, D.; Loibl, S.; Campone, M.; Mayer, I.A.; Rubovszky, G.; Yamashita, T.; Kaufman, B.; Lu, Y.S.; et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2–negative advanced breast cancer: Final overall survival results from SOLAR-1. Ann. Oncol. 2021, 32, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Bachelot, T.; Filleron, T.; Bieche, I.; Arnedos, M.; Campone, M.; Dalenc, F.; Coussy, F.; Sablin, M.-P.; Debled, M.; Lefeuvre-Plesse, C.; et al. Durvalumab compared to maintenance chemotherapy in metastatic breast cancer: The randomized phase II SAFIR02-BREAST IMMUNO trial. Nat. Med. 2021, 27, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Rasti, A.R.; Guimaraes-Young, A.; Datko, F.; Borges, V.F.; Aisner, D.L.; Shagisultanova, E. PIK3CA Mutations Drive Therapeutic Resistance in Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer. JCO Precis. Oncol. 2022, 6, e2100370. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhu, J.; Zhong, Y.; Geng, R.; Ji, Y.; Guan, Q.; Hong, C.; Wei, Y.; Min, N.; Qi, A.; et al. PIK3CA mutation confers resistance to chemotherapy in triple-negative breast cancer by inhibiting apoptosis and activating the PI3K/AKT/mTOR signaling pathway. Ann. Transl. Med. 2021, 9, 410. [Google Scholar] [CrossRef]
| PIK3CA-WT (n = 61) | PIK3CA-M (n = 42) | Total (n = 103) | p-Value | |
|---|---|---|---|---|
| Age at diagnosis [years] | 0.0338 | |||
| Mean (SD) | 54.6 (12.9) | 60.3 (14.0) | 56.9 (13.6) | |
| Median (Q1, Q3) | 55.0 (44.0, 63.0) | 64.0 (50.5, 70.0) | 59.0 (48.0, 64.5) | |
| Range | 25.0–83.0 | 25.0–91.0 | 25.0–91.0 | |
| Grade | 0.4901 | |||
| Missing data | 7 | 0 | 7 | |
