Current Status and Future of Artificial Intelligence in MM Imaging: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection
2.3. Data Extraction
2.4. Study Evaluation
3. Results
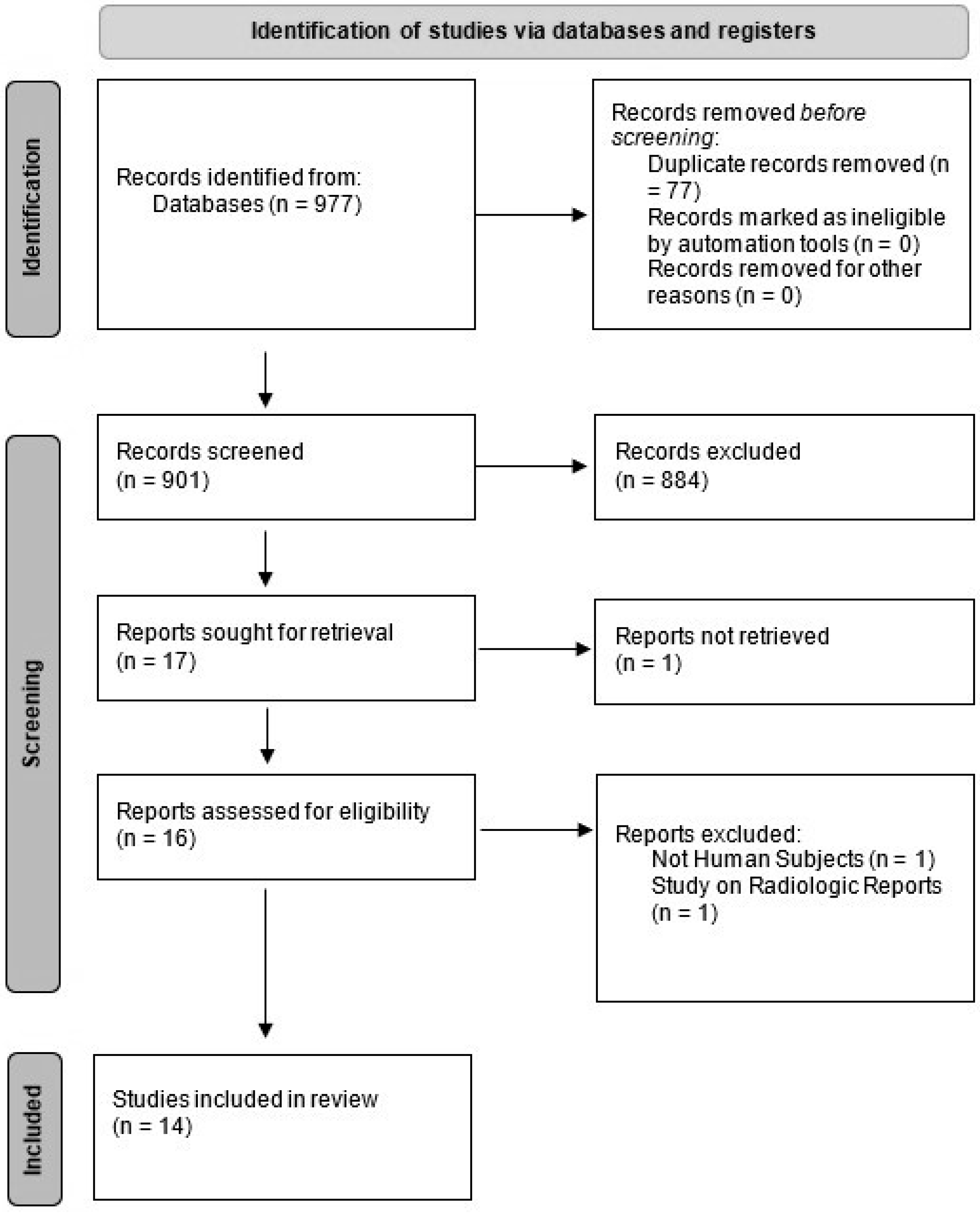
3.1. Study Selection
3.2. Characteristics of Included Studies
3.3. Radiomics Studies
3.4. Segmentation Studies
3.5. Other Types of Research
3.6. CLAIM Checklist Evaluation
4. Discussion
4.1. Radiomics in MM
4.2. ROI Segmentation in MM
4.3. Other Types of Studies
4.4. CLAIM Evaluation
4.5. Recommendations for Future Directions
4.6. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cowan, A.J.; Green, D.J.; Kwok, M. Diagnosis and Management of Multiple Myeloma: A Review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Ormond Filho, A.G.; Carneiro, B.C.; Pastore, D.; Silva, I.P.; Yamashita, S.R.; Consolo, F.D.; Hungria, V.T.M.; Sandes, A.F.; Rizzatti, E.G.; Nico, M.A.C. Whole-Body Imaging of Multiple Myeloma: Diagnostic Criteria. Radiographics 2019, 39, 1077–1097. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538-48. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Barsouk, A. Epidemiology, Staging, and Management of Multiple Myeloma. Med. Sci. 2021, 9, 3. [Google Scholar] [CrossRef]
- Nakaya, A.; Fujita, S.; Satake, A. Impact of CRAB Symptoms in Survival of Patients with Symptomatic Myeloma in Novel Agent Era. Hematol. Rep. 2017, 9, 6887. [Google Scholar] [CrossRef]
- Silbermann, R.; Roodman, G.D. Myeloma bone disease: Pathophysiology and management. J. Bone Oncol. 2013, 2, 59–69. [Google Scholar] [CrossRef]
- Kyle, R.A. Multiple myeloma: Review of 869 cases. Mayo Clin. Proc. 1975, 50, 29–40. [Google Scholar]
- Zamagni, E.; Tacchetti, P.; Cavo, M. Imaging in multiple myeloma: How? When? Blood 2019, 133, 644–651. [Google Scholar] [CrossRef]
- Mena, E.; Choyke, P.; Tan, E.; Landgren, O.; Kurdziel, K. Molecular imaging in myeloma precursor disease. Semin. Hematol. 2011, 48, 22–31. [Google Scholar] [CrossRef]
- Terpos, E.; Kleber, M.; Engelhardt, M. European Myeloma Network guidelines for the management of multiple myeloma-related complications. Haematologica 2015, 100, 1254–1266. [Google Scholar] [CrossRef]
- Raza, S.; Leng, S.; Lentzsch, S. The Critical Role of Imaging in the Management of Multiple Myeloma. Curr. Hematol. Malig. Rep. 2017, 12, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Derlin, T.; Bannas, P. Imaging of multiple myeloma: Current concepts. World J. Orthop. 2014, 5, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Gitto, S.; Cuocolo, R.; Albano, D. CT and MRI radiomics of bone and soft-tissue sarcomas: A systematic review of reproducibility and validation strategies. Insights Imaging 2021, 12, 68. [Google Scholar] [CrossRef]
- Xue, C.; Yuan, J.; Lo, G.G. Radiomics feature reliability assessed by intraclass correlation coefficient: A systematic review. Quant Imaging Med. Surg. 2021, 11, 4431–4460. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, E.J.; Post, M.; Mannil, M.; Witkam, R.L.; Ter Laan, M.; Patel, A.; Meijer, F.J.A.; Henssen, D. Performance of machine learning algorithms for glioma segmentation of brain MRI: A systematic literature review and meta-analysis. Eur. Radiol. 2021, 31, 9638–9653. [Google Scholar] [CrossRef]
