Rapid and Accurate Diagnosis of Dermatophyte Infections Using the DendrisCHIP® Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains
2.2. Clinical Specimen’s Provision
2.3. DNA Extraction and PCR Amplification
2.4. Design of Oligonucleotide Probes
2.5. Manufacture of the DendrisCHIP®DP
2.6. Hybridization Process and Reading of the DendrisCHIP®DP
2.7. Data Treatment Using Machine-Learning Methods and Statistical Analysis
2.8. Assessment of the Limit of Detection (LoD)
2.9. Diagnosis by Microbiological Cultures
2.10. Diagnostic Analysis Using RT-PCR and NGS
2.11. Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value
3. Results
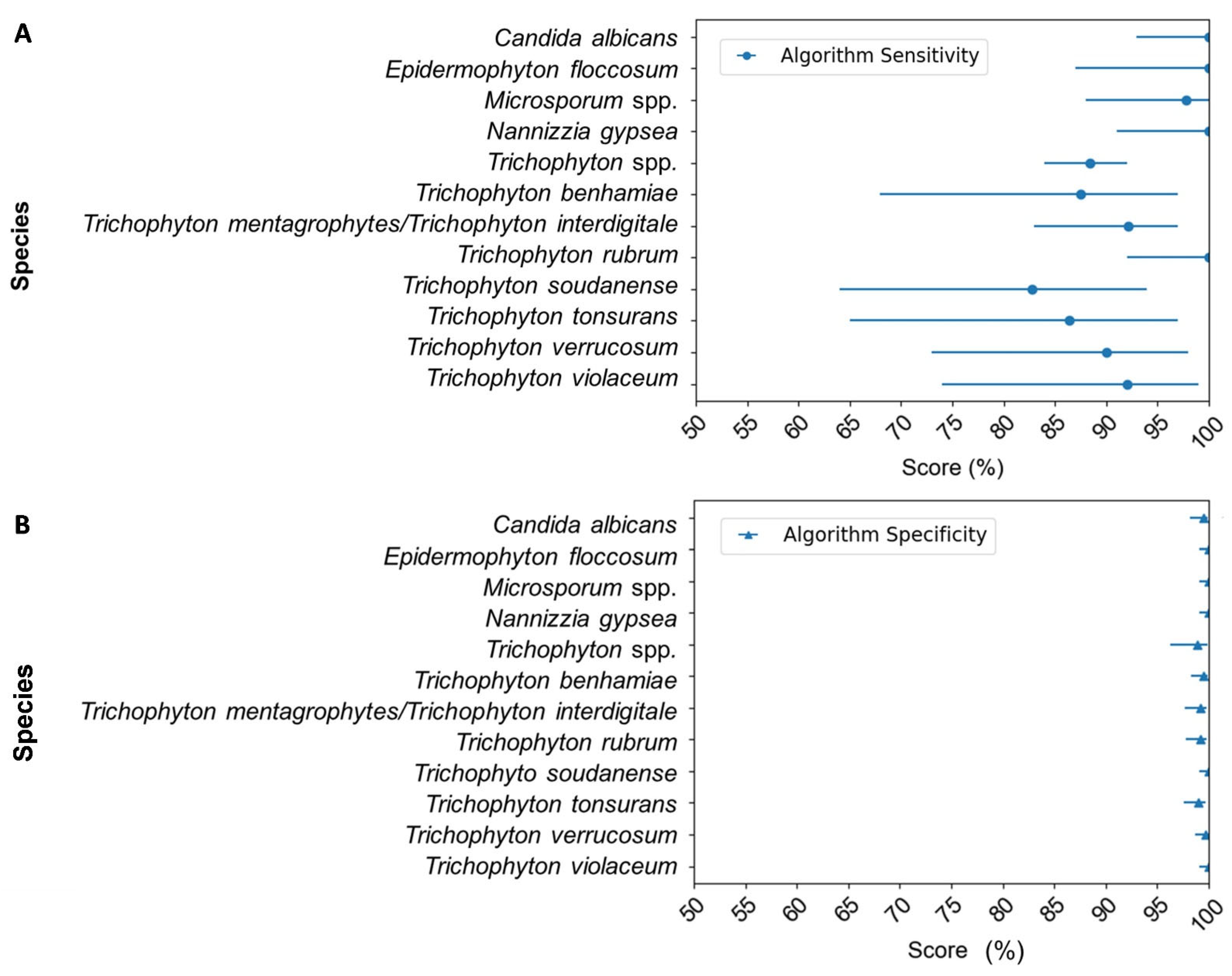
3.1. Construction and Validation of the DendrisCHIP®DP for the Dermatophyte Diagnosis
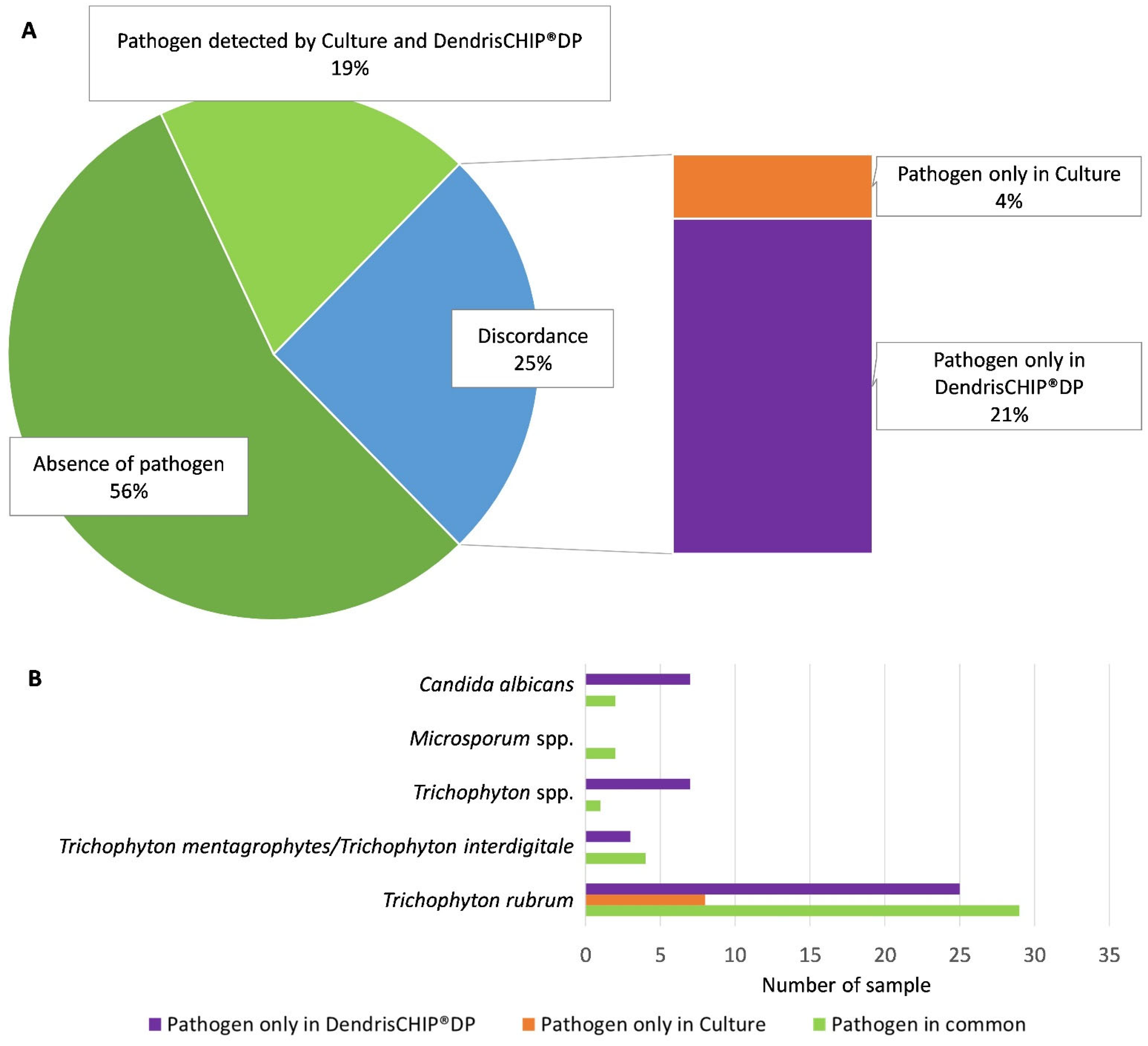
3.2. More Positive Clinical Samples Detected by the DendrisCHIP®DP Than by Microbiological Culture
3.3. Dermatophyte Diagnosis by DendrisCHIP®DP Is More Reliable Than by Microbiology Culture
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological Trends in Skin Mycoses Worldwide. Mycoses 2008, 51, 2–15. [Google Scholar] [CrossRef]
- Pires, C.A.A.; Cruz, N.F.S.D.; Lobato, A.M.; Sousa, P.O.D.; Carneiro, F.R.O.; Mendes, A.M.D. Clinical, Epidemiological, and Therapeutic Profile of Dermatophytosis. An. Bras. Dermatol. 2014, 89, 259–264. [Google Scholar] [CrossRef]
- Petrucelli, M.F.; Abreu, M.H.D.; Cantelli, B.A.M.; Segura, G.G.; Nishimura, F.G.; Bitencourt, T.A.; Marins, M.; Fachin, A.L. Epidemiology and Diagnostic Perspectives of Dermatophytoses. J. Fungi 2020, 6, 310. [Google Scholar] [CrossRef] [PubMed]
- Gräser, Y.; Monod, M.; Bouchara, J.-P.; Dukik, K.; Nenoff, P.; Kargl, A.; Kupsch, C.; Zhan, P.; Packeu, A.; Chaturvedi, V.; et al. New Insights in Dermatophyte Research. Med. Mycol. 2018, 56, S2–S9. [Google Scholar] [CrossRef] [PubMed]
- Weitzman, I.; Summerbell, R.C. The Dermatophytes. Clin. Microbiol. Rev. 1995, 8, 240–259. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Cooper, E.; Bunn, U.; Jamieson, F.; Gupta, A.K. Onychomycosis: A Critical Study of Techniques and Criteria for Confirming the Etiologic Significance of Nondermatophytes. Med. Mycol. 2005, 43, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Bosshard, P.P. Incubation of Fungal Cultures: How Long Is Long Enough?: Incubation of Fungal Cultures. Mycoses 2011, 54, e539–e545. [Google Scholar] [CrossRef]
