Automatized Detection of Periodontal Bone Loss on Periapical Radiographs by Vision Transformer Networks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Periapical Radiographs
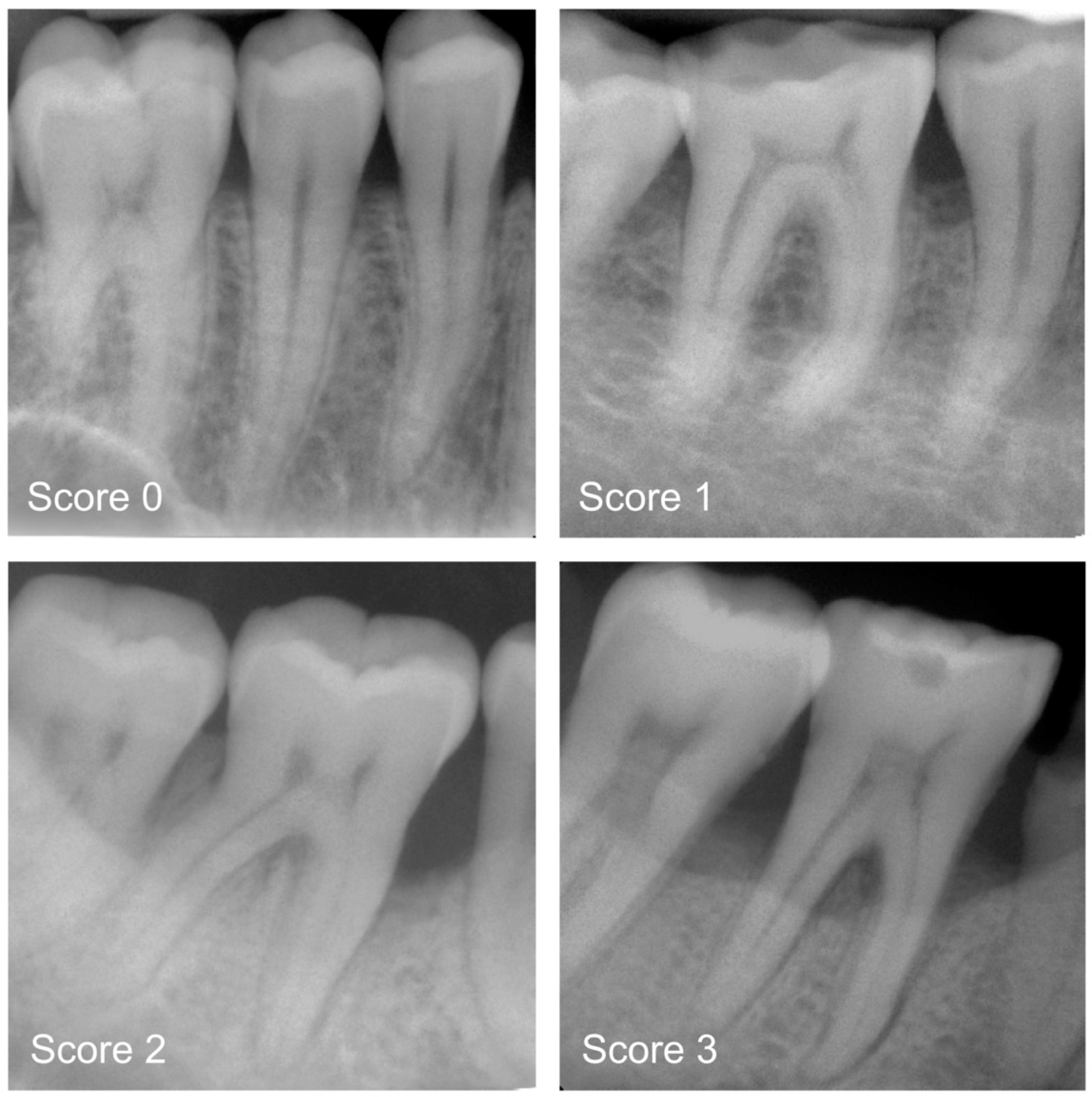
2.3. Categorisation of Periodontal Bone Loss (Reference Standard)
2.4. Training of the Deep Learning-Based Transformer Networks (Test Method)
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nazir, M.; Al-Ansari, A.; Al-Khalifa, K.; Alhareky, M.; Gaffar, B.; Almas, K. Global Prevalence of Periodontal Disease and Lack of Its Surveillance. Sci. World J. 2020, 2020, 2146160. [Google Scholar] [CrossRef] [PubMed]
- Frencken, J.E.; Sharma, P.; Stenhouse, L.; Green, D.; Laverty, D.; Dietrich, T. Global epidemiology of dental caries and severe periodontitis—A comprehensive review. J. Clin. Periodontol. 2017, 44 (Suppl. 18), S94–S105. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Jepsen, S.; Jin, L.; Otomo-Corgel, J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J. Clin. Periodontol. 2017, 44, 456–462. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Bernabe, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.; Marcenes, W. Global burden of severe periodontitis in 1990–2010: A systematic review and meta-regression. J. Dent. Res. 2014, 93, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Papapanou, P.N.; Susin, C. Periodontitis epidemiology: Is periodontitis under-recognized, over-diagnosed, or both? Periodontol. 2000 2017, 75, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Petersen, P.E.; Ogawa, H. The global burden of periodontal disease: Towards integration with chronic disease prevention and control. Periodontol. 2000 2012, 60, 15–39. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. 1), S173–S182. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. 1), S159–S172. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Sanz, M. Implementation of the new classification of periodontal diseases: Decision-making algorithms for clinical practice and education. J. Clin. Periodontol. 2019, 46, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Fiorellini, J.P.; Sourvanos, D.; Sarimento, H.; Karimbux, N.; Luan, K.W. Periodontal and Implant Radiology. Dent. Clin. N. Am. 2021, 65, 447–473. [Google Scholar] [CrossRef]
- Kong, Z.; Ouyang, H.; Cao, Y.; Huang, T.; Ahn, E.; Zhang, M.; Liu, H. Automated periodontitis bone loss diagnosis in panoramic radiographs using a bespoke two-stage detector. Comput. Biol. Med. 2023, 152, 106374. [Google Scholar] [CrossRef] [PubMed]