- Xie, T.; Wang, X.; Li, M.; Tong, T.; Yu, X.; Zhou, Z. Pancreatic ductal adenocarcinoma: A radiomics nomogram outperforms clinical model and TNM staging for survival estimation after curative resection. Eur. Radiol. 2020, 30, 2513–2524. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Yin, P. MRI-Based Bone Marrow Radiomics Nomogram for Prediction of Overall Survival in Patients With Multiple Myeloma. Front. Oncol. 2021, 11, 1929. [Google Scholar] [CrossRef]
- Peeken, J.C.; Neumann, J.; Asadpour, R.; Leonhardt, Y.; Moreira, J.R.; Hippe, D.S.; Klymenko, O.; Foreman, S.C.; von Schacky, C.E.; Spraker, M.B.; et al. Prognostic Assessment in High-Grade Soft-Tissue Sarcoma Patients: A Comparison of Semantic Image Analysis and Radiomics. Cancers 2021, 13, 1929. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Erdim, C.; Ates, E.; Kaya, O.K.; Kilickesmez, O. Radiomics of Renal Masses: Systematic Review of Reproducibility and Validation Strategies. Am. J. Roentgenol. 2020, 214, 129–136. [Google Scholar] [CrossRef]
- Allegra, A.; Tonacci, A.; Sciaccotta, R.; Genovese, S.; Musolino, C.; Pioggia, G.; Gangemi, S. Machine Learning and Deep Learning Applications in Multiple Myeloma Diagnosis, Prognosis, and Treatment Selection. Cancers 2022, 14, 606. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Mongan, J.; Moy, L.; Kahn, C.E., Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [PubMed]
- Schenone, D.; Dominietto, A.; Campi, C.; Frassoni, F.; Cea, M.; Aquino, S.; Angelucci, E.; Rossi, F.; Torri, L.; Bignotti, B.; et al. Radiomics and Artificial Intelligence for Outcome Prediction in Multiple Myeloma Patients Undergoing Autologous Transplantation: A Feasibility Study with CT Data. Diagnostics 2021, 11, 1759. [Google Scholar] [CrossRef] [PubMed]
- Fraenzle, A.; Bendl, R. Fully automated model positioning for shape-based bone segmentation. Radiother. Oncol. 2011, 99, S480–S481. [Google Scholar]
- Martínez-Martínez, F.; Kybic, J.; Lambert, L.; Mecková, Z. Fully automated classification of bone marrow infiltration in low-dose CT of patients with multiple myeloma based on probabilistic density model and supervised learning. Comput. Biol. Med. 2016, 71, 57–66. [Google Scholar] [CrossRef]
- Nishida, Y.; Kimura, S.; Mizobe, H.; Yamamichi, J.; Kojima, K.; Kawaguchi, A.; Fujisawa, M.; Matsue, K. Automatic Digital Quantification of Bone Marrow Myeloma Volume in Appendicular Skeletons-Clinical Implications and Prognostic Significance. Blood 2017, 130, 12885. [Google Scholar]
- Horger, M.; Ditt, H.; Liao, S.; Weisel, K.; Fritz, J.; Thaiss, W.M.; Kaufmann, S.; Nikolaou, K.; Kloth, C. Automated “Bone Subtraction” Image Analysis Software Package for Improved and Faster CT Monitoring of Longitudinal Spine Involvement in Patients with Multiple Myeloma. Acad. Radiol. 2017, 24, 623–632. [Google Scholar]
- Fervers, P.; Fervers, F.; Kottlors, J.; Lohneis, P.; Pollman-Schweckhorst, P.; Zaytoun, H.; Rinneburger, M.; Maintz, D.; Große Hokamp, N. Feasibility of artificial intelligence–supported assessment of bone marrow infiltration using dual-energy computed tomography in patients with evidence of monoclonal protein—A retrospective observational study. Eur. Radiol. 2021, 32, 2901–2911. [Google Scholar] [CrossRef]
- Satoh, Y.; Funayama, S.; Onishi, H.; Kirito, K. Semi-automated histogram analysis of normal bone marrow using (18)F-FDG PET/CT: Correlation with clinical indicators. BMC Med. Imaging 2022, 22, 31. [Google Scholar] [CrossRef]
