Detection and Classification of Melanoma Skin Cancer Using Image Processing Technique
Abstract
:1. Introduction
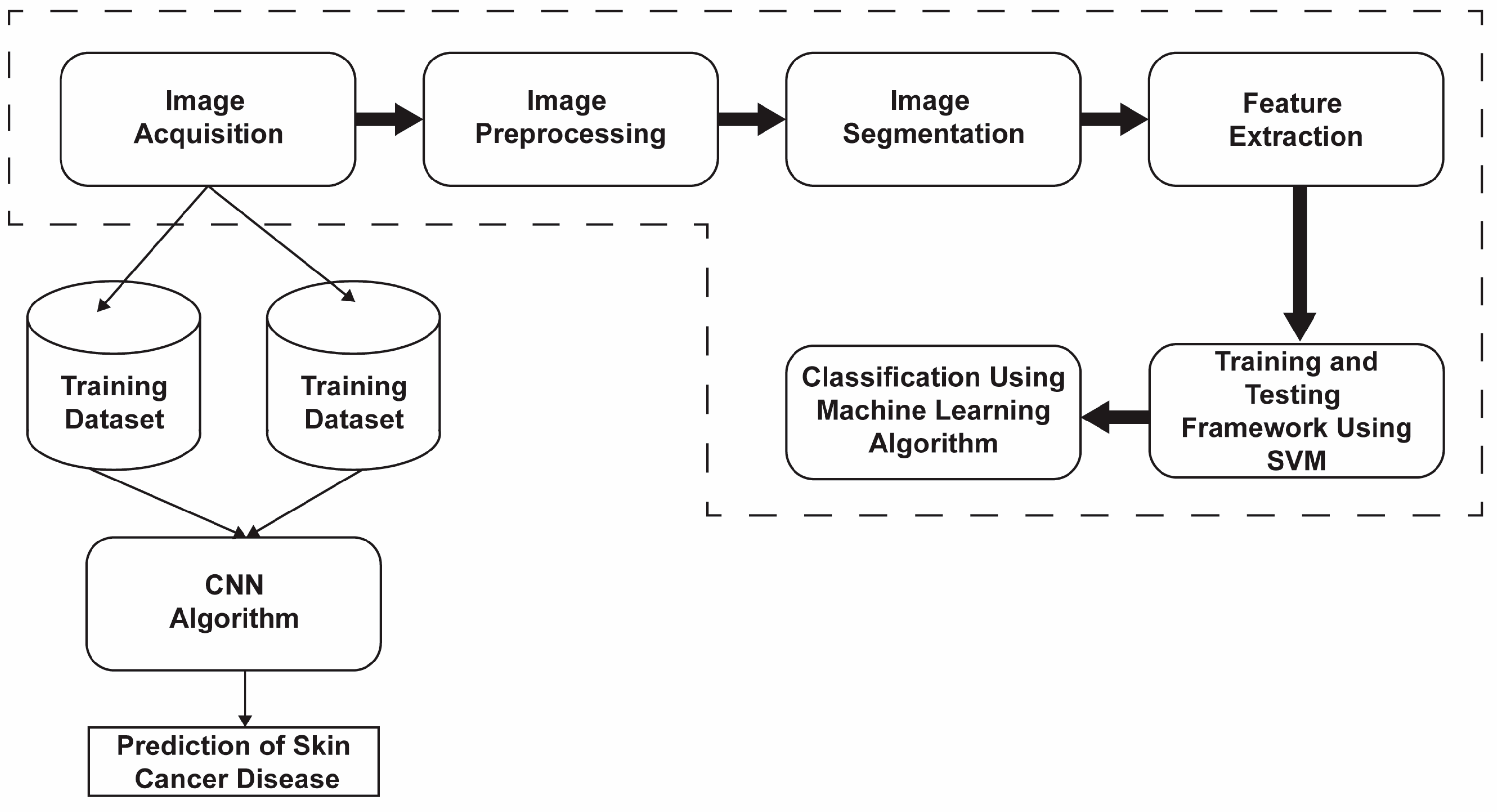
2. Materials and Methods
- Resize the fill in the image.
- Convert RGB images to grayscale images. Grayscale images enable the automatic processing that is often used in digital systems.
- Put the grayscale image through five filtering processes to reduce noise, determine the noise reduction target, detect and remove hair, and heal skin damage.
- Ng shows the quantity of the various power values.
- Ns shows the quantity of the individual zones.
- P(i,j) is the size zone framework.
- is the number of zones in the ROI.