- Danks, R.P.; Bano, S.; Orishko, A.; Tan, H.J.; Moreno Sancho, F.; D’Aiuto, F.; Stoyanov, D. Automating Periodontal bone loss measurement via dental landmark localisation. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Kabir, T.; Lee, C.T.; Chen, L.; Jiang, X.; Shams, S. A comprehensive artificial intelligence framework for dental diagnosis and charting. BMC Oral Health 2022, 22, 480. [Google Scholar] [CrossRef] [PubMed]
- Ertas, K.; Pence, I.; Cesmeli, M.S.; Ay, Z.Y. Determination of the stage and grade of periodontitis according to the current classification of periodontal and peri-implant diseases and conditions (2018) using machine learning algorithms. J. Periodontal. Implant Sci. 2022, 53, 38. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, D.; Cao, Z.; Wu, F.; Zhu, H.; Zhu, F. A two-stage deep learning architecture for radiographic staging of periodontal bone loss. BMC Oral Health 2022, 22, 106. [Google Scholar] [CrossRef] [PubMed]
- Zadrozny, L.; Regulski, P.; Brus-Sawczuk, K.; Czajkowska, M.; Parkanyi, L.; Ganz, S.; Mijiritsky, E. Artificial Intelligence Application in Assessment of Panoramic Radiographs. Diagnostics 2022, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, J.; Zhou, Y.; Chen, Q.; She, Y.; Gao, F.; Xu, Y.; Chen, J.; Gao, X. An Interpretable Computer-Aided Diagnosis Method for Periodontitis From Panoramic Radiographs. Front. Physiol. 2021, 12, 655556. [Google Scholar] [CrossRef]
- Thanathornwong, B.; Suebnukarn, S. Automatic detection of periodontal compromised teeth in digital panoramic radiographs using faster regional convolutional neural networks. Imaging Sci. Dent. 2020, 50, 169–174. [Google Scholar] [CrossRef]
- Chang, H.J.; Lee, S.J.; Yong, T.H.; Shin, N.Y.; Jang, B.G.; Kim, J.E.; Huh, K.H.; Lee, S.S.; Heo, M.S.; Choi, S.C.; et al. Deep Learning Hybrid Method to Automatically Diagnose Periodontal Bone Loss and Stage Periodontitis. Sci. Rep. 2020, 10, 7531. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.S.; Song, I.S.; Jung, K.H. DeNTNet: Deep Neural Transfer Network for the detection of periodontal bone loss using panoramic dental radiographs. Sci. Rep. 2019, 9, 17615. [Google Scholar] [CrossRef]
- Krois, J.; Ekert, T.; Meinhold, L.; Golla, T.; Kharbot, B.; Wittemeier, A.; Dorfer, C.; Schwendicke, F. Deep Learning for the Radiographic Detection of Periodontal Bone Loss. Sci. Rep. 2019, 9, 8495. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Wu, Y.F.; Aung, L.M.; Lin, J.C.; Ngo, S.T.; Su, J.N.; Lin, Y.M.; Chang, W.J. Automatic recognition of teeth and periodontal bone loss measurement in digital radiographs using deep-learning artificial intelligence. J. Dent. Sci. 2023, 18, 1301–1309. [Google Scholar] [CrossRef]