- Brescini, L.; Fioriti, S.; Morroni, G.; Barchiesi, F. Antifungal Combinations in Dermatophytes. J. Fungi 2021, 7, 727. [Google Scholar] [CrossRef] [PubMed]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M.; Osińska, M.; Sawicki, M. Detection and Identification of Dermatophytes Based on Currently Available Methods—A Comparative Study. J. Appl. Microbiol. 2021, 130, 278–291. [Google Scholar] [CrossRef]
- Jensen, R.H.; Arendrup, M.C. Molecular Diagnosis of Dermatophyte Infections. Curr. Opin. Infect. Dis. 2012, 25, 126–134. [Google Scholar] [CrossRef]
- Kim, J.Y.; Choe, Y.B.; Ahn, K.J.; Lee, Y.W. Identification of Dermatophytes Using Multiplex Polymerase Chain Reaction. Ann. Dermatol. 2011, 23, 304. [Google Scholar] [CrossRef] [PubMed]
- Brillowska-Dąbrowska, A.; Saunte, D.M.; Arendrup, M.C. Five-Hour Diagnosis of Dermatophyte Nail Infections with Specific Detection of Trichophyton rubrum. J. Clin. Microbiol. 2007, 45, 1200–1204. [Google Scholar] [CrossRef]
- Ebihara, M.; Makimura, K.; Sato, K.; Abe, S.; Tsuboi, R. Molecular Detection of Dermatophytes and Nondermatophytes in Onychomycosis by Nested Polymerase Chain Reaction Based on 28S Ribosomal RNA Gene Sequences. Br. J. Dermatol. 2009, 161, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Garg, J.; Tilak, R.; Singh, S.; Gulati, A.K.; Garg, A.; Prakash, P.; Nath, G. Evaluation of Pan-Dermatophyte Nested PCR in Diagnosis of Onychomycosis. J. Clin. Microbiol. 2007, 45, 3443–3445. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.L.; Shankland, G.S.; Carman, W.; Williams, C. Introduction of a Dermatophyte Polymerase Chain Reaction Assay to the Diagnostic Mycology Service in Scotland: Introduction of a T. Rubrum PCR Assay in Scotland. Br. J. Dermatol. 2011, 164, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Bergman, A.; Heimer, D.; Kondori, N.; Enroth, H. Fast and Specific Dermatophyte Detection by Automated DNA Extraction and Real-Time PCR. Clin. Microbiol. Infect. 2013, 19, E205–E211. [Google Scholar] [CrossRef] [PubMed]