- P(i,j) is the co-event framework.
- N is the number of dark levels.
- px+y(k) = Σ ΣP(i,j); px−y (k) = Σ ΣP(i,j); µx+y= Σkpx−y (k).
- p(i,j) is the normalized co-occurrence matrix and is equal to .
Model Evaluation
- The enhanced data are then used to create new images with rotation, cropping, scaling, transforming, and horizontal and vertical features. These mathematical changes were made without altering the original data, and they increased the accuracy of the model used. The rotation changes the image by rotating it clockwise or counterclockwise at random angles. Flipping is the process of turning an image along its horizontal or vertical axis. Translation moves the image along the x or y axis. The clipping replaces the image content with a random page. Resizing increases the size of the image. After all the transformations are performed, each image is normalized. When the dataset is increased, it is divided into training and testing sets. During training, 70% of the dataset is used for model learning, training, and validation, and 30% is used for model testing and evaluation. Image file creators help with this.
- Both the Conv2D layer and the max pooling layer are used to extract features and reduce the dimensions. The output channels of the two Conv2D layers are set to 32 and 64, respectively. ReLU is used in the activation function to mitigate the vanishing gradient problem and to expedite the learning process. The padding is set equally for all the layers to ensure that the output dimensions match the input dimensions.
- Following the pooling layer is a ReLU function that down-samples the convolutional feature map while preserving important feature information. A commonly used max-pooling technique employs an NxN pooling filter, sweeps across the data feature map, selects the highest value, and discards all other values to generate the output feature map.
- After the two Conv2D layers and pooling layers, a dropout layer with a dropout probability of 0.2 is added to constrain the composition and prevent overfitting. Finally, one even coat and two thick coats are applied. The dense layer contains 3 neurons corresponding to the patient’s skin.
- The project compared four models and chose the most reliable for web and mobile use. To make the comparison fair and efficient, each model was trained with a batch size of 32 and optimized using Adams optimization with categorical cross-entropy as a failover.
- The model was trained once more (100 times) to reduce the error rate. Each period includes training, validation, and training and validation. The prior learning is 0.0001, and the measurement accuracy is correct.
- The graph showing accuracy and loss is always seen. The success rate is approximately 91%. These criteria determine whether a skin lesion is benign, malignant, or normal over time. keras.models.save_model() is used to save the models and to deploy them to local servers and mobile applications using Django, a powerful web development framework. The overall working of the web and Android development is shown in Figure 3.