- Alotaibi, G.; Awawdeh, M.; Farook, F.F.; Aljohani, M.; Aldhafiri, R.M.; Aldhoayan, M. Artificial intelligence (AI) diagnostic tools: Utilizing a convolutional neural network (CNN) to assess periodontal bone level radiographically—A retrospective study. BMC Oral Health 2022, 22, 399. [Google Scholar] [CrossRef]
- Chang, J.; Chang, M.F.; Angelov, N.; Hsu, C.Y.; Meng, H.W.; Sheng, S.; Glick, A.; Chang, K.; He, Y.R.; Lin, Y.B.; et al. Application of deep machine learning for the radiographic diagnosis of periodontitis. Clin. Oral Investig. 2022, 26, 6629–6637. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T.; Kabir, T.; Nelson, J.; Sheng, S.; Meng, H.W.; Van Dyke, T.E.; Walji, M.F.; Jiang, X.; Shams, S. Use of the deep learning approach to measure alveolar bone level. J. Clin. Periodontol. 2022, 49, 260–269. [Google Scholar] [CrossRef]
- Tsoromokos, N.; Parinussa, S.; Claessen, F.; Moin, D.A.; Loos, B.G. Estimation of Alveolar Bone Loss in Periodontitis Using Machine Learning. Int. Dent. J. 2022, 72, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, H.; Zhao, Y.; Zhao, J.; Wang, Y. Dental disease detection on periapical radiographs based on deep convolutional neural networks. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 649–661. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, D.-h.; Jeong, S.-N.; Choi, S.-H. Diagnosis and prediction of periodontally compromised teeth using a deep learning-based convolutional neural network algorithm. J. Periodontal Implant. Sci. 2018, 48, 114–123. [Google Scholar] [CrossRef]
- Lin, P.L.; Huang, P.Y.; Huang, P.W. Automatic methods for alveolar bone loss degree measurement in periodontitis periapical radiographs. Comput. Methods Programs Biomed. 2017, 148, 1–11. [Google Scholar] [CrossRef]
- Lin, P.L.; Huang, P.W.; Huang, P.Y.; Hsu, H.C. Alveolar bone-loss area localization in periodontitis radiographs based on threshold segmentation with a hybrid feature fused of intensity and the H-value of fractional Brownian motion model. Comput. Methods Programs Biomed. 2015, 121, 117–126. [Google Scholar] [CrossRef]
- Patil, S.; Joda, T.; Soffe, B.; Awan, K.H.; Fageeh, H.N.; Tovani-Palone, M.R.; Licari, F.W. Efficacy of artificial intelligence in the detection of periodontal bone loss and classification of periodontal diseases: A systematic review. J. Am. Dent. Assoc. 2023, 154, 795–804.e791. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.; Biancardi, A.M.; Jones, O.; Andrew, D. Artificial Intelligence in Periodontology: A Scoping Review. Dent. J. 2023, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. Available online: https://arxiv.org/abs/2010.11929 (accessed on 23 October 2023).