- Takahashi, M.E.S.; Mosci, C.; Souza, E.M.; Brunetto, S.Q.; de Souza, C.; Pericole, F.V.; Lorand-Metze, I.; Ramos, C.D. Computed tomography-based skeletal segmentation for quantitative PET metrics of bone involvement in multiple myeloma. Nucl. Med. Commun. 2020, 41, 377–382. [Google Scholar] [CrossRef]
- Shi, K.; Xu, L.; Tetteh, G.; Gafita, A.; Eiber, M.; Buck, A.; Menze, B.H.; Rominger, A. Lesion detection for total-body PET imaging by means of deep learning. EJNMMI Phys. 2018, 5. [Google Scholar]
- Liu, J.; Wang, C.; Guo, W.; Zeng, P.; Liu, Y.; Lang, N.; Yuan, H. A preliminary study using spinal MRI-based radiomics to predict high-risk cytogenetic abnormalities in multiple myeloma. Radiol. Med. 2021, 126, 1226–1235. [Google Scholar] [PubMed]
- Wennmann, M.; Chmelik, J.; Bauer, F.; Klein, A.; Uhlenbrock, C.; Lochner, J.; Grözinger, M.; Rotkopf, L.; Sauer, S.; Bonekamp, D.; et al. P-012: Automatic bone marrow segmentation in whole-body magnetic resonance imaging: Towards comprehensive, objective MRI-phenotypic bone marrow characterization in multiple myeloma. Clin. Lymphoma Myeloma Leuk. 2021, 21, S45–S46. [Google Scholar] [CrossRef]
- Xiong, X.; Wang, J.; Hu, S.; Dai, Y.; Zhang, Y.; Hu, C. Differentiating Between Multiple Myeloma and Metastasis Subtypes of Lumbar Vertebra Lesions Using Machine Learning–Based Radiomics. Front. Oncol. 2021, 11, 601699. [Google Scholar] [PubMed]
- Wennmann, M.; Klein, A.; Bauer, F.; Chmelik, J.; Uhlenbrock, C.; Grözinger, M.; Rotkopf, L.; Sauer, S.; Thierjung, H.; Götz, M.; et al. P-018: Automatic analysis of magnetic resonance imaging in multiple myeloma patients: Deep-learning based pelvic bone marrow segmentation and radiomics analysis for prediction of plasma cell infiltration. Clin. Lymphoma Myeloma Leuk. 2021, 21, S49. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“How-to” guide and critical reflection. Insights Imaging. 2020, 11, 91. [Google Scholar] [CrossRef]
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A deep look into radiomics. Radiol Med. 2021, 126, 1296–1311. [Google Scholar] [CrossRef]
- The Precise4Q consortium; Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef]

| Authors (Year) | N | Imaging | Input * | Segmentation | Feature Reduction | Model/Analysis | Standard Test | Objective |
|---|---|---|---|---|---|---|---|---|
| Fervers et al., (2021) [28] | 35 | CT | CT values | Pretrained CNN | N/A | Multivariate regression | N/A | Estimate BM infiltration |
| Fraenzel et al., (2011) Abstract [24] | 14 | CT | Image | Threshold model | N/A | Threshold segmentation and flood fill algorithm | N/A | Automatic bone lesion detection |
| Horger et al., (2017) [27] | 188 | CT | N/A | N/A | N/A | N/A | Lab results and gold standard CT | Monitoring new bone lesions |
| Li et al., (2021) [17] | 121 | MRI | Image | Manual segmentation | Univariate regression + Spearman’s correlation + LASSO | Nomogram | N/A | To predict overall survival in MM patients using radiomics and clinical features |
| Liu et al., (2021) [32] | 50 | MRI | Radiomics | Manual segmentation | ICC + LASSO | Logistic regression | FISH | Detection of high-risk cytogenic abnormality |
| Martínez-Martínez et al., (2016) [25] | 127 | CT | Image | Shape model positioning | N/A | Probabilistic density model | Imaging diagnosis | Bone marrow infiltration |
| Nishida et al., (2017) [26] | 68 | CT | Cumulative CT values | Threshold model | N/A | Pearson’s correlation | Histology | BM infiltration |
| Satoh et al., (2022) [29] | 98 | PET/CT | Image | Deep learning-based automatic organ segmentation | N/A | Histogram analysis | N/A | Establish a standard for BM FDG uptake |