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Subramanian, R.R.; Dintakurthi, A.; Kumar, S.; Reddy, K.; Amara, S.; Chowdary, A. Skin Cancer Classification Using Convolutional Neural Networks. In Proceedings of the 2021 11th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Noida, India, 28–29 January 2021. [Google Scholar]
- Harsha, P.; Sahruthi, G.; Vaishnavi, A.; Varshini, P. Spotting Skin Cancer Using CNN. Int. J. Eng. Tech. 2022, 8, 47–51. [Google Scholar]
- Dubai, P.; Bhatt, S.; Joglekar, C.; Patii, S. Skin Cancer Detection and Classification. In Proceedings of the 2017 6th International Conference on Electrical Engineering and Informatics (ICEEI), Langkawi, Malaysia, 25–27 November 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Jana, E.; Subban, R.; Saraswathi, S. Research on Skin Cancer Cell Detection Using Image Processing. In Proceedings of the 2017 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Coimbatore, India, 14–16 December 2017. [Google Scholar]
- Dildar, M.; Akram, S.; Irfan, M.; Khan, H.U.; Ramzan, M.; Mahmood, A.R.; Alsaiari, S.A.; Saeed, A.H.M.; Alraddadi, M.O.; Mahnashi, M.H. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479. [Google Scholar] [CrossRef] [PubMed]
- Hosny, K.; Kassem, M.; Fouad, M. Skin Cancer Classification Using Deep Learning and Transfer Learning. In Proceedings of the 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), Cairo, Egypt, 20–22 December 2018. [Google Scholar]
- Ashraf, R.; Kiran, I.; Mahmood, T.; Butt, A.; Razzaq, N.; Farooq, Z. An Efficient Technique for Skin Cancer Classification Using Deep Learning. In Proceedings of the 2020 IEEE 23rd International Multitopic Conference (INMIC), Bahawalpur, Pakistan, 5–7 November 2020. [Google Scholar]
- Chaturvedi, S.S.; Tembhurne, J.V.; Diwan, T. A Multi-Class Skin Cancer Classification Using Deep Convolutional Neural Networks. Multimed. Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Marka, A.; Carter, J.B.; Toto, E.; Hassanpour, S. Automated Detection of Nonmelanoma Skin Cancer Using Digital Images: A Systematic Review. BMC Med. Imaging 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Jojoa Acosta, M.F.; Caballero Tovar, L.Y.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma Diagnosis Using Deep Learning Techniques on Dermatoscopic Images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef]
- Javaid, A.; Orakzai, M.; Akram, F. Skin Cancer Classification Using Image Processing and Machine Learning. In Proceedings of the 2021 International Bhurban Conference on Applied Sciences and Technologies (IBCAST), Islamabad, Pakistan, 12–16 January 2021. [Google Scholar]
- Hong, J.; Yu, S.C.H.; Chen, W. Unsupervised Domain Adaptation for Cross-Modality Liver Segmentation via Joint Adversarial Learning and Self-Learning. Appl. Soft Comput. 2022, 121, 108729. [Google Scholar] [CrossRef]