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C.; et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015, 351, h5527. [Google Scholar] [CrossRef]
- Schwendicke, F.; Singh, T.; Lee, J.H.; Gaudin, R.; Chaurasia, A.; Wiegand, T.; Uribe, S.; Krois, J. Artificial intelligence in dental research: Checklist for authors, reviewers, readers. J. Dent. 2021, 107, 103610. [Google Scholar] [CrossRef]
- Meusburger, T.; Wulk, A.; Kessler, A.; Heck, K.; Hickel, R.; Dujic, H.; Kühnisch, J. The Detection of Dental Pathologies on Periapical Radiographs-Results from a Reliability Study. J. Clin. Med. 2023, 12, 2224. [Google Scholar] [CrossRef]
- Bao, H.; Dong, L.; Piao, S.; Wei, F. BEiT: BERT Pre-Training of Image Transformers. arXiv 2022, arXiv:2106.08254v2. Available online: https://arxiv.org/abs/2106.08254 (accessed on 23 October 2023).
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training Data-Efficient Image Transformers & Distillation through Attention. arXiv 2021, arXiv:2012.12877v2. Available online: https://arxiv.org/abs/2012.12877 (accessed on 23 October 2023).
- Matthews, D.E.; Farewell, V.T. Using and Understanding Medical Statistics; S.Karger AG: Basel, Switzerland, 2015. [Google Scholar]
- Kurt, S.; Celik, O.; Bayrakdar, I.S.; Orhan, K.; Bilgir, E.; Odabas, A.; Aslan, A.F. Success of artificial intelligence system in determining alveolar bone loss from dental panoramic radiography images. Cumhuriyet. Dent. J. 2020, 23, 318–324. [Google Scholar] [CrossRef]
- Widyaningrum, R.; Candradewi, I.; Aji, N.; Aulianisa, R. Comparison of Multi-Label U-Net and Mask R-CNN for panoramic radiograph segmentation to detect periodontitis. Imaging Sci. Dent. 2022, 52, 383–391. [Google Scholar] [CrossRef]
- Lian, L.; Zhu, T.; Zhu, F.; Zhu, H. Deep Learning for Caries Detection and Classification. Diagnostics 2021, 11, 1672. [Google Scholar] [CrossRef]
- Moidu, N.P.; Sharma, S.; Chawla, A.; Kumar, V.; Logani, A. Deep learning for categorization of endodontic lesion based on radiographic periapical index scoring system. Clin. Oral Investig. 2022, 26, 651–658. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.H.; Jeong, S.N.; Choi, S.H. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J. Dent. 2018, 77, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Schonewolf, J.; Meyer, O.; Engels, P.; Schlickenrieder, A.; Hickel, R.; Gruhn, V.; Hesenius, M.; Kühnisch, J. Artificial intelligence-based diagnostics of molar-incisor-hypomineralization (MIH) on intraoral photographs. Clin. Oral Investig. 2022, 26, 5923–5930. [Google Scholar] [CrossRef] [PubMed]
