Machine Learning Augmented Interpretation of Chest X-rays: A Systematic Review
Abstract
:1. Introduction
2. Methods and Materials
2.1. Search Strategy
2.2. Eligibility Criteria
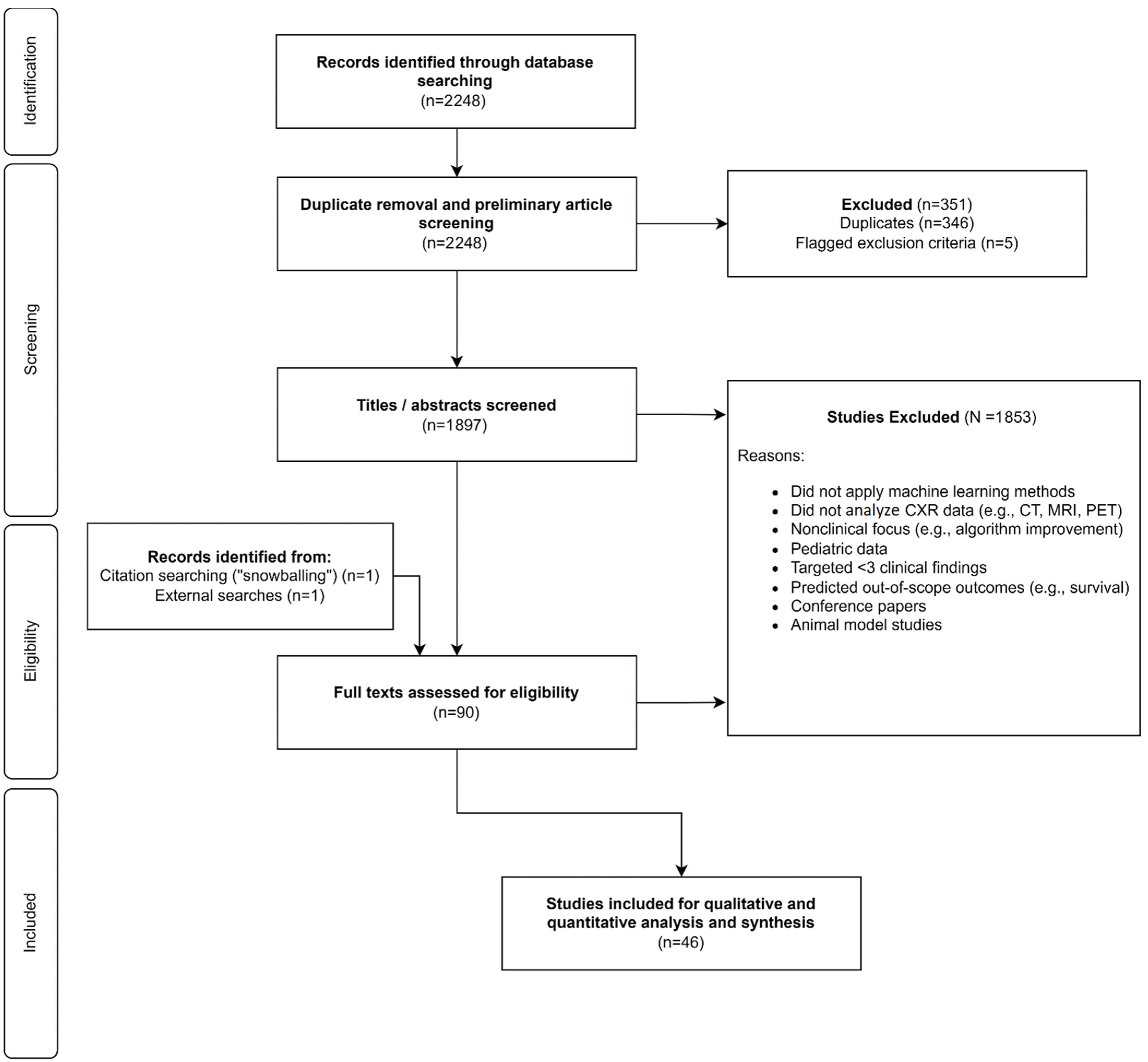
2.3. Study Selection Process
2.4. Data Extraction and Appraisal
2.5. Synthesis and Assessment
3. Results
3.1. Included Articles
3.2. Summary of Included Articles
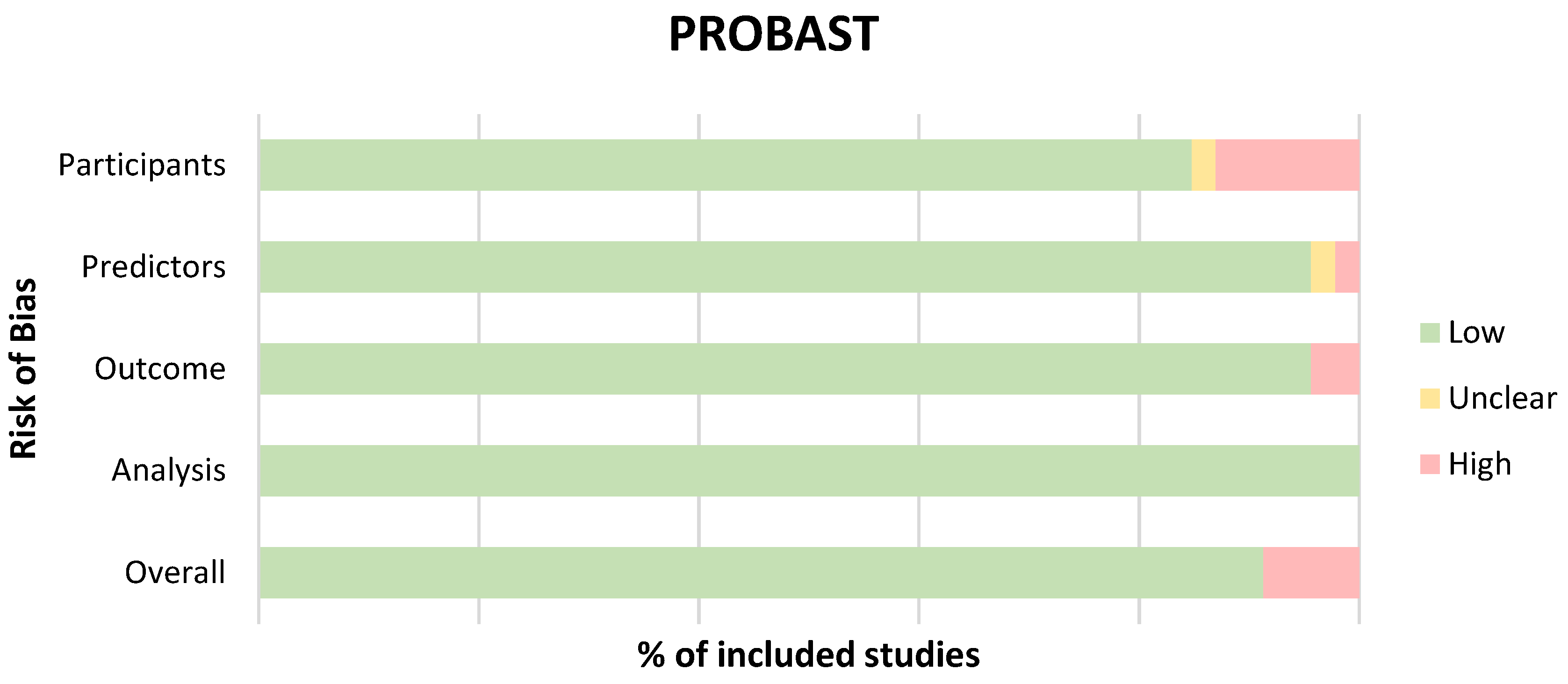
3.3. Quality Appraisal and Risk of Bias
3.4. Comprehensiveness and Algorithm Development
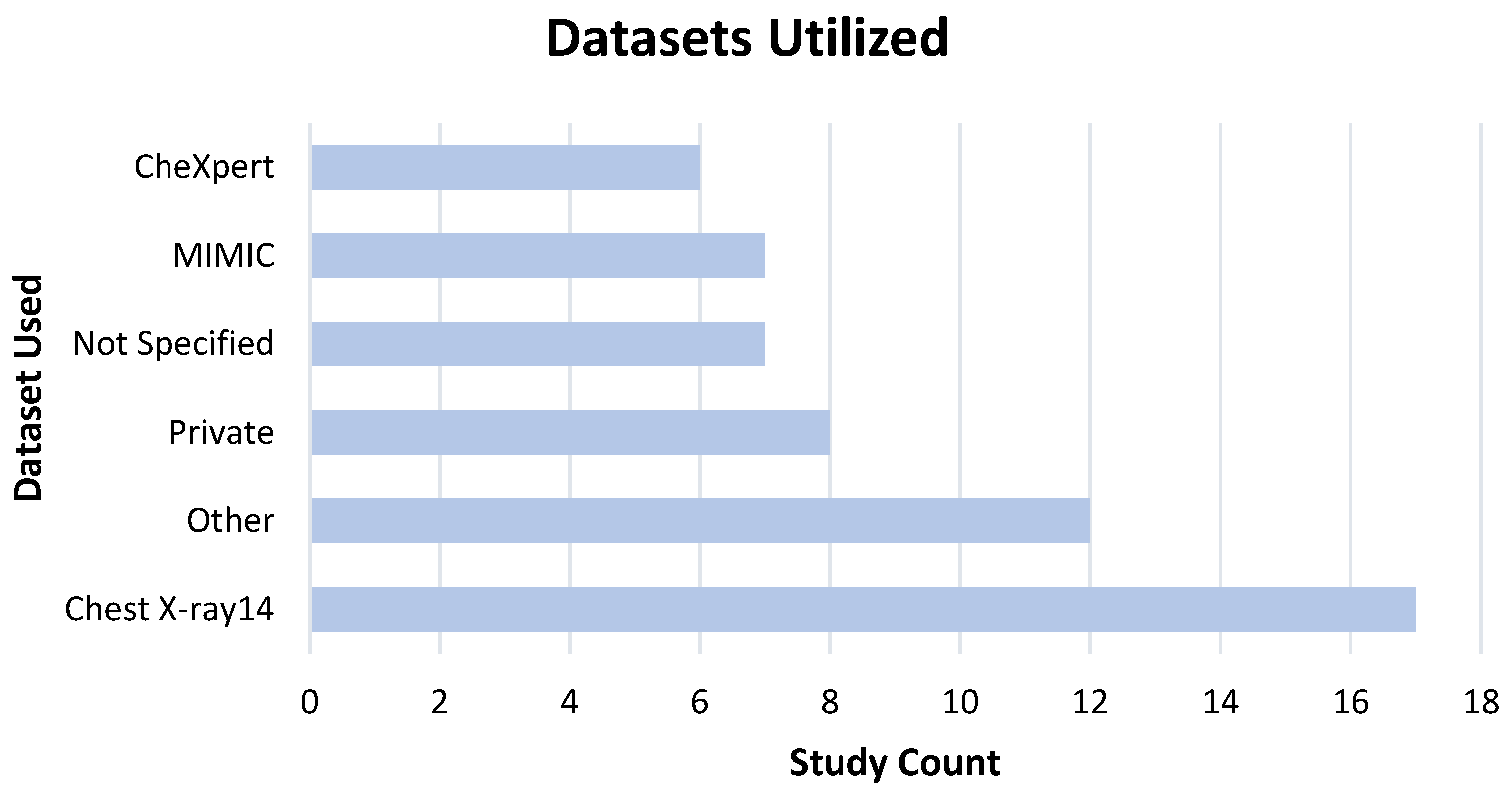
3.5. Data, Model Training, and Ground Truth Labeling
3.6. Performance and Safety
4. Discussion
4.1. Risk and Safety
4.2. Benefits
4.3. Study Strengths and Limitations
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A. Assessment Criteria for Quality and Risk of Bias
| PROBAST ROB Assessment | |||||
|---|---|---|---|---|---|
| Study | Participants | Predictors | Outcome | Analysis | Overall |
| Ahn et al., 2022 [43] | + | + | + | + | + |
| Albahli et al., 2021 [44] | + | + | + | + | + |
| Altaf et al., 2021 [46] | + | + | + | + | + |
| Baltruschat et al., 2021 [49] | + | + | + | + | + |
| Bharati et al., 2020 [50] | + | + | + | + | + |
| Chakravarty et al., 2020 [51] | + | + | + | + | + |
| Chen et al., 2020 [53] | + | + | + | + | + |
| Cho, Kim et al., 2020 [55] | + | + | + | + | + |
| Cho, Park et al., 2020 [56] | + | + | + | + | + |
| Choi et al., 2021 [57] | + | + | + | + | + |
| Fang et al., 2021 [58] | + | + | + | + | + |
| Gipson et al., 2022 [59] | + | + | + | + | + |
| Gündel et al., 2021 [60] | + | + | + | + | + |
| Han et al., 2022 [61] | + | + | + | + | + |
| Hwang et al., 2022 [62] | + | + | + | + | + |
| Jabbour et al., 2022 [63] | + | + | - | + | - |
| Jadhav et al., 2020 [64] | + | + | + | + | + |
| Jin et al., 2022 [66] | + | + | + | + | + |
| Jones et al., 2021 [42] | + | + | - | + | + |
| Kim et al., 2021 [67] | + | + | + | + | + |
| Kim et al., 2022 [68] | + | + | + | + | + |
| Kuo et al., 2021 [69] | + | + | + | + | + |
| Lee et al., 2022 [70] | + | + | + | + | + |
| Li et al., 2021 [71] | + | + | + | + | + |
| Majkowska et al., 2020 [72] | + | + | + | + | + |
| Mosquera et al., 2021 [73] | + | + | + | + | + |
| Nam et al., 2021 [74] | + | + | + | + | + |
| Niehues et al., 2021 [76] | + | + | + | + | + |
| Park et al., 2020 [77] | + | + | + | + | + |
| Paul et al., 2021 [78] | + | + | + | + | + |
| Pham et al., 2021 [80] | + | + | + | + | + |
| Rudolph et al., 2022 [81] | - | + | + | + | - |
| Rudolph et al., 2022 [82] | - | + | + | + | - |
| Seah et al., 2021 [83] | + | + | + | + | + |
| Senan et al., 2021 [84] | - | + | + | + | - |
| Sharma et al., 2020 [85] | + | + | + | - | - |
| Sung et al., 2021 [87] | + | + | + | + | + |
| Van Beek et al., 2022 [88] | + | + | + | + | + |
| Verma et al., 2020 [89] | + | + | + | ? | ? |
| Wang et al., 2021 [90] | - | + | + | + | - |
| Wang et al., 2020 [91] | + | + | + | + | + |
| Wang et al., 2021 [92] | + | + | + | + | + |
| Wang et al., 2020 [93] | + | + | + | + | + |
| Wu et al., 2020 [11] | + | + | + | + | + |
| Xu et al., 2020 [94] | + | + | + | + | + |
| Zhou et al., 2021 [95] | + | + | + | + | + |
References
- Mould, R.F. The Early History of X-ray Diagnosis with Emphasis on the Contributions of Physics 1895–1915. Phys. Med. Biol. 1995, 40, 1741–1787. [Google Scholar] [CrossRef] [PubMed]
- United Nations. Sources and Effects of Ionizing Radiation; United Nations: New York, NY, USA, 2011; ISBN 9789210544825. [Google Scholar]
- Lee, C.S.; Nagy, P.G.; Weaver, S.J.; Newman-Toker, D.E. Cognitive and System Factors Contributing to Diagnostic Errors in Radiology. AJR Am. J. Roentgenol. 2013, 201, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Del Ciello, A.; Franchi, P.; Contegiacomo, A.; Cicchetti, G.; Bonomo, L.; Larici, A.R. Missed Lung Cancer: When, Where, and Why? Diagn. Interv. Radiol. 2017, 23, 118–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, A.P. Error and Discrepancy in Radiology: Inevitable or Avoidable? Insights Imaging 2017, 8, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Nagarsheth, K.; Kurek, S. Ultrasound Detection of Pneumothorax Compared with Chest X-ray and Computed Tomography Scan. Am. Surg. 2011, 77, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Hayden, G.E.; Wrenn, K.W. Chest Radiograph vs. Computed Tomography Scan in the Evaluation for Pneumonia. J. Emerg. Med. 2009, 36, 266–270. [Google Scholar] [CrossRef]
- Vikgren, J.; Zachrisson, S.; Svalkvist, A.; Johnsson, A.A.; Boijsen, M.; Flinck, A.; Kheddache, S.; Båth, M. Comparison of Chest Tomosynthesis and Chest Radiography for Detection of Pulmonary Nodules: Human Observer Study of Clinical Cases. Radiology 2008, 249, 1034–1041. [Google Scholar] [CrossRef]
- Jones, C.M.; Buchlak, Q.D.; Oakden-Rayner, L.; Milne, M.; Seah, J.; Esmaili, N.; Hachey, B. Chest Radiographs and Machine Learning—Past, Present and Future. J. Med. Imaging Radiat. Oncol. 2021, 65, 538–544. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Wu, J.T.; Wong, K.C.L.; Gur, Y.; Ansari, N.; Karargyris, A.; Sharma, A.; Morris, M.; Saboury, B.; Ahmad, H.; Boyko, O.; et al. Comparison of Chest Radiograph Interpretations by Artificial Intelligence Algorithm vs Radiology Residents. JAMA Netw. Open 2020, 3, e2022779. [Google Scholar] [CrossRef]
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.-C.; Farrokhi, F.; Bennett, C.; Piccardi, M.; Sethi, R.K. Machine Learning Applications to Clinical Decision Support in Neurosurgery: An Artificial Intelligence Augmented Systematic Review. Neurosurg. Rev. 2020, 43, 1235–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.-C.; Bennett, C.; Farrokhi, F.; Piccardi, M. Machine Learning Applications to Neuroimaging for Glioma Detection and Classification: An Artificial Intelligence Augmented Systematic Review. J. Clin. Neurosci. 2021, 89, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Ben-Israel, D.; Jacobs, W.B.; Casha, S.; Lang, S.; Ryu, W.H.A.; de Lotbiniere-Bassett, M.; Cadotte, D.W. The Impact of Machine Learning on Patient Care: A Systematic Review. Artif. Intell. Med. 2020, 103, 101785. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the Accuracy of Human Readers versus Machine-Learning Algorithms for Pigmented Skin Lesion Classification: An Open, Web-Based, International, Diagnostic Study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Buchlak, Q.D.; Milne, M.R.; Seah, J.; Johnson, A.; Samarasinghe, G.; Hachey, B.; Esmaili, N.; Tran, A.; Leveque, J.-C.; Farrokhi, F.; et al. Charting the Potential of Brain Computed Tomography Deep Learning Systems. J. Clin. Neurosci. 2022, 99, 217–223. [Google Scholar] [CrossRef]
- Hwang, E.J.; Hong, J.H.; Lee, K.H.; Kim, J.I.; Nam, J.G.; Kim, D.S.; Choi, H.; Yoo, S.J.; Goo, J.M.; Park, C.M. Deep Learning Algorithm for Surveillance of Pneumothorax after Lung Biopsy: A Multicenter Diagnostic Cohort Study. Eur. Radiol. 2020, 30, 3660–3671. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.Y.; Kim, G.H.; Kang, D.; Kim, I.J.; Seo, J.; Andrews, J.R.; Park, C.M. Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness. J. Clin. Med. 2020, 9, 1981. [Google Scholar] [CrossRef]
- Khasawneh, N.; Fraiwan, M.; Fraiwan, L.; Khassawneh, B.; Ibnian, A. Detection of COVID-19 from Chest X-ray Images Using Deep Convolutional Neural Networks. Sensors 2021, 21, 5940. [Google Scholar] [CrossRef]