- Hong, J.; Zhang, Y.D.; Chen, W. Source-Free Unsupervised Domain Adaptation for Cross-Modality Abdominal Multi-Organ Segmentation. Knowl.-Based Syst. 2022, 250, 109155. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Ali, M.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M. An Enhanced Technique of Skin Cancer Classification Using Deep Convolutional Neural Network with Transfer Learning Models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Younis, H.; Bhatti, M.H.; Azeem, M. Classification of Skin Cancer Dermoscopy Images Using Transfer Learning. In Proceedings of the 2019 15th International Conference on Emerging Technologies (ICET), Peshawar, Pakistan, 2–3 December 2019; pp. 1–4. [Google Scholar]
- Dascalu, A.; Walker, B.N.; Oron, Y.; David, E.O. Non-Melanoma Skin Cancer Diagnosis: A Comparison between Dermoscopic and Smartphone Images by Unified Visual and Sonification Deep Learning Algorithms. J. Cancer Res. Clin. Oncol. 2022, 148, 2497–2505. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Hanaoka, J.; Ohshio, Y.; Okamoto, K.; Kaku, R.; Hayashi, K.; Shiratori, T.; Yoden, M. Sarcopenia Predicts Poor Postoperative Outcome in Elderly Patients with Lung Cancer. Gen. Thorac. Cardiovasc. Surg. 2019, 67, 949–954. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Meshram, A.A.; Abhimanyu Dutonde, A.G. A Review of Skin Melanoma Detection Based on Machine Learning. Int. J. New Pract. Manag. Eng. 2022, 11, 15–23. [Google Scholar] [CrossRef]
- Babu, G.N.K.; Peter, V.J. Skin Cancer Detection Using Support Vector Machine with Histogram of Oriented Gradients Features. ICTACT J. Soft Comput. 2021, 6956, 2301–2305. [Google Scholar] [CrossRef]
- Faiza, F.; Irfan Ullah, S.; Salam, A.; Ullah, F.; Imad, M.; Hassan, M. Diagnosing of Dermoscopic Images Using Machine Learning Approaches for Melanoma Detection. In Proceedings of the 2020 IEEE 23rd International Multitopic Conference (INMIC), Bahawalpur, Pakistan, 5–7 November 2020. [Google Scholar]
- Majumder, S.; Ullah, M.A. Feature Extraction from Dermoscopy Images for Melanoma Diagnosis. SN Appl. Sci. 2019, 1, 753. [Google Scholar] [CrossRef]
- Chaitanya, K.N.; Pavani, I. Melanoma Early Detection Using Dual Classifier. Int. J. Sci. Eng. Technol. Res. 2015, 1, 1–6. [Google Scholar]
- Bernstein, M.N.; Gladstein, A.; Latt, K.Z.; Clough, E.; Busby, B.; Dillman, A. Jupyter Notebook-Based Tools for Building Structured Datasets from the Sequence Read Archive. F1000Research 2020, 9, 376. [Google Scholar] [CrossRef]
- Fan, L.; Zhang, F.; Fan, H.; Zhang, C. Brief Review of Image Denoising Techniques. Vis. Comput. Ind. Biomed. Art 2019, 2, 7. [Google Scholar] [CrossRef]
- Ansari, U.; Sarode, T.K. Skin Cancer Detection Using Image Processing. In Proceedings of the WSCG 2021: Full Papers Proceedings: 29. International Conference in Central Europe on Computer Graphics, Visualization and Computer Vision, Pilsen, Czech Republic, 17–21 May 2021. [Google Scholar]
- Senan, E.; Jadhav, M. Analysis of Dermoscopy Images by Using ABCD Rule for Early Detection of Skin Cancer. Glob. Transitions Proc. 2021, 2, 1–7. [Google Scholar] [CrossRef]
- Mabrouk, M.; Sheha, M.; Sharawi, A. Automatic Detection of Melanoma Skin Cancer Using Texture Analysis. Int. J. Comput. Appl. 2012, 42, 22–26. [Google Scholar] [CrossRef]
- Seeja, R.D.; Suresh, A. Deep Learning Based Skin Lesion Segmentation and Classification of Melanoma Using Support Vector Machine (SVM). Asian Pac. J. Cancer Prev. 2019, 20, 1555–1561. [Google Scholar] [CrossRef]