- Bergmans, A.M.C.; Van Der Ent, M.; Klaassen, A.; Böhm, N.; Andriesse, G.I.; Wintermans, R.G.F. Evaluation of a Single-Tube Real-Time PCR for Detection and Identification of 11 Dermatophyte Species in Clinical Material. Clin. Microbiol. Infect. 2010, 16, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, Y.; Satoh, K.; Uchida, T.; Yamada, T.; Abe, M.; Watanabe, S.; Makimura, M.; Makimura, K. Rapid Real-Time Diagnostic PCR for Trichophyton Rubrum and Trichophyton Mentagrophytes in Patients with Tinea Unguium and Tinea Pedis Using Specific Fluorescent Probes. J. Dermatol. Sci. 2013, 69, 229–235. [Google Scholar] [CrossRef]
- Wisselink, G.J.; Van Zanten, E.; Kooistra-Smid, A.M.D. Trapped in Keratin; a Comparison of Dermatophyte Detection in Nail, Skin and Hair Samples Directly from Clinical Samples Using Culture and Real-Time PCR. J. Microbiol. Methods 2011, 85, 62–66. [Google Scholar] [CrossRef]
- Elavarashi, E. Optimization of PCR–RFLP Directly from the Skin and Nails in Cases of Dermatophytosis, Targeting the ITS and the 18S Ribosomal DNA Regions. JCDR 2013, 7, 646. [Google Scholar] [CrossRef]
- Verrier, J.; Krähenbühl, L.; Bontems, O.; Fratti, M.; Salamin, K.; Monod, M. Dermatophyte Identification in Skin and Hair Samples Using a Simple and Reliable Nested Polymerase Chain Reaction Assay: Dermatophyte PCR Identification in Skin and Hair Samples. Br. J. Dermatol. 2013, 168, 295–301. [Google Scholar] [CrossRef]
- Bergmans, A.M.C.; Schouls, L.M.; Van Der Ent, M.; Klaassen, A.; Böhm, N.; Wintermans, R.G.F. Validation of PCR–Reverse Line Blot, a Method for Rapid Detection and Identification of Nine Dermatophyte Species in Nail, Skin and Hair Samples. Clin. Microbiol. Infect. 2008, 14, 778–788. [Google Scholar] [CrossRef][Green Version]
- Beifuss, B.; Bezold, G.; Gottlöber, P.; Borelli, C.; Wagener, J.; Schaller, M.; Korting, H.C. Direct Detection of Five Common Dermatophyte Species in Clinical Samples Using a Rapid and Sensitive 24-h PCR-ELISA Technique Open to Protocol Transfer: PCR-ELISA Technique for Dermatophyte Detection. Mycoses 2011, 54, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Pankewitz, F.; Nenoff, P.; Uhrlaß, S.; Bezold, G.; Winter, I.; Gräser, Y. Development of a Novel Polymerase Chain Reaction-Enzyme-Linked Immunosorbent Assay for the Diagnosis of Trichophyton rubrum Onychomycosis: Onychomycosis PCR-ELISA. Br. J. Dermatol. 