- Nasiri, H.; Hasani, S. Automated Detection of COVID-19 Cases from Chest X-ray Images Using Deep Neural Network and XGBoost. Radiography (London) 2022, 28, 732–738. [Google Scholar] [CrossRef]
- Khan, S.H.; Sohail, A.; Khan, A.; Lee, Y.-S. COVID-19 Detection in Chest X-ray Images Using a New Channel Boosted CNN. Diagnostics 2022, 12, 267. [Google Scholar] [CrossRef]
- Aboutalebi, H.; Pavlova, M.; Shafiee, M.J.; Sabri, A.; Alaref, A.; Wong, A. COVID-Net CXR-S: Deep Convolutional Neural Network for Severity Assessment of COVID-19 Cases from Chest X-ray Images. Res. Sq. 2021, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Ezzoddin, M.; Nasiri, H.; Dorrigiv, M. Diagnosis of COVID-19 Cases from Chest X-ray Images Using Deep Neural Network and LightGBM. arXiv 2022, arXiv:2203.14275. [Google Scholar]
- Nasiri, H.; Kheyroddin, G.; Dorrigiv, M.; Esmaeili, M.; Nafchi, A.R.; Ghorbani, M.H.; Zarkesh-Ha, P. Classification of COVID-19 in Chest X-ray Images Using Fusion of Deep Features and LightGBM. arXiv 2022, arXiv:2206.04548. [Google Scholar]
- Wang, X.; Yu, J.; Zhu, Q.; Li, S.; Zhao, Z.; Yang, B.; Pu, J. Potential of Deep Learning in Assessing Pneumoconiosis Depicted on Digital Chest Radiography. Occup. Environ. Med. 2020, 77, 597–602. [Google Scholar] [CrossRef]
- Qin, Z.Z.; Sander, M.S.; Rai, B.; Titahong, C.N.; Sudrungrot, S.; Laah, S.N.; Adhikari, L.M.; Carter, E.J.; Puri, L.; Codlin, A.J.; et al. Using Artificial Intelligence to Read Chest Radiographs for Tuberculosis Detection: A Multi-Site Evaluation of the Diagnostic Accuracy of Three Deep Learning Systems. Sci. Rep. 2019, 9, 15000. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.; Song, H.; Shin, Y.J.; Kim, J.; Kim, J.; Lee, K.W.; Lee, S.S.; Lee, W.; Lee, S.; Lee, K.H. Deep Learning-Based Automatic Detection Algorithm for Reducing Overlooked Lung Cancers on Chest Radiographs. Radiology 2020, 296, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Gordienko, Y.; Gang, P.; Hui, J.; Zeng, W.; Kochura, Y.; Alienin, O.; Rokovyi, O.; Stirenko, S. Deep Learning with Lung Segmentation and Bone Shadow Exclusion Techniques for Chest X-ray Analysis of Lung Cancer. In Advances in Intelligent Systems and Computing; Advances in intelligent systems and computing; Springer International Publishing: Cham, Switzerland, 2019; pp. 638–647. ISBN 9783319910079. [Google Scholar]
- Singh, V.; Danda, V.; Gorniak, R.; Flanders, A.; Lakhani, P. Assessment of Critical Feeding Tube Malpositions on Radiographs Using Deep Learning. J. Digit. Imaging 2019, 32, 651–655. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Kalra, M.K.; Nitiwarangkul, C.; Patti, J.A.; Homayounieh, F.; Padole, A.; Rao, P.; Putha, P.; Muse, V.V.; Sharma, A.; et al. Deep Learning in Chest Radiography: Detection of Findings and Presence of Change. PLoS ONE 2018, 13, e0204155. [Google Scholar] [CrossRef] [Green Version]
- Nam, J.G.; Park, S.; Hwang, E.J.; Lee, J.H.; Jin, K.-N.; Lim, K.Y.; Vu, T.H.; Sohn, J.H.; Hwang, S.; Goo, J.M.; et al. Development and Validation of Deep Learning-Based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology 2019, 290, 218–228. [Google Scholar] [CrossRef] [Green Version]
- Cicero, M.; Bilbily, A.; Colak, E.; Dowdell, T.; Gray, B.; Perampaladas, K.; Barfett, J. Training and Validating a Deep Convolutional Neural Network for Computer-Aided Detection and Classification of Abnormalities on Frontal Chest Radiographs. Investig. Radiol. 2017, 52, 281–287. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human-Computer Collaboration for Skin Cancer Recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xia, Y. ChestNet: A Deep Neural Network for Classification of Thoracic Diseases on Chest Radiography. arXiv 2018, arXiv:1807.03058. [Google Scholar]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key Challenges for Delivering Clinical Impact with Artificial Intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [Green Version]
- Geis, J.R.; Brady, A.P.; Wu, C.C.; Spencer, J.; Ranschaert, E.; Jaremko, J.L.; Langer, S.G.; Borondy Kitts, A.; Birch, J.; Shields, W.F.; et al. Ethics of Artificial Intelligence in Radiology: Summary of the Joint European and North American Multisociety Statement. Radiology 2019, 293, 436–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeGrave, A.J.; Janizek, J.D.; Lee, S.-I. AI for Radiographic COVID-19 Detection Selects Shortcuts over Signal. Nat. Mach. Intell. 2021, 3, 610–619. [Google Scholar] [CrossRef]
- Finding What Works in Health Care: Standards for Systematic Reviews; Eden, J.; Levit, L.; Berg, A.; Morton, S. (Eds.) National Academies Press: Washington, DC, USA, 2011; ISBN 9781283151757. [Google Scholar]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Int. J. Surg. 2010, 8, 336–341. [Google Scholar] [CrossRef] [Green Version]
- Wolff, R.F.; Moons, K.G.M.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 Explanation and Elaboration: Updated Guidance and Exemplars for Reporting Systematic Reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Jones, C.M.; Danaher, L.; Milne, M.R.; Tang, C.; Seah, J.; Oakden-Rayner, L.; Johnson, A.; Buchlak, Q.D.; Esmaili, N. Assessment of the Effect of a Comprehensive Chest Radiograph Deep Learning Model on Radiologist Reports and Patient Outcomes: A Real-World Observational Study. BMJ Open 2021, 11, e052902. [Google Scholar] [CrossRef]
- Ahn, J.S.; Ebrahimian, S.; McDermott, S.; Lee, S.; Naccarato, L.; Di Capua, J.F.; Wu, M.Y.; Zhang, E.W.; Muse, V.; Miller, B.; et al. Association of Artificial Intelligence-Aided Chest Radiograph Interpretation With Reader Performance and Efficiency. JAMA Netw. Open 2022, 5, e2229289. [Google Scholar] [CrossRef]
- Albahli, S.; Yar, G.N.A.H. Fast and Accurate Detection of COVID-19 along with 14 Other Chest Pathologies Using a Multi-Level Classification: Algorithm Development and Validation Study. J. Med. Internet Res. 2021, 23, e23693. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.P.; Morrison, P.; Dao, L.; Roth, K.; Duong, T.Q.; Ghassemi, M. COVID-19 Image Data Collection: Prospective Predictions Are the Future. arXiv 2020, arXiv:2006.11988. [Google Scholar]
- Altaf, F.; Islam, S.M.S.; Janjua, N.K. A Novel Augmented Deep Transfer Learning for Classification of COVID-19 and Other Thoracic Diseases from X-rays. Neural Comput. Appl. 2021, 33, 14037–14048. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-Scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 Image Data Collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Baltruschat, I.; Steinmeister, L.; Nickisch, H.; Saalbach, A.; Grass, M.; Adam, G.; Knopp, T.; Ittrich, H. Smart Chest X-ray Worklist Prioritization Using Artificial Intelligence: A Clinical Workflow Simulation. Eur. Radiol. 2021, 31, 3837–3845. [Google Scholar] [CrossRef] [PubMed]
- Bharati, S.; Podder, P.; Mondal, M.R.H. Hybrid Deep Learning for Detecting Lung Diseases from X-ray Images. Inform. Med. Unlocked 2020, 20, 100391. [Google Scholar] [CrossRef]
- Chakravarty, A.; Sarkar, T.; Ghosh, N.; Sethuraman, R.; Sheet, D. Learning Decision Ensemble Using a Graph Neural Network for Comorbidity Aware Chest Radiograph Screening. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2020, 2020, 1234–1237. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Proc. Conf. AAAI Artif. Intell. 2019, 33, 590–597. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Miao, S.; Xu, D.; Hager, G.D.; Harrison, A.P. Deep Hiearchical Multi-Label Classification Applied to Chest X-ray Abnormality Taxonomies. Med. Image Anal. 2020, 66, 101811. [Google Scholar] [CrossRef]