- Do, T.T.; Hoang, T.; Pomponiu, V.; Zhou, Y.; Chen, Z.; Cheung, N.M.; Koh, D.; Tan, A.; Tan, S.H. Accessible Melanoma Detection Using Smartphones and Mobile Image Analysis. IEEE Trans. Multimed. 2017, 20, 2849–2864. [Google Scholar] [CrossRef]
- Ozkan, I.A.; Koklu, M. Skin Lesion Classification Using Machine Learning Algorithms. Int. J. Intell. Syst. Appl. Eng. 2017, 5, 285–289. [Google Scholar] [CrossRef]
- Murugan, A.; Nair, A.; Kumar, K.P. Research on SVM and KNN Classifiers for Skin Cancer Detection. Int. J. Eng. Adv. Technol. 2020, 9, 4627–4632. [Google Scholar] [CrossRef]
- Mijwil, M.M. Skin Cancer Disease Images Classification Using Deep Learning Solutions. Multimed. Tools Appl. 2021, 80, 26255–26271. [Google Scholar] [CrossRef]
- Naeem, A.; Farooq, S.; Khelifi, A.; Abid, A. Malignant Melanoma Classification Using Deep Learning: Datasets, Performance Measurements, Challenges and Opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar] [CrossRef]
- Junayed, M.S.; Anjum, N.; Noman, A.; Islam, B. A Deep CNN Model for Skin Cancer Detection and Classification. In Proceedings of the WSCG 2021: Full Papers Proceedings: 29. International Conference in Central Europe on Computer Graphics, Visualization and Computer Vision, Pilsen, Czech Republic, 17–21 May 2021. [Google Scholar]


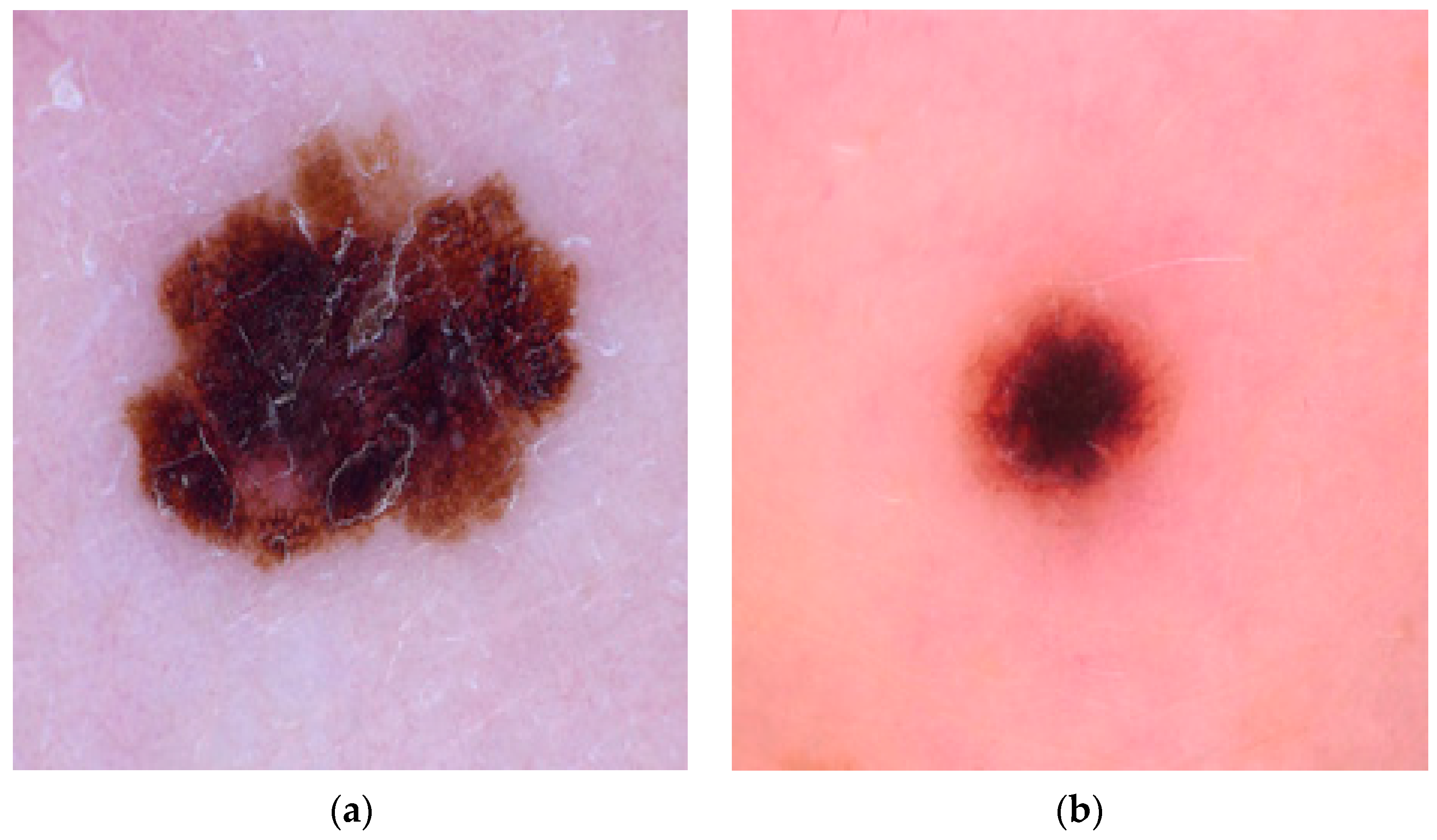





















| S. No. | Images | Threshold | Entropy |
|---|---|---|---|
| 1 | 1 | 164 | 4.415 |
| 2 | 2 | 160 | 3.977 |
| S. No. | Features | Benign | Malignant |
|---|---|---|---|
| 1 | Mean | 67.9958 ± 18.841 | 75.926 ± 25.194 |
| 2 | Median | 66.85 ± 22.43216 | 69.69 ± 28.15571 |
| 3 | Area | 6727.06 ± 3149.772 | 13,009.76 ± 6649.319 |
| 4 | Perimeter | 289.52 ± 92.624 | 405.02 ± 119.0192 |
| 5 | Coefficient of variation | 0.0815 ± 0.009398 | 0.00204 ± 0.016354 |
| 6 | Inverse difference moment | 0.205417 ± 0.070323 | 0.1841 ± 0.0543 |
| 7 | Sum average | 139.131 ± 47.795 | 150.46 ± 60.2164 |
| 8 | Gray level variance | 20.23 ± 15.1 | 8.221 ± 5.54 |
| 9 | Zone size entropy | −7.2965 ± 0.21483 | −7.51093 ± 0.29246 |
| 10 | Difference entropy | −3.7532 ± 0.76304 | −3.9180 ± 0.71125 |
| 11 | Histogram width | 69.85 ± 17.658 | 72.852 ± 22.28 |
| 12 | Maximum gray level intensity | 162.3 ± 18.04381 | 185.46 ± 31.8422 |
| 13 | Minimum gray level intensity | 31.5 ± 15.13106 | 21.6078 ± 18.0431 |
| 14 | Coefficient of variation | 0.0815 ± 0.009398 | 0.00204 ± 0.016354 |
| 15 | Inverse difference moment | 0.205417 ± 0.070323 | 0.1841 ± 0.0543 |
| 16 | Sum average | 139.131 ± 47.795 | 150.46 ± 60.2164 |
| 17 | Gray level variance | 20.23 ± 15.1 | 8.221 ± 5.54 |
| 18 | Zone size entropy | −7.2965 ± 0.21483 | −7.51093 ± 0.29246 |
| 19 | Difference entropy | −3.7532 ± 0.76304 | −3.9180 ± 0.71125 |
| 20 | Histogram width | 69.85 ± 17.658 | 72.852 ± 22.28 |
| 21 | Maximum gray level intensity | 162.3 ± 18.04381 | 185.46 ± 31.8422 |
| Classification Report of Support Vector Classifier | |||||
|---|---|---|---|---|---|
| S. No. | Precision | Recall | F1-Score | Support | |
| 1. | Benign | 0.82 | 0.93 | 0.87 | 15 |
| 2. | Malignant | 0.92 | 0.80 | 0.86 | 15 |