| G1 | 17 (31.5%) | 15 (35.7%) | 32 (33.3%) | |
| G2 | 24 (44.4%) | 21 (50.0%) | 45 (46.9%) | |
| G3 | 13 (24.1%) | 6 (14.3%) | 19 (19.8%) | |
| ER | 0.1133 | |||
| Negative (−) | 13 (21.3%) | 4 (9.5%) | 17 (16.5%) | |
| Positive (+, ++, +++) | 48 (78.7%) | 38 (90.5%) | 86 (83.5%) | |
| PR | 0.0465 | |||
| Negative (−) | 21 (34.4%) | 7 (16.7%) | 28 (27.2%) | |
| Positive (+, ++, +++) | 40 (65.6%) | 35 (83.3%) | 75 (72.8%) | |
| HER2 1 | 0.1648 | |||
| Negative | 51 (83.6%) | 39 (92.9%) | 90 (87.4%) | |
| Positive | 10 (16.4%) | 3 (7.1%) | 13 (12.6%) | |
| Ki67 [%] | 0.0408 | |||
| Missing data | 2 | 0 | 2 | |
| Mean (SD) | 37.1 (32.4) | 24.9 (26.8) | 32.0 (30.7) | |
| Median (Q1, Q3) | 25.0 (10.0, 70.0) | 17.5 (5.0, 30.0) | 20.0 (10.0, 50.0) | |
| Range | 1.0–100.0 | 1.0–95.0 | 1.0–100.0 | |
| Ki67 groups | 0.0556 | |||
| Missing data | 2 | 0 | 2 | |
| High (≥20%) | 31 (52.5%) | 14 (33.3%) | 45 (44.6%) | |
| Low (<20%) | 28 (47.5%) | 28 (66.7%) | 56 (55.4%) | |
| Molecular subtype 2 | 0.2349 | |||
| Borderline | 0 (0.0%) | 1 (2.4%) | 1 (1.0%) | |
| Luminal A | 26 (42.6%) | 23 (54.8%) | 49 (47.6%) | |
| Luminal B, HER2-negative | 19 (31.1%) | 14 (33.3%) | 33 (32.0%) | |
| Luminal B, HER2 positive | 4 (6.6%) | 0 (0.0%) | 4 (3.9%) | |
| HER2-positive, non-luminal | 4 (6.6%) | 2 (4.8%) | 6 (5.8%) | |
| TNBC | 8 (13.1%) | 2 (4.8%) | 10 (9.7%) | |
| TNBC | 0.1937 | |||
| no | 53 (86.9%) | 40 (95.2%) | 93 (90.3%) | |
| yes | 8 (13.1%) | 2 (4.8%) | 10 (9.7%) | |
| cT | 0.4094 | |||
| Missing data | 3 | 0 | 3 | |
| T1 | 9 (15.5%) | 3 (7.1%) | 12 (12.0%) | |
| T2 | 27 (46.6%) | 19 (45.2%) | 46 (46.0%) | |
| T3 | 10 (17.2%) | 12 (28.6%) | 22 (22.0%) | |