- Gohagan, J.K.; Prorok, P.C.; Hayes, R.B.; Kramer, B.-S. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: History, Organization, and Status. Control. Clin. Trials 2000, 21, 251S–272S. [Google Scholar] [CrossRef]
- Cho, Y.; Kim, Y.-G.; Lee, S.M.; Seo, J.B.; Kim, N. Reproducibility of Abnormality Detection on Chest Radiographs Using Convolutional Neural Network in Paired Radiographs Obtained within a Short-Term Interval. Sci. Rep. 2020, 10, 17417. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Park, B.; Lee, S.M.; Lee, K.H.; Seo, J.B.; Kim, N. Optimal Number of Strong Labels for Curriculum Learning with Convolutional Neural Network to Classify Pulmonary Abnormalities in Chest Radiographs. Comput. Biol. Med. 2021, 136, 104750. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Park, S.; Kim, M.; Park, J.; Choi, Y.R.; Jin, K.N. Evaluation of a Deep Learning-Based Computer-Aided Detection Algorithm on Chest Radiographs: Case-Control Study. Medicine (Baltimore) 2021, 100, e25663. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Xu, Y.; Zhao, Y.; Yan, Y.; Liu, J.; Liu, J. Weighing Features of Lung and Heart Regions for Thoracic Disease Classification. BMC Med. Imaging 2021, 21, 99. [Google Scholar] [CrossRef]
- Gipson, J.; Tang, V.; Seah, J.; Kavnoudias, H.; Zia, A.; Lee, R.; Mitra, B.; Clements, W. Diagnostic Accuracy of a Commercially Available Deep-Learning Algorithm in Supine Chest Radiographs Following Trauma. Br. J. Radiol. 2022, 95, 20210979. [Google Scholar] [CrossRef]
- Gündel, S.; Setio, A.A.A.; Ghesu, F.C.; Grbic, S.; Georgescu, B.; Maier, A.; Comaniciu, D. Robust Classification from Noisy Labels: Integrating Additional Knowledge for Chest Radiography Abnormality Assessment. Med. Image Anal. 2021, 72, 102087. [Google Scholar] [CrossRef]
- Han, Y.; Chen, C.; Tang, L.; Lin, M.; Jaiswal, A.; Wang, S.; Tewfik, A.; Shih, G.; Ding, Y.; Peng, Y. Using Radiomics as Prior Knowledge for Thorax Disease Classification and Localization in Chest X-rays. AMIA Annu. Symp. Proc. 2022, 2021, 546–555. [Google Scholar]
- Hwang, E.J.; Park, J.; Hong, W.; Lee, H.-J.; Choi, H.; Kim, H.; Nam, J.G.; Goo, J.M.; Yoon, S.H.; Lee, C.H.; et al. Artificial Intelligence System for Identification of False-Negative Interpretations in Chest Radiographs. Eur. Radiol. 2022, 32, 4468–4478. [Google Scholar] [CrossRef]
- Jabbour, S.; Fouhey, D.; Kazerooni, E.; Wiens, J.; Sjoding, M.W. Combining Chest X-rays and Electronic Health Record (EHR) Data Using Machine Learning to Diagnose Acute Respiratory Failure. J. Am. Med. Inform. Assoc. 2022, 29, 1060–1068. [Google Scholar] [CrossRef]
- Jadhav, A.; Wong, K.C.L.; Wu, J.T.; Moradi, M.; Syeda-Mahmood, T. Combining Deep Learning and Knowledge-Driven Reasoning for Chest X-ray Findings Detection. AMIA Annu. Symp. Proc. 2020, 2020, 593–601. [Google Scholar]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a Freely Accessible Critical Care Database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, K.N.; Kim, E.Y.; Kim, Y.J.; Lee, G.P.; Kim, H.; Oh, S.; Kim, Y.S.; Han, J.H.; Cho, Y.J. Diagnostic Effect of Artificial Intelligence Solution for Referable Thoracic Abnormalities on Chest Radiography: A Multicenter Respiratory Outpatient Diagnostic Cohort Study. Eur. Radiol. 2022, 32, 3469–3479. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Kim, Y.J.; Choi, W.-J.; Lee, G.P.; Choi, Y.R.; Jin, K.N.; Cho, Y.J. Correction: Performance of a Deep-Learning Algorithm for Referable Thoracic Abnormalities on Chest Radiographs: A Multicenter Study of a Health Screening Cohort. PLoS ONE 2021, 16, e0251045. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Kim, Y.J.; Choi, W.-J.; Jeon, J.S.; Kim, M.Y.; Oh, D.H.; Jin, K.N.; Cho, Y.J. Concordance Rate of Radiologists and a Commercialized Deep-Learning Solution for Chest X-ray: Real-World Experience with a Multicenter Health Screening Cohort. PLoS ONE 2022, 17, e0264383. [Google Scholar] [CrossRef]
- Kuo, P.-C.; Tsai, C.C.; López, D.M.; Karargyris, A.; Pollard, T.J.; Johnson, A.E.W.; Celi, L.A. Recalibration of Deep Learning Models for Abnormality Detection in Smartphone-Captured Chest Radiograph. NPJ Digit. Med. 2021, 4, 25. [Google Scholar] [CrossRef]
- Lee, M.S.; Han, S.W. DuETNet: Dual Encoder Based Transfer Network for Thoracic Disease Classification. Pattern Recognit. Lett. 2022, 161, 143–153. [Google Scholar] [CrossRef]
- Li, F.; Shi, J.-X.; Yan, L.; Wang, Y.-G.; Zhang, X.-D.; Jiang, M.-S.; Wu, Z.-Z.; Zhou, K.-Q. Lesion-Aware Convolutional Neural Network for Chest Radiograph Classification. Clin. Radiol. 2021, 76, 155.e1–155.e14. [Google Scholar] [CrossRef]
- Majkowska, A.; Mittal, S.; Steiner, D.F.; Reicher, J.J.; McKinney, S.M.; Duggan, G.E.; Eswaran, K.; Cameron Chen, P.-H.; Liu, Y.; Kalidindi, S.R.; et al. Chest Radiograph Interpretation with Deep Learning Models: Assessment with Radiologist-Adjudicated Reference Standards and Population-Adjusted Evaluation. Radiology 2020, 294, 421–431. [Google Scholar] [CrossRef]
- Mosquera, C.; Diaz, F.N.; Binder, F.; Rabellino, J.M.; Benitez, S.E.; Beresñak, A.D.; Seehaus, A.; Ducrey, G.; Ocantos, J.A.; Luna, D.R. Chest X-ray Automated Triage: A Semiologic Approach Designed for Clinical Implementation, Exploiting Different Types of Labels through a Combination of Four Deep Learning Architectures. Comput. Methods Programs Biomed. 2021, 206, 106130. [Google Scholar] [CrossRef]
- Nam, J.G.; Kim, M.; Park, J.; Hwang, E.J.; Lee, J.H.; Hong, J.H.; Goo, J.M.; Park, C.M. Development and Validation of a Deep Learning Algorithm Detecting 10 Common Abnormalities on Chest Radiographs. Eur. Respir. J. 2021, 57, 2003061. [Google Scholar] [CrossRef]
- Bustos, A.; Pertusa, A.; Salinas, J.-M.; de la Iglesia-Vayá, M. PadChest: A Large Chest X-ray Image Dataset with Multi-Label Annotated Reports. Med. Image Anal. 2020, 66, 101797. [Google Scholar] [CrossRef]
- Niehues, S.M.; Adams, L.C.; Gaudin, R.A.; Erxleben, C.; Keller, S.; Makowski, M.R.; Vahldiek, J.L.; Bressem, K.K. Deep-Learning-Based Diagnosis of Bedside Chest X-ray in Intensive Care and Emergency Medicine. Investig. Radiol. 2021, 56, 525–534. [Google Scholar] [CrossRef]
- Park, S.; Lee, S.M.; Lee, K.H.; Jung, K.-H.; Bae, W.; Choe, J.; Seo, J.B. Deep Learning-Based Detection System for Multiclass Lesions on Chest Radiographs: Comparison with Observer Readings. Eur. Radiol. 2020, 30, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Tang, Y.-X.; Shen, T.C.; Summers, R.M. Discriminative Ensemble Learning for Few-Shot Chest X-ray Diagnosis. Med. Image Anal. 2021, 68, 101911. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. Preparing a Collection of Radiology Examinations for Distribution and Retrieval. J. Am. Med. Inform. Assoc. 2016, 23, 304–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, H.H.; Le, T.T.; Tran, D.Q.; Ngo, D.T.; Nguyen, H.Q. Interpreting Chest X-rays via CNNs That Exploit Hierarchical Disease Dependencies and Uncertainty Labels. Neurocomputing 2021, 437, 186–194. [Google Scholar] [CrossRef]
- Rudolph, J.; Huemmer, C.; Ghesu, F.-C.; Mansoor, A.; Preuhs, A.; Fieselmann, A.; Fink, N.; Dinkel, J.; Koliogiannis, V.; Schwarze, V.; et al. Artificial Intelligence in Chest Radiography Reporting Accuracy: Added Clinical Value in the Emergency Unit Setting Without 24/7 Radiology Coverage. Investig. Radiol. 2022, 57, 90–98. [Google Scholar] [CrossRef]