| S. No. | Precision (%) | Recall (%) | F1 Score (%) | |
|---|---|---|---|---|
| 1. | Benign | 82 | 93 | 87 |
| 2. | Malignant | 92 | 80 | 86 |
| S. No. | Parameters | Overall % |
|---|---|---|
| 1 | Accuracy | 86.6 |
| 2 | Sensitivity | 82.3 |
| 3 | Specificity | 92.3 |
| S. No. | Image Count | Classes | |
|---|---|---|---|
| 1. | Training | 470 | 3 |
| 2. | Testing | 203 | 3 |
| S. No. | Training Data for Malignant | |
|---|---|---|
| Images in: Data/Train/Malignant | ||
| 1. | images_count: | 178 |
| 2. | min_width: | 224 |
| 3. | max_width: | 224 |
| 4. | min_height: | 224 |
| 5. | max_height: | 224 |
| S. No. | Training Data for Benign | |
|---|---|---|
| Images in: Data/Train/Benign | ||
| 1. | images_count: | 178 |
| 2. | min_width: | 224 |
| 3. | max_width: | 224 |
| 4. | min_height: | 224 |
| 5. | max_height: | 224 |
| S. No. | Training Data for Normal | |
|---|---|---|
| Images in: Data/Train/Normal | ||
| 1. | images_count: | 114 |
| 2. | min_width: | 224 |
| 3. | max_width: | 224 |
| 4. | min_height: | 224 |
| 5. | max_height: | 224 |
| Model: “Sequential” | |||
|---|---|---|---|
| S. No. | Layer (Type) | Output Shape | Parameter |
| 1. | conv2d (Conv2D) | (None, 56, 56, 32) | 896 |
| 2. | activation (Activation) | (None, 56, 56, 32) | 0 |
| 3. | max_pooling2d (MaxPooling2D) | (None, 28, 28, 32) | 0 |
| 4. | conv2d_1 (Conv2D) | (None, 12, 12, 64) | 51,264 |
| 5. | activation (Activation) | (None, 12, 12, 64) | 0 |
| 6. | max_pooling2d_1 (MaxPooling2D) | (None, 6, 6, 64) | 0 |
| 7. | flatten (Flatten) | (None, 2304) | 0 |
| 8. | dense (Dense) | (None, 38) | 87,590 |
| 9. | dropout (Dropout) | (None, 38) | 0 |
| 10. | Dense_1 (Dense) | (None, 3) | 117 |
| Performance of the Trained Results | ||||||||
|---|---|---|---|---|---|---|---|---|
| S. No. | Models | Database | Epochs | Batch Size | Training Accuracy (%) | Training Loss (%) | Testing Accuracy (%) | Testing Loss (%) |
| 1. | ALEX NET | ISIC | 100 | 32 | 91 | 23 | 84 | 78 |
| 2. | LENET | ISIC | 100 | 32 | 90 | 22 | 81 | 59 |
| 3. | VGG16 | ISIC | 100 | 32 | 38 | 107 | 37 | 108 |
| 4. | DESIGNED NETWORK | ISIC | 100 | 32 | 94 | 15 | 91 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viknesh, C.K.; Kumar, P.N.; Seetharaman, R.; Anitha, D. Detection and Classification of Melanoma Skin Cancer Using Image Processing Technique. Diagnostics 2023, 13, 3313. https://doi.org/10.3390/diagnostics13213313
Viknesh CK, Kumar PN, Seetharaman R, Anitha D. Detection and Classification of Melanoma Skin Cancer Using Image Processing Technique. Diagnostics. 2023; 13(21):3313. https://doi.org/10.3390/diagnostics13213313
Chicago/Turabian StyleViknesh, Chandran Kaushik, Palanisamy Nirmal Kumar, Ramasamy Seetharaman, and Devasahayam Anitha. 2023. "Detection and Classification of Melanoma Skin Cancer Using Image Processing Technique" Diagnostics 13, no. 21: 3313. https://doi.org/10.3390/diagnostics13213313
APA StyleViknesh, C. K., Kumar, P. N., Seetharaman, R., & Anitha, D. (2023). Detection and Classification of Melanoma Skin Cancer Using Image Processing Technique. Diagnostics, 13(21), 3313. https://doi.org/10.3390/diagnostics13213313