2013, 168, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Paterson, R.R.M.; Venâncio, A.; Lima, N. Filamentous Fungal Characterizations by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. J. Appl. Microbiol. 2010, 108, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Li, H.C.; Bouchara, J.-P.; Hsu, M.M.-L.; Barton, R.; Chang, T.C. Identification of Dermatophytes by an Oligonucleotide Array. J. Clin. Microbiol. 2007, 45, 3160–3166. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Takayanagi, A.; Nagao, K.; Tomatsu, N.; Fukui, T.; Kawaguchi, M.; Kudoh, J.; Amagai, M.; Yamamoto, N.; Shimizu, N. Simple PCR-Based DNA Microarray System To Identify Human Pathogenic Fungi in Skin. J. Clin. Microbiol. 2010, 48, 2357–2364. [Google Scholar] [CrossRef]
- Senescau, A.; Kempowsky, T.; Bernard, E.; Messier, S.; Besse, P.; Fabre, R.; François, J. Innovative DendrisChips® Technology for a Syndromic Approach of In Vitro Diagnosis: Application to the Respiratory Infectious Diseases. Diagnostics 2018, 8, 77. [Google Scholar] [CrossRef]
- Bernard, E.; Peyret, T.; Plinet, M.; Contie, Y.; Cazaudarré, T.; Rouquet, Y.; Bernier, M.; Pesant, S.; Fabre, R.; Anton, A.; et al. The DendrisCHIP® Technology as a New, Rapid and Reliable Molecular Method for the Diagnosis of Osteoarticular Infections. Diagnostics 2022, 12, 1353. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Papanikolaou, Y.; Tsoumakas, G.; Katakis, I. Hierarchical Partitioning of the Output Space in Multi-Label Data. Data Knowl. Eng. 2018, 116, 42–60. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE General FASTA Release for Fungi. 2022. Available online: https://unite.ut.ee/index.php (accessed on 16 October 2022).
- Rivolli, A.; de Carvalho, A.C. The Utiml Package: Multi-Label Classification in R. R J. 2019, 10, 24. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 92–96. [Google Scholar]
- Kim, S.; Lee, W. Does McNemar’s Test Compare the Sensitivities and Specificities of Two Diagnostic Tests? Stat. Methods Med. Res. 2017, 26, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Sabou, M. Épidémiologie, répartition géographique et modes de contamination des dermatophytes. Rev. Francoph. Des. Lab. 2022, 2022, 31–40. [Google Scholar] [CrossRef]