- Rudolph, J.; Schachtner, B.; Fink, N.; Koliogiannis, V.; Schwarze, V.; Goller, S.; Trappmann, L.; Hoppe, B.F.; Mansour, N.; Fischer, M.; et al. Clinically Focused Multi-Cohort Benchmarking as a Tool for External Validation of Artificial Intelligence Algorithm Performance in Basic Chest Radiography Analysis. Sci. Rep. 2022, 12, 12764. [Google Scholar] [CrossRef]
- Seah, J.C.Y.; Tang, C.H.M.; Buchlak, Q.D.; Holt, X.G.; Wardman, J.B.; Aimoldin, A.; Esmaili, N.; Ahmad, H.; Pham, H.; Lambert, J.F.; et al. Effect of a Comprehensive Deep-Learning Model on the Accuracy of Chest X-ray Interpretation by Radiologists: A Retrospective, Multireader Multicase Study. Lancet Digit. Health 2021, 3, e496–e506. [Google Scholar] [CrossRef]
- Senan, E.M.; Alzahrani, A.; Alzahrani, M.Y.; Alsharif, N.; Aldhyani, T.H.H. Automated Diagnosis of Chest X-ray for Early Detection of COVID-19 Disease. Comput. Math. Methods Med. 2021, 2021, 6919483. [Google Scholar] [CrossRef]
- Sharma, A.; Rani, S.; Gupta, D. Artificial Intelligence-Based Classification of Chest X-ray Images into COVID-19 and Other Infectious Diseases. Int. J. Biomed. Imaging 2020, 2020, 8889023. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.-X.J.; Lu, P.-X.; Thoma, G. Two Public Chest X-ray Datasets for Computer-Aided Screening of Pulmonary Diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [PubMed]
- Sung, J.; Park, S.; Lee, S.M.; Bae, W.; Park, B.; Jung, E.; Seo, J.B.; Jung, K.-H. Added Value of Deep Learning-Based Detection System for Multiple Major Findings on Chest Radiographs: A Randomized Crossover Study. Radiology 2021, 299, 450–459. [Google Scholar] [CrossRef] [PubMed]
- van Beek, E.J.R.; Ahn, J.S.; Kim, M.J.; Murchison, J.T. Validation Study of Machine-Learning Chest Radiograph Software in Primary and Emergency Medicine. Clin. Radiol. 2022, 78, 1–7. [Google Scholar] [CrossRef]
- Verma, D.; Bose, C.; Tufchi, N.; Pant, K.; Tripathi, V.; Thapliyal, A. An Efficient Framework for Identification of Tuberculosis and Pneumonia in Chest X-ray Images Using Neural Network. Procedia Comput. Sci. 2020, 171, 217–224. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, W. MARnet: Multi-Scale Adaptive Residual Neural Network for Chest X-ray Images Recognition of Lung Diseases. Math. Biosci. Eng. 2021, 19, 331–350. [Google Scholar] [CrossRef]
- Wang, H.; Jia, H.; Lu, L.; Xia, Y. Thorax-Net: An Attention Regularized Deep Neural Network for Classification of Thoracic Diseases on Chest Radiography. IEEE J. Biomed. Health Inform. 2020, 24, 475–485. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Y.-Y.; Pan, Y.; Han, P.; Li, Z.-X.; Huang, H.-G.; Zhu, S.-Z. Detecting Thoracic Diseases via Representation Learning with Adaptive Sampling. Neurocomputing 2020, 406, 354–360. [Google Scholar] [CrossRef]
- Wang, H.; Wang, S.; Qin, Z.; Zhang, Y.; Li, R.; Xia, Y. Triple Attention Learning for Classification of 14 Thoracic Diseases Using Chest Radiography. Med. Image Anal. 2021, 67, 101846. [Google Scholar] [CrossRef]
- Xu, J.; Li, H.; Li, X. MS-ANet: Deep Learning for Automated Multi-Label Thoracic Disease Detection and Classification. PeerJ Comput. Sci. 2021, 7, e541. [Google Scholar] [CrossRef]
- Zhou, L.; Yin, X.; Zhang, T.; Feng, Y.; Zhao, Y.; Jin, M.; Peng, M.; Xing, C.; Li, F.; Wang, Z.; et al. Detection and Semiquantitative Analysis of Cardiomegaly, Pneumothorax, and Pleural Effusion on Chest Radiographs. Radiol. Artif. Intell. 2021, 3, e200172. [Google Scholar] [CrossRef] [PubMed]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Oakden-Rayner, L. Exploring Large-Scale Public Medical Image Datasets. Acad. Radiol. 2020, 27, 106–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard, K.; Roudsari, A.; Wyatt, J.C. Automation Bias: A Systematic Review of Frequency, Effect Mediators, and Mitigators. J. Am. Med. Inform. Assoc. 2012, 19, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosby, J.; Rhines, T.; Li, F.; MacMahon, H.; Giger, M. Deep Convolutional Neural Networks in the Classification of Dual-Energy Thoracic Radiographic Views for Efficient Workflow: Analysis on over 6500 Clinical Radiographs. J. Med. Imaging (Bellingham) 2020, 7, 016501. [Google Scholar] [CrossRef]
- Huang, S.-C.; Pareek, A.; Zamanian, R.; Banerjee, I.; Lungren, M.P. Multimodal Fusion with Deep Neural Networks for Leveraging CT Imaging and Electronic Health Record: A Case-Study in Pulmonary Embolism Detection. Sci. Rep. 2020, 10, 22147. [Google Scholar] [CrossRef]
- Morrison, A.; Polisena, J.; Husereau, D.; Moulton, K.; Clark, M.; Fiander, M.; Mierzwinski-Urban, M.; Clifford, T.; Hutton, B.; Rabb, D. The Effect of English-Language Restriction on Systematic Review-Based Meta-Analyses: A Systematic Review of Empirical Studies. Int. J. Technol. Assess. Health Care 2012, 28, 138–144. [Google Scholar] [CrossRef]
- Parikh, J.R.; Wolfman, D.; Bender, C.E.; Arleo, E. Radiologist Burnout According to Surveyed Radiology Practice Leaders. J. Am. Coll. Radiol. 2020, 17, 78–81. [Google Scholar] [CrossRef]
- Rosenkrantz, A.B.; Hughes, D.R.; Duszak, R., Jr. The U.S. Radiologist Workforce: An Analysis of Temporal and Geographic Variation by Using Large National Datasets. Radiology 2016, 279, 175–184. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.-Y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-Identified Publicly Available Database of Chest Radiographs with Free-Text Reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollura, D.J.; Azene, E.M.; Starikovsky, A.; Thelwell, A.; Iosifescu, S.; Kimble, C.; Polin, A.; Garra, B.S.; DeStigter, K.K.; Short, B.; et al. White Paper Report of the RAD-AID Conference on International Radiology for Developing Countries: Identifying Challenges, Opportunities, and Strategies for Imaging Services in the Developing World. J. Am. Coll. Radiol. 2010, 7, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Candemir, S.; Antani, S. A Review on Lung Boundary Detection in Chest X-rays. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 563–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.-C.; Bennett, C.; Piccardi, M.; Farrokhi, F. Ethical Thinking Machines in Surgery and the Requirement for Clinical Leadership. Am. J. Surg. 2020, 220, 1372–1374. [Google Scholar] [CrossRef]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The Practical Implementation of Artificial Intelligence Technologies in Medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef]
- Strohm, L.; Hehakaya, C.; Ranschaert, E.R.; Boon, W.P.C.; Moors, E.H.M. Implementation of Artificial Intelligence (AI) Applications in Radiology: Hindering and Facilitating Factors. Eur. Radiol. 2020, 30, 5525–5532. [Google Scholar] [CrossRef]
- Moradi, M.; Madani, A.; Karargyris, A.; Syeda-Mahmood, T.F. Chest X-ray Generation and Data Augmentation for Cardiovascular Abnormality Classification. In Proceedings of the Medical Imaging 2018: Image Processing, Houston, TX, USA, 10–15 February 2018; Angelini, E.D., Landman, B.A., Eds.; SPIE: San Francisco, CA, USA, 2018. [Google Scholar]
- Liu, W.-T.; Lin, C.-S.; Tsao, T.-P.; Lee, C.-C.; Cheng, C.-C.; Chen, J.-T.; Tsai, C.-S.; Lin, W.-S.; Lin, C. A Deep-Learning Algorithm-Enhanced System Integrating Electrocardiograms and Chest X-rays for Diagnosing Aortic Dissection. Can. J. Cardiol. 2022, 38, 160–168. [Google Scholar] [CrossRef]