| Schenone et al., (2021) [23] | 33 | CT | Clinical data + Image | Manual Segmentation | PCA, Pearson’s correlation | FCM, Extended version of HTF | Clinical diagnosis | Prognosis |
| Shi et al., (2018) Abstract [31] | 12 | PET/CT | Image | Manual segmentation | N/A | V-Net and W-Net | Manual segmentation | Computer-aided lesion detection |
| Takahashi et al., (2020) [30] | 58 | PET/CT | Standardized uptake values | Fully automated segmentation | N/A | Generalized estimating equation | Visual analysis | Standardizing VOI determination in PET/CT scans |
| Wennmann et al., (2021) Abstract [33] | 66 | MRI | Image | Manual segmentation | N/A | nnU-Net | Manual segmentation | BM segmentation |
| Wennmann et al., (2021) Abstract [35] | 270 | MRI | Radiomics | nn-Unet | N/A | Random Forest | Manual segmentation, biopsy | Estimate plasma cell infiltration |
| Xiong et al., (2021) [34] | 107 | MRI | Radiomics | Manual segmentation | ICC + LASSO | ANN | Clinical diagnosis | Differentiation of metastatic lesions from MM |
| Radiomics | ||||
|---|---|---|---|---|
| Authors (Year) | Methods | Performance | Conclusion | |
| Schenone et al., (2021) [23] | FCM, Extended version of HTF | Critical success index: 0.52 ± 0.1 | Radiomic has the potential for the stratification of relapsed and non-relapsed MM patients. | |
| Xiong et al., (2021) [34] | LASSO + Artificial Neural Networks | MCC for ANN on T2WI: 0.605 | Machine learning on radiomics features extracted from conventional MRI sequences can help differentiate newly diagnosed MM lesions from other cancer metastatic lesions in lumbar vertebrae. | |
| Li et al., (2021) [17] | LASSO + Nomogram | Radiomics nomogram c-index on validation cohort: 0.81 (95% CI: 0.70–0.92) | The development of a radiomics nomogram can help with the prediction of overall survival in MM patients. | |
| Liu et al., (2021) [32] | LASSO + Logistic Regression | Radiomics test AUC: 0.863 Combined test AUC: 0.870 | Radiomics features in conventional MRI images of MM patients can help distinguish HRCAs and non-HRCAs. | |
| Wennmann et al., (2021) [35] | Random Forest | MAE: 14.3 compared to biopsy | The tool has an accuracy comparable to a radiologist in predicting plasma cell infiltration percentage. | |
| Segmentation | ||||
| Authors (year) | ROI | Classifier | Performance | Conclusion |
| Fraenzel et al., (2011) [24] | Medullary Cavity of Bones | Random Forest | Classification accuracy: Femur: 90%, Tibia: 79%, Fibula: 79%, Humerus: 93%, Radius: 69%, Ulna: 46%, Other: 99% | Classification of medullary cavities enables the identification of long bone structures in whole-body CT scans. |
| Shi et al., (2018) [31] | Bone Lesions | V-Net, W-Net | V-Net: Sensitivity: 71.1%, Specificity: 99.5%, W-Net: Sensitivity 73.5%, Specificity: 99.6% | W-Nets have superior performance compared to traditional ML models in the detection of bone lesions. |
| Takahashi et al., (2020) [30] | Bone | Threshold | SUVmean was correlated with visual assessment, OR: 10.52 (95% CI, 5.68–19.48), p < 0.0001 | CT–based skeletal segmentation allows for the automated and therefore reproducible calculation of PET quantitative parameters of bone involvement in MM patients. |
| Wennmann et al., (2021) Abstract [33] | Bone Marrow | nn-Unet | Mean Dice scores ranging from 0.80 to 0.97 | A Unet model can be used to segment out bone marrow structures of the body with high accuracy. |