- Engels, P.; Meyer, O.; Schonewolf, J.; Schlickenrieder, A.; Hickel, R.; Hesenius, M.; Gruhn, V.; Kühnisch, J. Automated detection of posterior restorations in permanent teeth using artificial intelligence on intraoral photographs. J. Dent. 2022, 121, 104124. [Google Scholar] [CrossRef] [PubMed]
- Kühnisch, J.; Meyer, O.; Hesenius, M.; Hickel, R.; Gruhn, V. Caries Detection on Intraoral Images Using Artificial Intelligence. J. Dent. Res. 2022, 101, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, Y.; Li, W.; Liu, C.; Gu, D.; Sun, W.; Miao, L. Development and evaluation of deep learning for screening dental caries from oral photographs. Oral Dis. 2022, 28, 173–181. [Google Scholar] [CrossRef]
- Schlickenrieder, A.; Meyer, O.; Schoenewolf, J.; Engels, P.; Hickel, R.; Gruhn, V.; Hesenius, M.; Kühnisch, J. Automatized Detection and Categorization of Fissure Sealants from Intraoral Digital Photographs Using Artificial Intelligence. Diagnostics 2021, 11, 1608. [Google Scholar] [CrossRef]
- Zhou, X.; Yu, G.; Yin, Q.; Yang, J.; Sun, J.; Lv, S.; Shi, Q. Tooth Type Enhanced Transformer for Children Caries Diagnosis on Dental Panoramic Radiographs. Diagnostics 2023, 13, 689. [Google Scholar] [CrossRef]
- Gao, S.; Li, X.; Li, X.; Li, Z.; Deng, Y. Transformer based tooth classification from cone-beam computed tomography for dental charting. Comput. Biol. Med. 2022, 148, 105880. [Google Scholar] [CrossRef]
- Ying, S.; Wang, B.; Zhu, H.; Liu, W.; Huang, F. Caries segmentation on tooth X-ray images with a deep network. J. Dent. 2022, 119, 104076. [Google Scholar] [CrossRef]

| Region of Periapical Radiograph | Healthy Periodontium (Score 0) | Mild PBL (Score 1) | Moderate PBL (Score 2) | Severe PBL (Score 3) | Total (N) |
|---|---|---|---|---|---|
| 1st Quadrant | 1701 (35.8%) | 1826 (38.5%) | 851 (18.0%) | 367 (7.7%) | 4745 |
| 2nd Quadrant | 1231 (26.1%) | 2080 (44.1%) | 1093 (23.2%) | 312 (6.6%) | 4716 |
| 3rd Quadrant | 1477 (34.7%) | 2033 (47.7%) | 593 (13.9%) | 157 (3.7%) | 4260 |
| 4th Quadrant | 1282 (30.8%) | 2027 (48.7%) | 713 (17.1%) | 143 (3.4%) | 4165 |
| Maxillary anteriors | 653 (33.6%) | 661 (34.0%) | 433 (22.3%) | 197 (10.1%) | 1944 |
| Mandibular anteriors | 202 (10.2%) | 676 (34.0%) | 786 (39.5%) | 325 (16.3%) | 1989 |
| ViT-Base (Google) | ViT-Large (Google) | BEiT-Base (Microsoft) | BEiT-Large (Microsoft) | DeiT-Base (Facebook/Meta) | |
|---|---|---|---|---|---|