- Nishimori, M.; Kiuchi, K.; Nishimura, K.; Kusano, K.; Yoshida, A.; Adachi, K.; Hirayama, Y.; Miyazaki, Y.; Fujiwara, R.; Sommer, P.; et al. Accessory Pathway Analysis Using a Multimodal Deep Learning Model. Sci. Rep. 2021, 11, 8045. [Google Scholar] [CrossRef]
- Lewis, A.; Mahmoodi, E.; Zhou, Y.; Coffee, M.; Sizikova, E. Improving Tuberculosis (TB) Prediction Using Synthetically Generated Computed Tomography (CT) Images. arXiv 2021, arXiv:2109.11480. [Google Scholar]

| Region | Modality | Methodology | Task | Performance | |
|---|---|---|---|---|---|
| Keyword | chest thora * cardiorespiratory | CXR X-ray radiograph | artificial intelligence machine learning neural net * radiomics supervised learning random forest naive bayes CNN convolution * | diagnosis image interpretation radiographic image interpretation decision Support system classif * screen detect * interpret * identifi * | diagnos * prognos * inferiority validat * superiority predict * reader * decision * clinical * risk * classif * performance bootstrapping split sample area under the curve ROC AUC performance sensitiv * accura * |
| MeSH Terms | machine learning artificial intelligence Neural Networks, Computer | diagnosis roc curve sensitivity and specificity triage |
| Study | Study Aim | Study Design | Datasets Used | Dataset Size | Number of Pathologies Investigated |
|---|---|---|---|---|---|
| Ahn et al., 2022 [43] | Evaluate whether a deep-learning–based AI engine used concurrently can improve reader performance and efficiency in interpreting CXR abnormalities | Retrospective reader study | Two sources: MIMIC-CXR (public) and MGH (private) | MIMIC-CXR: 247 images; MGH: 250 images | 4 |
| Albahli et al., 2021 [44] | To achieve a fast and more accurate diagnosis of COVID-19 | Retrospective | COVID Chest X-ray dataset [45] | 112,812 | 15 |
| Altaf et al., 2021 [46] | Classify thoracic pathologies | Retrospective | Chest X-ray14 [47], COVID-19 CXRs [48] | 112,777 | 14 |
| Baltruschat et al., 2021 [49] | Evaluate whether smart worklist prioritization by AI can optimize radiology workflow and reduce report turnaround times (RTATs) for critical findings in CXRs | Retrospective workflow simulation study | Chest X-ray14 (public) (112,120), Open-I dataset (public) (3125) | 112,120 + 3125 images | 8 |
| Bharati et al., 2020 [50] | Develop a new hybrid deep learning algorithm suitable for predicting lung disease from CXR images | Retrospective | Chest X-ray14 [47] | 112,120 | 14 |
| Chakravarty et al., 2020 [51] | Develop a CXR pathology classifier | Retrospective | CheXpert [52] | 223,648 | 13 |
| Chen et al., 2020 [53] | Present a deep hierarchical multi-label classification approach for CXRs | Retrospective | PLCO dataset [54] | 198,000 | 19 |
| Cho et al., 2020 [55] | Evaluate the reproducibility of CADs with a CNN on CXRs of abnormal pulmonary patterns in patients | Retrospective | - | 9792 | 5 |
| Cho et al., 2020 [56] | Develop a convolutional neural network to differentiate normal and five types of pulmonary abnormalities in CXRs | Retrospective | - | 9534 | 5 |
| Choi et al., 2021 [57] | Evaluate the deep-learning-based CAD algorithm for detecting and localizing three major thoracic abnormalities on CXRs and compare the performance of physicians with and without the assistance of the algorithm | Reader study using retrospective data | - | 244 | 3 |
| Fang et al., 2021 [58] | Propose a deep learning framework to explore discriminative information from lung and heart regions | Retrospective | Chest X-ray14 [47] | 112,120 | 14 |
| Gipson et al., 2022 [59] | Evaluate the performance of a commercially available deep CNN for detection of traumatic injuries on supine CXRs | Retrospective | Internal dataset (private) | 1404 patients/images | 7 |
| Gündel et al., 2021 [60] | Train high performing CXR abnormality classifiers | Retrospective | Chest X-ray14 [47], PLCO [54] | 297,541 | 17 |
| Han et al., 2022 [61] | Develop ChexRadiNet to utilize radiomics features to improve abnormality classification performance | Retrospective | Chest X-ray14 | 112,120 images | 14 |
| Hwang et al., 2022 [62] | Investigate the efficacy of utilizing AI for the identification and correction of false-negative interpretations in consecutive CXRs that were initially read as normal by radiologists | Retrospective feasibility study | Dataset from Seoul National University Hospital (private) | 4208 images | 3 |
| Jabbour et al., 2022 [63] | Validate a model to act as a diagnostic aid in the evaluation of patients with acute respiratory failure combining CXR and EHR data | Retrospective | CheXpert and MIMIC-CXR-DICOM (public) | 1618 patients | 3 |
| Jadhav et al., 2020 [64] | Predict a large set of CXR findings using a deep neural network classifier and improve prediction outcomes using a knowledge-driven reasoning algorithm | Retrospective | MIMIC [65] | 339,558 | 54 |
| Jin et al., 2022 [66] | Evaluate a commercial AI solution on a multicenter cohort of CXRs and compare physicians’ ability to detect and localize referable thoracic abnormalities with and without AI assistance | Retrospective reader study | Dataset from respiratory outpatient clinics (private) | 6006 patients/images | 3 |
| Jones et al., 2021 [42] | Evaluate the real-world usefulness of the model as a diagnostic assistance device for radiologists | Real-world prospective reader study | Internal dataset (private) | 2972 cases | 124 |
| Kim et al., 2021 [67] | Test the performance of a commercial algorithm | Retrospective generalizability study | - | 5887 | 3 |
| Kim et al., 2022 [68] | Evaluate the concordance rate of radiologists and a commercially available AI for thoracic abnormalities in a multicenter health screening cohort | Retrospective reader study | Health screening dataset (private) | 3113 patients/images | 3 |
| Kuo et al., 2021 [69] | Explore combining deep learning and smartphones for CXR-finding detection | Retrospective generalizability study | CheXpert [52], MIMIC [65] | 6453 | 6 |
| Lee et al., 2022 [70] | Create a model that counters the effects of memory inefficiency caused by input size and treats high class imbalance | Retrospective | ChestX-ray14 and MIMIC-CXR (public) | Training—77,871 images, Testing—25,596 + 227,827 images | 14 |
| Li et al., 2021 [71] | Investigate the performance of a deep learning approach termed lesion-aware CNN to identify 14 different thoracic diseases on CXRs | Retrospective | Chest X-ray14 [47] | 10,738 | 14 |
| Majkowska et al., 2020 [72] | Develop and evaluate deep learning models for CXR interpretation by using radiologist-adjudicated reference standards | Retrospective | Chest X-ray14 [47] | 871,731 | 4 |
| Mosquera et al., 2021 [73] | Present a deep learning method based on the fusion of different convolutional architectures that allows training with heterogeneous data with a simple implementation and evaluates its performance on independent test data | Retrospective | Chest X-ray14 [47] | 5440 | 4 |
| Nam et al., 2021 [74] | Develop a deep learning algorithm detecting 10 common abnormalities on CXRs and evaluate its impact on diagnostic accuracy, timeliness of reporting, and workflow efficacy | Reader study using retrospective data | PadChest [75] | 146,717 | 10 |
| Niehues et al., 2021 [76] | Develop and evaluate deep learning models for the identification of clinically relevant abnormalities in bedside CXRs | Retrospective | - | 18,361 | 8 |