- Paugam, A.; L’Ollivier, C.; Viguié, C.; Anaya, L.; Mary, C.; De Ponfilly, G.; Ranque, S. Comparison of Real-Time PCR with Conventional Methods to Detect Dermatophytes in Samples from Patients with Suspected Dermatophytosis. J. Microbiol. Methods 2013, 95, 218–222. [Google Scholar] [CrossRef]
- Safari, S.; Baratloo, A.; Elfil, M.; Negida, A. Evidence Based Emergency Medicine Part 2: Positive and Negative Predictive Values of Diagnostic Tests. Emergency 2015, 3, 87–88. [Google Scholar] [PubMed]
- Gräser, Y.; Saunte, D. A Hundred Years of Diagnosing Superficial Fungal Infections: Where Do We Come From, Where Are We Now and Where Would We Like To Go? Acta Derm.-Venereol. 2020, 100, adv00111-224. [Google Scholar] [CrossRef]
- Brillowska-Dabrowska, A.; Nielsen, S.S.; Nielsen, H.V.; Arendrup, M.C. Optimized 5-Hour Multiplex PCR Test for the Detection of Tinea Unguium: Performance in a Routine PCR Laboratory. Med. Mycol. 2010, 48, 828–831. [Google Scholar] [CrossRef]
- Chandran, N.S.; Pan, J.-Y.; Pramono, Z.A.; Tan, H.-H.; Seow, C.-S. Complementary Role of a Polymerase Chain Reaction Test in the Diagnosis of Onychomycosis: Onychomycosis: Complementary Role of PCR. Australas. J. Dermatol. 2013, 54, 105–108. [Google Scholar] [CrossRef]
- Kondori, N.; Abrahamsson, A.-L.; Ataollahy, N.; Wennerås, C. Comparison of a New Commercial Test, Dermatophyte-PCR Kit, with Conventional Methods for Rapid Detection and Identification of Trichophyton rubrum in Nail Specimens. Med. Mycol. 2010, 48, 1005–1008. [Google Scholar] [CrossRef] [PubMed]
- Bontems, O.; Hauser, P.M.; Monod, M. Evaluation of a Polymerase Chain Reaction-Restriction Fragment Length Polymorphism Assay for Dermatophyte and Nondermatophyte Identification in Onychomycosis. Br. J. Dermatol. 2009, 161, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Brasch, J. Pathogenesis of Tinea: Pathogenesis of Tinea. J. Der Dtsch. Dermatol. Ges. 2010, 8, 780–786. [Google Scholar] [CrossRef]
- Mehlig, L.; Garve, C.; Ritschel, A.; Zeiler, A.; Brabetz, W.; Weber, C.; Bauer, A. Clinical Evaluation of a Novel Commercial Multiplex-Based PCR Diagnostic Test for Differential Diagnosis of Dermatomycoses. Mycoses 2014, 57, 27–34. [Google Scholar] [CrossRef] [PubMed]


| Fungal Microorganisms | Taxonomy | Source | Accession Number |
|---|---|---|---|