| Neural network | Vision transformer | Bidirectional encoder representation from image transformers | Data-efficient image transformer | ||
| Epochs | 5 | 5 | 5 | 5 | 5 |
| Learning rate | 0.00005 | 0.00005 | 0.00005 | 0.00005 | 0.00005 |
| FLOS | 7.280 × 1015 | 25.735 × 1015 | 7.277 × 1015 | 25.744 × 1015 | 7.280 × 1015 |
| Samples per second | 298.6 | 111.7 | 274.4 | 102.9 | 298.5 |
| Parameter count | 85.8 × 106 | 303.3 × 106 | 85.7 × 106 | 303.4 × 106 | 85.8 × 106 |
| All Apical Radiographs | True Positive (TP) | True Negative (TN) | False Positive (FP) | False Negative (FN) | Diagnostic Performance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | ACC | SE | SP | NPV | PPV | AUC | |
| ViT-base | 1884 | 62.8 | 673 | 22.4 | 230 | 7.7 | 213 | 7.1 | 85.2 | 89.8 | 74.5 | 76.0 | 89.1 | 0.918 |
| ViT-large | 1831 | 61.0 | 671 | 22.4 | 232 | 7.7 | 266 | 8.9 | 83.4 | 87.3 | 74.3 | 71.6 | 88.8 | 0.899 |
| BEiT-base | 1885 | 62.8 | 649 | 21.6 | 254 | 8.5 | 212 | 7.1 | 84.5 | 89.9 | 71.9 | 75.4 | 88.1 | 0.914 |
| BEiT-large | 1914 | 63.8 | 631 | 21.0 | 272 | 9.1 | 183 | 6.1 | 84.8 | 91.3 | 69.9 | 77.5 | 87.6 | 0.907 |
| DeiT-base | 1879 | 62.6 | 646 | 21.5 | 257 | 8.6 | 218 | 7.3 | 84.2 | 89.6 | 71.5 | 74.8 | 88.0 | 0.908 |
| True Positive (TP) | True Negative (TN) | False Positive (FP) | False Negative (FN) | Diagnostic Performance | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | ACC | SE | SP | NPV | PPV | AUC | ||
| Mandibular anterior teeth | ViT-base | 240 | 88.2 | 16 | 5.9 | 9 | 3.3 | 7 | 2.6 | 94.1 | 97.2 | 64.0 | 69.6 | 96.4 | 0.944 |
| ViT-large | 241 | 88.6 | 18 | 6.6 | 7 | 2.6 | 6 | 2.2 | 95.2 | 97.6 | 72.0 | 75.0 | 97.2 | 0.960 | |
| BEiT-base | 242 | 89.0 | 21 | 7.7 | 4 | 1.5 | 5 | 1.8 | 96.7 | 98.0 | 84.0 | 80.8 | 98.4 | 0.963 | |
| BEiT-large | 245 | 90.1 | 18 | 6.6 | 7 | 2.6 | 2 | 0.7 | 96.7 | 99.2 | 72.0 | 90.0 | 97.2 | 0.952 | |
| DeiT-base | 242 | 89.0 | 15 | 5.5 | 10 | 3.7 | 5 | 1.8 | 94.5 | 98.0 | 60.0 | 75.0 | 96.0 | 0.970 | |
| Mandibular posterior teeth | ViT-base | 700 | 61.6 | 287 | 25.3 | 78 | 6.9 | 70 | 6.2 | 87.0 | 90.9 | 78.6 | 80.4 | 90.0 | 0.937 |
| ViT-large | 687 | 60.5 | 285 | 25.1 | 80 | 7.1 | 83 | 7.3 | 85.6 | 89.2 | 78.1 | 77.4 | 89.6 | 0.913 | |
| BEiT-base | 704 | 62.0 | 277 | 24.4 | 88 | 7.8 | 66 | 5.8 | 86.4 | 91.4 | 75.9 | 80.8 | 88.9 | 0.933 | |
| BEiT-large | 711 | 62.6 | 279 | 24.6 | 86 | 7.6 | 59 | 5.2 | 87.2 | 92.3 | 76.4 | 82.5 | 89.2 | 0.923 | |
| DeiT-base | 694 | 61.1 | 281 | 24.8 | 84 | 7.4 | 76 | 6.7 | 85.9 | 90.1 | 77.0 | 78.7 | 89.2 | 0.927 | |