| Park et al., 2020 [77] | Investigate the feasibility of a deep-learning–based detection system for multiclass lesions on CXRs, in comparison with observers | Reader study using retrospective data | - | 15,809 | 4 |
| Paul et al., 2021 [78] | Propose a method for few-shot diagnosis of diseases and conditions from CXRs using discriminative ensemble learning | Retrospective | Chest X-ray14 [47], Openi [79] | >112,000 | 14 |
| Pham et al., 2021 [80] | Present a supervised multi-label classification framework based on CNNs for predicting the presence of 14 common thoracic diseases | Retrospective | CheXpert [52] | 224,316 | 13 |
| Rudolph et al., 2022 [81] | Develop an AI system that aims to mimic board-certified radiologists’ performance and support non–radiology residents in clinical settings lacking 24/7 radiology coverage | Retrospective reader study | EU CXR dataset (private) | 563 images | 4 |
| Rudolph et al., 2022 [82] | Investigate multiple clinically relevant aspects that might influence algorithm performance, considering patient positioning, reference standards, and comparison to medical expert performance | Retrospective reader study | 3 cohorts (private) | 3 cohorts: 563 images, 6258 images, and 166 patients, respectively | 7 |
| Seah et al., 2021 [83] | Assess the accuracy of radiologists with and without the assistance of a deep learning model | Reader study using retrospective data | MIMIC [65], PadChest [75], Chest X-ray14 [47], CheXpert [52] | 821,681 | 124 |
| Senan et al., 2021 [84] | Introduce two deep learning models, ResNet-50 and AlexNet, to diagnose X-ray datasets collected from many sources | Retrospective | Chest X-ray dataset comprising images from several public sources | 21,165 images | 4 |
| Sharma et al., 2020 [85] | Create efficient deep learning models, trained with CXR images, for rapid screening of COVID-19 patients | Retrospective | Montgomery County X-ray Set [86] | 352 | 4 |
| Sung et al., 2021 [87] | Evaluate effects of a deep learning system on radiologist pathology detection | Reader study using retrospective data | - | 228 | 5 |
| Van Beek et al., 2022 [88] | Evaluate the performance of a machine-learning-based algorithm tool for CXRs, applied to a consecutive cohort of historical clinical cases, in comparison to expert radiologists | Retrospective reader study | Internal training dataset (private) from primary care and ED settings | Training—168,056 images, Testing—1960 images | 10 |
| Verma et al., 2020 [89] | Implementation of computer-aided image analysis for identifying and discriminating tuberculosis, bacterial pneumonia, and viral pneumonia | Retrospective | Shenzhen chest X-ray set [86] | 5894 | 3 |
| Wang et al., 2021 [90] | Construct a multi-scale adaptive residual neural network (MARnet) to identify CXR images of lung diseases and compare MARnet with classical neural networks | Retrospective | Chest X-ray14 | 13,382 images | 4 |
| Wang et al., 2020 [91] | Propose a novel deep convolutional neural network called Thorax-Net to diagnose 14 thorax diseases using CXRs | Retrospective | Chest X-ray14 [47] | 112,120 | 14 |
| Wang et al., 2021 [92] | Propose the triple-attention learning (A 3 Net) model | Retrospective | Chest X-ray14 [47] | 112,120 | 14 |
| Wang et al., 2020 [93] | Use deep learning techniques to develop a multi-class CXR classifier | Retrospective | Chest X-ray14 [47] | 112,120 | 14 |
| Wu et al., 2020 [11] | Assess the performance of AI algorithms in realistic radiology workflows by performing an objective comparative evaluation of the preliminary reads of AP CXRs performed by an AI algorithm and radiology residents | Reader study using retrospective data | CheXpert [52], MIMIC [65] | 342,126 | 72 |
| Xu et al., 2020 [94] | Explore a multi-label classification algorithm for medical images to help doctors identify lesions | Retrospective | Chest X-ray14 [47] | 112,120 | 14 |
| Zhou et al., 2021 [95] | Develop and evaluate deep learning models for the detection and semiquantitative analysis of cardiomegaly, pneumothorax, and pleural effusion on chest radiographs | Retrospective | Montgomery County Department of Health and Human Services, Shenzhen No. 3 People’s Hospital [86] | 2838 | 3 |
| Study | Appropriate Study Design | Appropriate Comparators | Appropriate Training Dataset | Appropriate Validation Methods | Appropriate Sample Size | Appropriate Metric Used to Measure Performance | Appropriate Statistics Methods Used to Measure Performance | Study Quality Score |
|---|---|---|---|---|---|---|---|---|
| Ahn et al., 2022 [43] | Yes | Yes | Yes | NA | Yes | Yes | Yes | 70 |
| Albahli et al., 2021 [44] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Altaf et al., 2021 [46] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Baltruschat et al., 2021 [49] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Bharati et al., 2020 [50] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Chakravarty et al., 2020 [51] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Chen et al., 2020 [53] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Cho et al., 2020 [55] | Yes | Yes | NA | Yes | Yes | Yes | Yes | 70 |
| Cho et al., 2020 [56] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Choi et al., 2021 [57] | Yes | Yes | NA | Yes | Yes | Yes | Yes | 70 |
| Fang et al., 2021 [58] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Gipson et al., 2022 [59] | Yes | Yes | Yes | NA | Yes | Yes | Yes | 70 |
| Gündel et al., 2021 [60] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Han et al., 2022 [61] | Yes | Yes | Yes | NA | Yes | Yes | Yes | 70 |
| Hwang et al., 2022 [62] | Yes | Yes | NA | NA | Yes | Yes | Yes | 70 |
| Jabbour et al., 2022 [63] | No | No | No | NA | Yes | Yes | No | 30 |
| Jadhav et al., 2020 [64] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Jin et al., 2022 [66] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Jones et al., 2021 [42] | Yes | Yes | Yes | NA | Yes | Yes | Yes | 70 |
| Kim et al., 2021 [67] | Yes | No | NA | NA | Yes | Yes | Yes | 60 |
| Kim et al., 2022 [68] | Yes | Yes | Yes | NA | Yes | Yes | Yes | 70 |
| Kuo et al., 2021 [69] | Yes | Yes | NA | Yes | Yes | Yes | Yes | 70 |
| Lee et al., 2022 [70] | Yes | Yes | Yes | Yes | Yes | Yes | No | 60 |
| Li et al., 2021 [71] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Majkowska et al., 2020 [72] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Mosquera et al., 2021 [73] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Nam et al., 2021 [74] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Niehues et al., 2021 [76] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Park et al., 2020 [77] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Paul et al., 2021 [78] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Pham et al., 2021 [80] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Rudolph et al., 2022 [81] | Yes | Yes | No | Yes | Yes | Yes | Yes | 60 |
| Rudolph et al., 2022 [82] | Yes | No | No | Yes | No | Yes | Yes | 40 |
| Seah et al., 2021 [83] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Senan et al., 2021 [84] | Yes | No | Yes | NA | Yes | Yes | Yes | 60 |
| Sharma et al., 2020 [85] | Yes | No | No | Yes | Yes | Yes | Yes | 50 |
| Sung et al., 2021 [87] | Yes | Yes | NA | NA | Yes | Yes | Yes | 70 |