| Other | ||||
| Authors (year) | Measure | Performance | Conclusion | |
| Martínez-Martínez et al. (2016) [25] | Diagnosis accuracy | Infiltration vs. Healthy group: SVM, AUC: 0.996 ± 0.009; Infiltration vs. All other: k-NN, AUC: 0.894 ± 0.070 | Classification based on features extracted from the probabilistic density model using k-NN, allowing differentiation of patients with infiltration from others. | |
| Nishida et al., (2017) [26] | Correlation with clinical features | cCTv is correlated with higher R-ISS stage and serum or urine M-protein cCTv is inversely correlated with therapy and serum albumin | cCTv demonstrated a relationship with disease aggressiveness and had prognostic value. | |
| Horger et al. (2017) [27] | Diagnosis accuracy | Performance of radiologist while using the tool: Sensitivity: 97.8%, Specificity: 96.7%, Accuracy: 97.7% | Accuracy was slightly increased and reading time was significantly reduced when using subtraction maps. | |
| Satoh et al., (2022) [29] | N/A | SLUmean in men: 0.79 (95% CI 0.78–0.90), SLUmean in women: 0.75 (95% CI 0.74–0.76), SLUmean inversely correlated with age | A normal FDG uptake pattern was demonstrated by simplified FDG PET/CT bone marrow quantification. | |
| Fervers et al., (2021) [28] | Diagnostic accuracy | AUROC for osteolytic lesions: 0.70 (95% CI, 0.49–0.90), AUROC for MM diagnosis: 0.71 (95% CI 0.54–0.89) | Automated, AI-supported attenuation assessment of the spine in DECT after VNCa is feasible to predict BM infiltration in MM. | |
| Category | Subcategory | Item | Percentage of Articles That Were Compliant |
|---|---|---|---|
| Title/Abstract | Identification as a study of AI methodology, specifying the category of technology used (e.g., deep learning) | 87 | |
| Title/Abstract | Structured summary of study design, methods, results, and conclusions | 62 | |
| Introduction | Scientific and clinical background, including the intended use and clinical role of the AI approach | 100 | |
| Introduction | Study objectives and hypotheses | 100 | |
| Methods | Study design | Prospective or retrospective study | 75 |
| Methods | Study design | Study goals, such as model creation, exploratory study, feasibility study, and non-inferiority trial | 87 |
| Methods | Data | Data sources | 100 |
| Methods | Data | Eligibility criteria: how, where, and when potentially eligible participants or studies were identified. (e.g., symptoms, results from previous tests, inclusion in the registry, patient-care setting, location, and dates) | 100 |
| Methods | Data | Data pre-processing steps | 75 |
| Methods | Data | Selection of data subsets, if applicable | 62 |
| Methods | Data | Definitions of data elements, with references to common data elements | 12 |
| Methods | Data | De-identification methods | 0 |
| Methods | Data | How missing data were handled | 50 |
| Methods | Ground Truth | Definition of ground truth reference standard, in sufficient detail to allow replication | 100 |
| Methods | Ground Truth | Rationale for choosing the reference standard (if alternatives exist) | 75 |
| Methods | Ground Truth | Source of ground-truth annotations; qualifications and preparation of annotators | 100 |
| Methods | Ground Truth | Annotation tools | 100 |
| Methods | Ground Truth | Measurement of inter- and intrarater variability; methods to mitigate variability and/or resolve discrepancies | 75 |