| Maxillary anterior teeth | ViT-base | 157 | 59.5 | 81 | 30.7 | 18 | 6.8 | 8 | 3.0 | 90.2 | 95.2 | 81.8 | 91.0 | 89.7 | 0.958 |
| ViT-large | 156 | 59.1 | 77 | 29.2 | 22 | 8.3 | 9 | 3.4 | 88.3 | 94.5 | 77.8 | 89.5 | 87.6 | 0.948 | |
| BEiT-base | 158 | 59.8 | 73 | 27.7 | 26 | 9.8 | 7 | 2.7 | 87.5 | 95.8 | 73.7 | 91.3 | 85.9 | 0.954 | |
| BEiT-large | 157 | 59.5 | 73 | 27.7 | 26 | 9.8 | 8 | 3.0 | 87.1 | 95.2 | 73.7 | 90.1 | 85.8 | 0.954 | |
| DeiT-base | 154 | 58.3 | 75 | 28.4 | 24 | 9.1 | 11 | 4.2 | 86.7 | 93.3 | 75.8 | 87.2 | 86.5 | 0.954 | |
| Maxillary posterior teeth | ViT-base | 787 | 59.2 | 289 | 21.8 | 125 | 9.4 | 128 | 9.6 | 81.0 | 86.0 | 69.8 | 69.3 | 86.3 | 0.875 |
| ViT-large | 747 | 56.2 | 291 | 21.9 | 123 | 9.3 | 168 | 12.6 | 78.1 | 81.6 | 70.3 | 63.4 | 85.9 | 0.851 | |
| BEiT-base | 781 | 58.8 | 278 | 20.9 | 136 | 10.2 | 134 | 10.1 | 79.7 | 85.4 | 67.1 | 67.5 | 85.2 | 0.865 | |
| BEiT-large | 801 | 60.3 | 261 | 19.6 | 153 | 11.5 | 114 | 8.6 | 79.9 | 87.5 | 63.0 | 69.6 | 84.0 | 0.861 | |
| DeiT-base | 789 | 59.4 | 275 | 20.7 | 139 | 10.4 | 126 | 9.5 | 80.1 | 86.2 | 66.4 | 68.6 | 85.0 | 0.860 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dujic, H.; Meyer, O.; Hoss, P.; Wölfle, U.C.; Wülk, A.; Meusburger, T.; Meier, L.; Gruhn, V.; Hesenius, M.; Hickel, R.; et al. Automatized Detection of Periodontal Bone Loss on Periapical Radiographs by Vision Transformer Networks. Diagnostics 2023, 13, 3562. https://doi.org/10.3390/diagnostics13233562
Dujic H, Meyer O, Hoss P, Wölfle UC, Wülk A, Meusburger T, Meier L, Gruhn V, Hesenius M, Hickel R, et al. Automatized Detection of Periodontal Bone Loss on Periapical Radiographs by Vision Transformer Networks. Diagnostics. 2023; 13(23):3562. https://doi.org/10.3390/diagnostics13233562
Chicago/Turabian StyleDujic, Helena, Ole Meyer, Patrick Hoss, Uta Christine Wölfle, Annika Wülk, Theresa Meusburger, Leon Meier, Volker Gruhn, Marc Hesenius, Reinhard Hickel, and et al. 2023. "Automatized Detection of Periodontal Bone Loss on Periapical Radiographs by Vision Transformer Networks" Diagnostics 13, no. 23: 3562. https://doi.org/10.3390/diagnostics13233562
APA StyleDujic, H., Meyer, O., Hoss, P., Wölfle, U. C., Wülk, A., Meusburger, T., Meier, L., Gruhn, V., Hesenius, M., Hickel, R., & Kühnisch, J. (2023). Automatized Detection of Periodontal Bone Loss on Periapical Radiographs by Vision Transformer Networks. Diagnostics, 13(23), 3562. https://doi.org/10.3390/diagnostics13233562