| Van Beek et al., 2022 [88] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Verma et al., 2020 [89] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Wang et al., 2021 [90] | Yes | No | Yes | Yes | Yes | Yes | No | 50 |
| Wang et al., 2020 [91] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Wang et al., 2021 [92] | Yes | No | Yes | Yes | Yes | Yes | Yes | 60 |
| Wang et al., 2020 [93] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Wu et al., 2020 [11] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Xu et al., 2020 [94] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Zhou et al., 2021 [95] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 70 |
| Deep Learning Model | Model Architecture | Model Validation Process | Model Performance and Study Results |
|---|---|---|---|
| Ahn et al., 2022 [43] | Not specified | Not specified—commercial | AI was associated with higher sensitivity for all findings compared with readers (nodule, 0.816 vs. 0.567; pneumonia, 0.887 vs. 0.673; pleural effusion, 0.872 vs. 0.889; pneumothorax, 0.988 vs. 0.792) |
| Albahli et al., 2021 [44] | Unet, NasNetLarge, Xception, Inception-V3, Inception-ResNetV2, ResNet50 | Train, test | Test accuracy: 0.66 and 0.62 |
| Altaf et al., 2021 [46] | DenseNet-201, ResNet50, Inception-V3, VGG-16 | Train, test with cross-validation | Specificity 0.95, sensitivity 0.65, F1 0.53, accuracy 0.91 |
| Baltruschat et al., 2021 [49] | ResNet-50 | 5-fold resampling scheme—7:1:2 (training, validation, testing) | Average RTAT for all critical findings was significantly reduced in all prioritization simulations compared to the first-in-first-out simulation, while the maximum RTAT for most findings increased. Pneumothorax (Min/Max) 80.1/890 vs. 35.6/1178, congestion 80.5/916 vs. 45.3/2018, pleural effusion 80.5/932 vs. 54.6/2144, infiltrate 80.3/916 vs. 59.1/2144, atelectasis 80.4/906 vs. 61.7/1958, cardiomegaly 80.5/932 vs. 62.5/1698, mass 81.0/902 vs. 64.3/1556, foreign object 80.4/930 vs. 80.6/2093, normal 80.2/940 vs. 113.9/2093 |
| Bharati et al., 2020 [50] | VDSNet | Train, test | Accuracy 0.73 |
| Chakravarty et al., 2020 [51] | DenseNet-121 | Train, test | Average AUC 0.82 |
| Chen et al., 2020 [53] | DenseNet-121 | Train, test | Average AUC 0.89 |
| Cho et al., 2020 [55] | eDenseYOLO | Train, tune, test (7:1:2) | Percent positive agreement: 83.39%, 74.14%, 95.12%, 96.84%, and 84.58% |
| Cho et al., 2020 [56] | ResNet-50 | Train, tune, test (7:1:2) | Accuracy: 0.90, 0.90, 0.91, 0.92, and 0.93 |
| Choi et al., 2021 [57] | Insight CXR, Lunit | - | Average AUC 0.99, sensitivity 0.97, specificity 0.93, and accuracy of 0.96. The model outperformed board-certified radiologists, non-radiology physicians, and general practitioners. Average AUC of physicians was 0.87 without model assistance and 0.91 with model assistance |
| Fang et al., 2021 [58] | CXR-IRNet | Train, test, validation | Average AUC 0.83 |
| Gipson et al., 2022 [59] | EfficientNet architecture, segmentation CNN based on U-Net/EfficientNet backbone | Not specified | AI superior to radiologists for pneumothorax (AI AUC = 0.926, sens. = 39.2%, spec. = 99.8%, FP n = 2, p = 0.007) and lobar/segmental collapse (AI AUC = 0.917, sens. = 36.1%, spec. = 98.5%, FP n = 21, p = 0.012). AI inferior for clavicle (AI AUC = 0.831, sens. = 55.7%, spec. = 97.2%, FP n = 37, p = 0.002), humerus (AI AUC = 0.836, sens. = 32.3%, spec. = 99.4%, FP n = 8, p < 0.001), and scapular fracture (AI AUC = 0.855, sens. = 34.6%, spec. = 95.2%, FP n = 64, p = 0.014). No sig. diff. for rib fracture (AI AUC = 0.749, sens. = 41.1%, spec. = 92.9%, FP n = 75, k = 0.39) and pneumomediastinum (AUC = 0.872, sens. = 11.1%, spec. = 100%, FP n = 0, k = 0.19) |
| Gündel et al., 2021 [60] | DenseNet architecture | Train, test, validation | Average AUC 0.88 |
| Han et al., 2022 [61] | ChexRadiNet | Not specified | The model achieved AUC scores of 0.831, 0.934, 0.817, 0.906, 0.892, 0.925, 0.798, 0.882, 0.734, 0.846, 0.748, 0.867, 0.737, and 0.889, respectively, for the pathologies (atelectasis, cardiomegaly, consolidation, edema, effusion, emphysema, fibrosis, hernia, infiltration, mass, nodule, pleural thickening, pneumonia, and pneumothorax) |
| Hwang et al., 2022 [62] | Not specified | Not specified—commercial | 16.5% of scans initially labeled normal classified abnormal by model. 103/591 were clinically relevant (488 false positives). 13.3% of detected abnormalities accepted by radiologist. Situation (a) AI as the advisor: detection yield = 1.2%, FRR = 0.97%, PPV = 55.4%. Situation (b) AI as the final consultant: detection yield = 2.4%, FRR = 14%, PPV = 14.8%. Higher net benefit of AI as an advisor |
| Jabbour et al., 2022 [63] | CNN with DenseNet-121 architecture | External validation | Pneumonia: (k = 0.47) (AUC: combined int. = 0.71, ext. = 0.65) (Combined sens. = 81%, spec. = 60%), heart failure: (k = 0.48) (AUC: combined int. = 0.82, ext. = 0.82) (Combined sens. = 62%, spec. = 83%), COPD: (k = 0.56) (AUC: combined int. = 0.76, ext. = 0.86) (Combined sens. = 68%, spec. = 94%). Combined model sensitivity higher than both other models, lower specificity than both other models. Combined model AUROC higher than physician for heart failure (0.79 vs. 0.77) and COPD (0.89 vs. 0.78), lower for pneumonia (0.74 vs. 0.75) |
| Jadhav et al., 2020 [64] | VGGNet (16 layers), ResNet (50 layers) | Train, validation, test (7:1:2) | Precision 0.85, recall 0.83, F1 0.84 |
| Jin et al., 2022 [66] | Not specified | Not specified—commercial | Standalone model performance average sensitivity, specificity, and AUC of 0.885, 0.723, and 0.867, respectively. For readers, average AUC and AUAFROC significantly increased with AI assistance (from 0.861 to 0.886; p = 0.003 and from 0.797 to 0.822; p = 0.003, respectively) |
| Jones et al., 2021 [42] | EfficientNet architecture, segmentation CNN based on U-Net/EfficientNet backbone | Not specified | 90% of radiologists reported increased reporting accuracy with model by radiologists w/ (a) 5> yrs. experience, (b) 6–10 yrs. exp., (c) 10+ yrs. Exp., (a) 5% (b) 1.3% (c) 1.6% rate of sig. report change with model, (a) 2.4% (b) 0.4% (c) 0.9% patient management change, and (a) 1.5% (b) 0.5% (c) 0.6% increase in recommendations for further imaging. No sig. impact of radiologist experience on these rates |
| Kim et al., 2021 [67] | Insight CXR, Lunit | - | Sensitivity 0.83, specificity 0.79 |
| Kim et al., 2022 [68] | ResNet-34 based architecture | Not specified—commercial | Thoracic abnormalities were found in 343 cases (11.0%) based on the CXR radiology reports and 621 (20.1%) based on the Lunit results. The concordance rate was 86.8% (accept: 85.3%, edit: 0.9%, and add: 0.6%), and the discordance rate was 13.2%. The median reading time increased after the clinical integration of Lunit (median, 19 s vs. 14 s, p < 0.001) |
| Kuo et al., 2021 [69] | - | Train, test, validation and external validation | Average AUC 0.75 |
| Lee et al., 2022 [70] | DuETNet: DenseNet backbone, dual encoder | Train, validation, test | Model superior to all other models. AUC: atelectasis = 0.7711, cardiomegaly = 0.914, effusion = 0.8197, infiltration = 0.7096, mass = 0.8582, nodule = 0.8223, pneumonia = 0.8928, pneumothorax = 0.8805, consolidation = 0.7976, edema = 0.8892, emphysema = 0.9331, fibrosis = 0.93, PT = 0.8493, hernia = 0.997, average AUC = 0.8617 |