| T4 | 12 (20.7%) | 8 (19.0%) | 20 (20.0%) | |
| cT | 0.3326 | |||
| Missing data | 3 | 0 | 3 | |
| T1 or T2 | 36 (62.1%) | 22 (52.4%) | 58 (58.0%) | |
| T3 or T4 | 22 (37.9%) | 20 (47.6%) | 42 (42.0%) | |
| cN | 0.2126 | |||
| Missing data | 4 | 0 | 4 | |
| N0 | 33 (57.9%) | 19 (45.2%) | 52 (52.5%) | |
| other than N0 | 24 (42.1%) | 23 (54.8%) | 47 (47.5%) | |
| cN | 0.5883 | |||
| Missing data | 4 | 0 | 4 | |
| N0 | 33 (57.9%) | 19 (45.2%) | 52 (52.5%) | |
| N1 | 19 (33.3%) | 19 (45.2%) | 38 (38.4%) | |
| N2 | 3 (5.3%) | 2 (4.8%) | 5 (5.1%) | |
| N3 | 2 (3.5%) | 2 (4.8%) | 4 (4.0%) | |
| cM | 0.0046 | |||
| M0 | 58 (95.1%) | 32 (76.2%) | 90 (87.4%) | |
| M1 | 3 (4.9%) | 10 (23.8%) | 13 (12.6%) | |
| Stage | 0.0329 | |||
| I | 9 (14.8%) | 3 (7.1%) | 12 (11.7%) | |
| II | 31 (50.8%) | 20 (47.6%) | 51 (49.5%) | |
| III | 18 (29.5%) | 9 (21.4%) | 27 (26.2%) | |
| IV | 3 (4.9%) | 10 (23.8%) | 13 (12.6%) | |
| Stage | 0.2686 | |||
| I or II | 40 (65.6%) | 23 (54.8%) | 63 (61.2%) | |
| III or IV | 21 (34.4%) | 19 (45.2%) | 40 (38.8%) | |
| Stage | 0.0046 | |||
| I–III | 58 (95.1%) | 32 (76.2%) | 90 (87.4%) | |
| IV | 3 (4.9%) | 10 (23.8%) | 13 (12.6%) | |
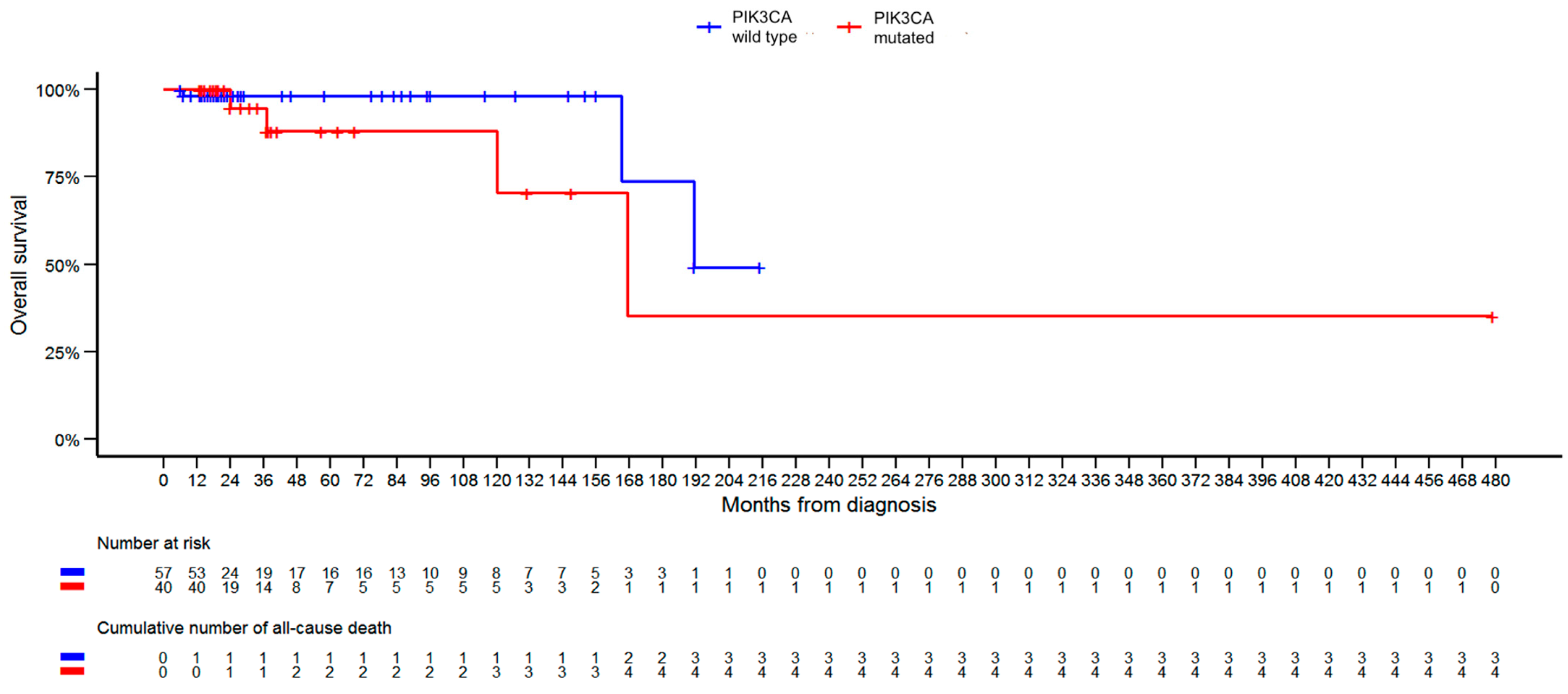
| OS [months] | 0.7934 | |||
| Mean (SD) | 49.8 (54.0) | 48.5 (77.5) | 49.2 (64.3) | |
| Median (Q1, Q3) | 21.0 (17.0, 76.0) | 23.0 (15.5, 41.0) | 21.0 (16.5, 51.5) | |
| Range | 6.0–215.0 | 13.0–479.0 | 6.0–479.0 | |
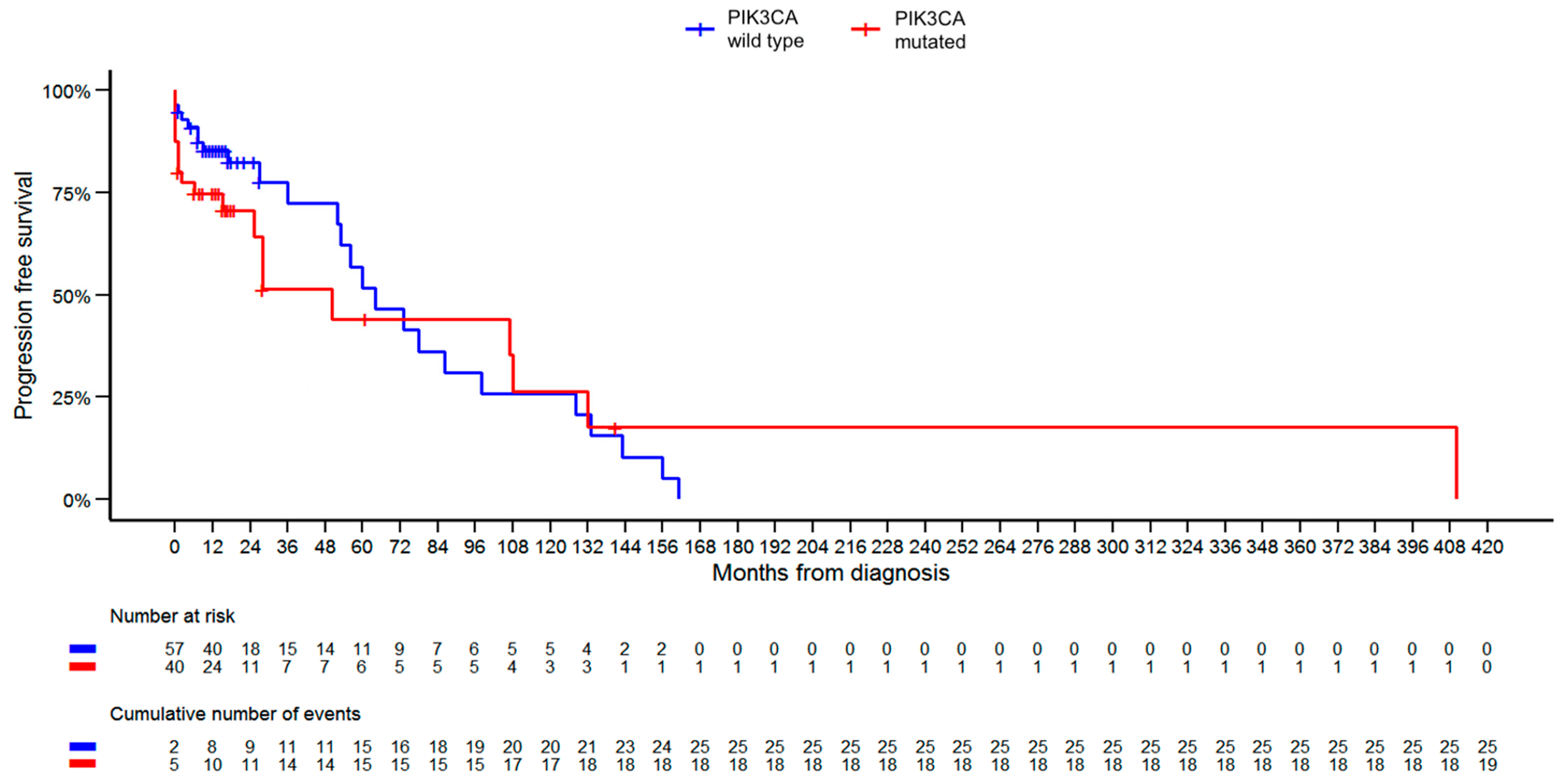
| PFS [months] | 0.2591 | |||
| Mean (SD) | 32.7 (40.7) | 34.0 (68.9) | 33.2 (53.7) | |
| Median (Q1, Q3) | 16.0 (9.0, 36.0) | 14.0 (6.0, 28.0) | 16.0 (7.0, 28.0) | |