| Candida albicans | species | Vircell ATCC 90028 | LC388876.1 |
| Epidermophyton floccosum | species | CECT 2769 | LC317573.1 |
| Microsporum spp. | genus | CHU Purpan | AJ006251.1/LC53030.1 |
| Nannizzia gypsea | species | BCCM/IHEM 25157 | LC170561.1 |
| Trichophyton spp. | genus | CHU Purpan | - |
| Trichophyton benhamiae | species | CECT 2892 | LT897802.1 |
| Trichophyton mentagrophytes/Trichophyton interdigitale | species | CECT 2901/CECT 2793 | LC317813.1/LC413778.1 |
| Trichophyton rubrum | species | CECT 2794 | LC404119.1 |
| Trichophyton soudanense | species | BCCM/IHEM 20772 | MF173063.1 |
| Trichophyton tonsurans | species | BCCM/IHEM 24955 | AB220044.1 |
| Trichophyton verrucosum | species | CECT 2992 | AB491473.1 |
| Trichophyton violaceum | species | BCCM/IHEM 26519 | AB430482.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anton, A.; Plinet, M.; Peyret, T.; Cazaudarré, T.; Pesant, S.; Rouquet, Y.; Tricoteaux, M.-A.; Bernier, M.; Bayette, J.; Fournier, R.; et al. Rapid and Accurate Diagnosis of Dermatophyte Infections Using the DendrisCHIP® Technology. Diagnostics 2023, 13, 3430. https://doi.org/10.3390/diagnostics13223430
Anton A, Plinet M, Peyret T, Cazaudarré T, Pesant S, Rouquet Y, Tricoteaux M-A, Bernier M, Bayette J, Fournier R, et al. Rapid and Accurate Diagnosis of Dermatophyte Infections Using the DendrisCHIP® Technology. Diagnostics. 2023; 13(22):3430. https://doi.org/10.3390/diagnostics13223430
Chicago/Turabian StyleAnton, Aurore, Mathilde Plinet, Thomas Peyret, Thomas Cazaudarré, Stéphanie Pesant, Yannick Rouquet, Marie-Andrée Tricoteaux, Matthieu Bernier, Jérémy Bayette, Remi Fournier, and et al. 2023. "Rapid and Accurate Diagnosis of Dermatophyte Infections Using the DendrisCHIP® Technology" Diagnostics 13, no. 22: 3430. https://doi.org/10.3390/diagnostics13223430
APA StyleAnton, A., Plinet, M., Peyret, T., Cazaudarré, T., Pesant, S., Rouquet, Y., Tricoteaux, M.-A., Bernier, M., Bayette, J., Fournier, R., Marguerettaz, M., Rolland, P., Bayol, T., Abbaoui, N., Berry, A., Iriart, X., Cassaing, S., Chauvin, P., Bernard, E., ... François, J.-M. (2023). Rapid and Accurate Diagnosis of Dermatophyte Infections Using the DendrisCHIP® Technology. Diagnostics, 13(22), 3430. https://doi.org/10.3390/diagnostics13223430