| Li et al., 2021 [71] | ResNet-38 | Train, test, validation with 10-fold cross-validation | The model generated statistically significant higher AUC performance compared with radiologists on atelectasis, mass, and nodule, with AUC values of 0.83, 0.96, and 0.93, respectively. For the other 11 pathologies, there were no statistically significant differences |
| Majkowska et al., 2020 [72] | - | Train, validation, test | AUCs 0.94, 0.91, 0.94, and 0.81 |
| Mosquera et al., 2021 [73] | RetinaNet, Inception-ResnetV2, AlbuNet-34 | Train, test, external validation | AUCs 0.75 and 0.87, sensitivity 0.86, specificity 0.88 |
| Nam et al., 2021 [74] | ResNet34 | - | Model AUCs 0.90–1.00 (CT-confirmed dataset) and 0.91–1.00 (PadChest dataset). The model correctly classified significantly more critical abnormalities (95%) than radiologists (84%). Radiologists detected significantly more critical (71% vs. 29%) and urgent (83% vs. 78%) abnormalities when aided by the model |
| Niehues et al., 2021 [76] | - | Train, test | AUCs 0.90, 0.95, 0.85, 0.92, 0.99, 0.99, 0.98, and 0.99 |
| Park et al., 2020 [77] | - | Train, validation, test | AUC 0.99 vs. 0.96 |
| Paul et al., 2021 [78] | DenseNet | Train, test, external validation | AUCs 0.55–0.79 |
| Pham et al., 2021 [80] | - | Train, validation, test | Average AUC 0.94 (validation set), 0.93 (test set) |
| Rudolph et al., 2022 [81] | Not specified | External validation | AUC of 0.940 (pneumothorax), 0.953 (pleural effusion), 0.883 (lung lesions), and 0.847 (consolidation). The AI system matched radiology residents’ performance, and significantly outperformed non-radiology residents’ diagnostic accuracy for pneumothorax, pleural effusion, and lung lesions |
| Rudolph et al., 2022 [82] | CheXNet | External validation | CheXNet was similar to radiology resident (RR) detection of suspicious lung nodules (cohort, AUC AI/RR: 0.851/0.839, p = 0.793), basal pneumonia (cohort, AUC AI/reader consensus: 0.825/0.782, p = 0.390), and basal pleural effusion (cohort, AUC AI/reader consensus: 0.762/0.710, p = 0.336) |
| Seah et al., 2021 [83] | EfficientNet | Train, test | Average AUC of the model 0.96. Average AUC of unassisted radiologists 0.72. Average AUC when radiologists used the model 0.81. Model use significantly improved accuracy for 102 (80%) clinical findings |
| Senan et al., 2021 [84] | ResNet-50 and AlexNet | Not specified | The ResNet-50 network reached average accuracy, sensitivity, specificity, and AUC of 95%, 94.5%, 98%, and 97.10%, respectively |
| Sharma et al., 2020 [85] | Not reported | Train, test, external validation | Accuracies 1.00, 1.00, 0.95, 0.00, 0.94, and 0.00 |
| Sung et al., 2021 [87] | Med-Chest X-ray | - | AUC of radiologists using the model (from 0.93 to 0.98), sensitivity (from 0.83 to 0.89), and specificity (from 0.89 to 0.97) |
| Van Beek et al., 2022 [88] | ResNet34 basis, AutoAugment + Attend and Compare modules, binary cross-entropy loss function | Not specified—commercial | Atelectasis (AUC = 0.914, 0.891, sens. = 0.816, 0.55, spec. = 0.887, 0.961), calcification (AUC = 0.92, 0.922, sens. = 0.765, 0.692, spec. = 0.887, 0.919), cardiomegaly (AUC = 0.943, 0.97, sens. = 0.88, 0.85, spec. = 0.884, 0.962), consolidation (AUC = 0.903, 0.881, sens. = 0.886, 0.922, spec. = 0.792, 0.674), fibrosis (AUC = 0.948, 0.92, sens. = 0.933, 0.714, spec. = 0.895, 0.924), mediastinal widening (AUC = 0.909, 0.998, sens. = 0.8, 1, spec. = 0.97, 0.993), nodule (AUC = 0.881, 0.905, sens. = 0.794, 0.833, spec. = 0.848, 0.886), pleural effusion (AUC = 0.954, 0.988, sens. = 0.784, 0.837, spec. = 0.942, 0.986), pneumoperitoneum (AUC = 0.999, insuff. case no., sens. = 1, --, spec. = 0.975, 0.996), pneumothorax (AUC = 0.954, insuff. case no., sens. = 0.833, 1, spec. = 0.978, 0.992). Non-significant difference of performance in acute and non-acute sources; model outperformed radiologists for all findings |
| Verma et al., 2020 [89] | Not reported | Train, test | Accuracy 0.99 |
| Wang et al., 2021 [90] | MARnet | 5-fold cross-validation | AUC: nodule 0.90, atelectasis 0.93, normal 0.99, infection 1.00. MARnet outperformed all other CNNs |
| Wang et al., 2020 [91] | Thorax-Net | Train, test, validation | Average AUC 0.79 and 0.90 |
| Wang et al., 2021 [92] | DenseNet-121 | Train, test, validation | Average AUC 0.83 |
| Wang et al., 2020 [93] | DenseNet-121 | Train, validation | Average AUC 0.82 |
| Wu et al., 2020 [11] | ResNet50, HVGG16 S | Train, validation, test | Average AUC: model 0.77, residents 0.72. PPV: model 0.73, residents 0.68. Specificity: model 0.98, residents 0.97 |
| Xu et al., 2020 [94] | MS-ANet | Train, test, validation | Average AUCs 0.85 and 0.82 |
| Zhou et al., 2021 [95] | - | Training, test (9:1) | Accuracy for cardiomegaly 0.98, pneumothorax 0.71, and pleural effusion 0.78 |
| Identified Benefit | Clinical Setting | Associated Clinical Outcomes | Reference |
|---|---|---|---|
| Improved radiologist accuracy in detecting pathology on the medical image | All clinical settings | Reduced unnecessary and increased appropriate follow-up CT examinations or earlier detection of findings, leading to improved patient outcomes. Reduction in false positives | Choi et al., 2021 [57] Nam et al., 2021 [74] Seah et al., 2021 [83] Sung et al., 2021 [87] Jin et al., 2022 [66] Jones et al., 2021 [42] Hwang et al., 2022 [62] Ahn et al., 2022 [43] |
| Reduced time to report studies that contain critical pathology | All clinical settings | Reduction in report turnaround time for sensitive findings such as pneumothorax and rib fracture allowing correct patient management and earlier treatment | Nam et al., 2021 [74] Baltruschat et al., 2021 [49] |
| Reduced per-study reporting time | Inpatient or outpatient or screening | Increased reporting efficiency | Nam et al., 2021 [74] Sung et al., 2021 [87] Ahn et al., 2022 [43] |
| Consistent detection accuracy across variations in image quality | Inpatient or outpatient or screening or emergency | Accurate detection of pathology on CXRs regardless of imaging source or quality of the acquisition | Choi et al., 2021 [57] Gündel et al., 2021 [60] Rudolph et al., 2022 [82] van Beek et al., 2022 [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, H.K.; Milne, M.R.; Buchlak, Q.D.; Ektas, N.; Sanderson, G.; Chamtie, H.; Karunasena, S.; Chiang, J.; Holt, X.; Tang, C.H.M.; et al. Machine Learning Augmented Interpretation of Chest X-rays: A Systematic Review. Diagnostics 2023, 13, 743. https://doi.org/10.3390/diagnostics13040743
Ahmad HK, Milne MR, Buchlak QD, Ektas N, Sanderson G, Chamtie H, Karunasena S, Chiang J, Holt X, Tang CHM, et al. Machine Learning Augmented Interpretation of Chest X-rays: A Systematic Review. Diagnostics. 2023; 13(4):743. https://doi.org/10.3390/diagnostics13040743
Chicago/Turabian StyleAhmad, Hassan K., Michael R. Milne, Quinlan D. Buchlak, Nalan Ektas, Georgina Sanderson, Hadi Chamtie, Sajith Karunasena, Jason Chiang, Xavier Holt, Cyril H. M. Tang, and et al. 2023. "Machine Learning Augmented Interpretation of Chest X-rays: A Systematic Review" Diagnostics 13, no. 4: 743. https://doi.org/10.3390/diagnostics13040743
APA StyleAhmad, H. K., Milne, M. R., Buchlak, Q. D., Ektas, N., Sanderson, G., Chamtie, H., Karunasena, S., Chiang, J., Holt, X., Tang, C. H. M., Seah, J. C. Y., Bottrell, G., Esmaili, N., Brotchie, P., & Jones, C. (2023). Machine Learning Augmented Interpretation of Chest X-rays: A Systematic Review. Diagnostics, 13(4), 743. https://doi.org/10.3390/diagnostics13040743