| Methods | Data Partitions | Intended sample size and how it was determined | 0 |
| Methods | Data Partitions | How data were assigned to partitions; specify proportions | 50 |
| Methods | Data Partitions | Level at which partitions are disjoint (e.g., image, study, patient, and institution) | 50 |
| Methods | Model | Detailed description of the model, including inputs, outputs, all intermediate layers, and connections | 87 |
| Methods | Model | Software libraries, frameworks, and packages | 62 |
| Methods | Model | Initialization of model parameters (e.g., randomization, transfer learning) | 0 |
| Methods | Training | Details of the training approach, including data augmentation, hyperparameters, and number of models trained | 50 |
| Methods | Training | Method of selecting the final model | 62 |
| Methods | Training | Ensembling techniques, if applicable | 0 |
| Methods | Evaluation | Metrics of model performance | 87 |
| Methods | Evaluation | Statistical measures of significance and uncertainty (e.g., confidence intervals) | 75 |
| Methods | Evaluation | Robustness or sensitivity analysis | 62 |
| Methods | Evaluation | Methods for explainability or interpretability (e.g., saliency maps), and how they were validated | 62 |
| Methods | Evaluation | Validation or testing on external data | 0 |
| Results | Data | Flow of participants or cases, using a diagram to indicate inclusion and exclusion | 37 |
| Results | Data | Demographic and clinical characteristics of cases in each partition | 62 |
| Results | Model Performance | Performance metrics for optimal model(s) on all data partitions | 87 |
| Results | Model Performance | Estimates of diagnostic accuracy and their precision (such as 95% confidence intervals) | 75 |
| Results | Model Performance | Failure analysis of incorrectly classified cases | 0 |
| Discussion | Study limitations, including potential bias, statistical uncertainty, and generalizability | 87 | |
| Discussion | Implications for practice, including the intended use and/or clinical role | 100 | |
| Other Information | Registration number and name of registry | 25 | |
| Other Information | Where the full study protocol can be accessed | 12 | |
| Other Information | Sources of funding and other support; role of funders | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alipour, E.; Pooyan, A.; Shomal Zadeh, F.; Darbandi, A.D.; Bonaffini, P.A.; Chalian, M. Current Status and Future of Artificial Intelligence in MM Imaging: A Systematic Review. Diagnostics 2023, 13, 3372. https://doi.org/10.3390/diagnostics13213372
Alipour E, Pooyan A, Shomal Zadeh F, Darbandi AD, Bonaffini PA, Chalian M. Current Status and Future of Artificial Intelligence in MM Imaging: A Systematic Review. Diagnostics. 2023; 13(21):3372. https://doi.org/10.3390/diagnostics13213372
Chicago/Turabian StyleAlipour, Ehsan, Atefe Pooyan, Firoozeh Shomal Zadeh, Azad Duke Darbandi, Pietro Andrea Bonaffini, and Majid Chalian. 2023. "Current Status and Future of Artificial Intelligence in MM Imaging: A Systematic Review" Diagnostics 13, no. 21: 3372. https://doi.org/10.3390/diagnostics13213372
APA StyleAlipour, E., Pooyan, A., Shomal Zadeh, F., Darbandi, A. D., Bonaffini, P. A., & Chalian, M. (2023). Current Status and Future of Artificial Intelligence in MM Imaging: A Systematic Review. Diagnostics, 13(21), 3372. https://doi.org/10.3390/diagnostics13213372