| Range | 0.0–161.0 | 0.0–410.0 | 0.0–410.0 | |
| Death | 1 | |||
| no | 56 (91.8%) | 38 (90.5%) | 94 (91.3%) | |
| yes | 5 (8.2%) | 4 (9.5%) | 9 (8.7%) |
| OR | 95% CI | p–Value | ||
|---|---|---|---|---|
| Age | 1) <50 years | |||
| 2) ≥50 years | 1.45 | 0.59–3.54 | 0.417 | |
| Grade | 1) G1 | |||
| 2) G2 or G3 | 0.83 | 0.35–1.94 | 0.6627 | |
| ER | 1) Negative (−) | |||
| 2) Positive (+, ++, +++) | 2.57 | 0.78–8.53 | 0.1223 | |
| PR | 1) Negative (−) | |||
| 2) Positive (+, ++, +++) | 2.62 | 1–6.91 | 0.0508 | |
| HER2 1 | 1) Negative (−) | |||
| 2) Positive (+, ++, +++) | 0.39 | 0.1–1.52 | 0.1762 | |
| Ki67 | 1) Low (<20%) | |||
| 2) High (≥20%) | 0.45 | 0.2–1.03 | 0.0575 | |
| TNBC | 1) no | |||
| 2) yes | 0.33 | 0.07–1.65 | 0.1767 | |
| cT | 1) T1 or T2 | |||
| 2) T3 or T4 | 1.49 | 0.67–3.33 | 0.3335 | |
| cN | 1) N0 | |||
| 2) Other than N0 | 1.66 | 0.75–3.72 | 0.2139 | |
| cM | 1) M0 | |||
| 2) M1 | 6.04 | 1.55–23.55 | 0.0096 | |
| Stage | I | |||
| II | 1.94 | 0.47–8.03 | 0.3629 | |
| III | 1.5 | 0.32–6.94 | 0.604 | |
| IV | 10 | 1.59–62.73 | 0.014 | |
| Stage | 1) I or II | |||
| 2) III or IV | 1.57 | 0.7–3.52 | 0.2698 | |
| Stage | 1) I or II or III | |||
| 2) IV | 6.04 | 1.55–23.55 | 0.0096 | |
| Death | 1) no | |||
| 2) yes | 1.18 | 0.3–4.68 | 0.8148 |
| No. | PIK3CA Status | Age at Diagnosis | HER2 Status | Metastasis Sites |
|---|---|---|---|---|
| 1 | PIK3CA-WT | 62 | Negative | Bone |
| 2 | PIK3CA-WT | 46 | Negative | Bone, liver |
| 3 | PIK3CA-WT | 57 | Negative | Bone, lung |
| 4 | PIK3CA-M | 70 | Negative | Lung |
| 5 | PIK3CA-M | 60 | Negative | Bone, pleura, ovaries |
| 6 | PIK3CA-M | 75 | Negative | Lung, bone |
| 7 | PIK3CA-M | 50 | Negative | Bone |
| 8 | PIK3CA-M | 38 | Negative | Liver |
| 9 | PIK3CA-M | 69 | Positive | Lung, pleura |
| 10 | PIK3CA-M | 57 | Negative | Bone |
| 11 | PIK3CA-M | 64 | Negative | Bone |
| 12 | PIK3CA-M | 70 | Negative | Liver |
| 13 | PIK3CA-M | 71 | Negative | Bone |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smok-Kalwat, J.; Chmielewski, G.; Stando, R.; Sadowski, J.; Macek, P.; Kowalik, A.; Nowak-Ozimek, E.; Góźdź, S. Next-Generation Sequencing-Based Analysis of Clinical and Pathological Features of PIK3CA-Mutated Breast Cancer. Diagnostics 2023, 13, 2887. https://doi.org/10.3390/diagnostics13182887
Smok-Kalwat J, Chmielewski G, Stando R, Sadowski J, Macek P, Kowalik A, Nowak-Ozimek E, Góźdź S. Next-Generation Sequencing-Based Analysis of Clinical and Pathological Features of PIK3CA-Mutated Breast Cancer. Diagnostics. 2023; 13(18):2887. https://doi.org/10.3390/diagnostics13182887
Chicago/Turabian StyleSmok-Kalwat, Jolanta, Grzegorz Chmielewski, Rafał Stando, Jacek Sadowski, Paweł Macek, Artur Kowalik, Ewelina Nowak-Ozimek, and Stanisław Góźdź. 2023. "Next-Generation Sequencing-Based Analysis of Clinical and Pathological Features of PIK3CA-Mutated Breast Cancer" Diagnostics 13, no. 18: 2887. https://doi.org/10.3390/diagnostics13182887
APA StyleSmok-Kalwat, J., Chmielewski, G., Stando, R., Sadowski, J., Macek, P., Kowalik, A., Nowak-Ozimek, E., & Góźdź, S. (2023). Next-Generation Sequencing-Based Analysis of Clinical and Pathological Features of PIK3CA-Mutated Breast Cancer. Diagnostics, 13(18), 2887. https://doi.org/10.3390/diagnostics13182887







