Performance Evaluation of a BZ COVID-19 NALF Assay for Rapid Diagnosis of SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Study Design
2.2. BZ COVID-19 NALF Assay
2.3. Allplex™ SARS-CoV-2 Assay
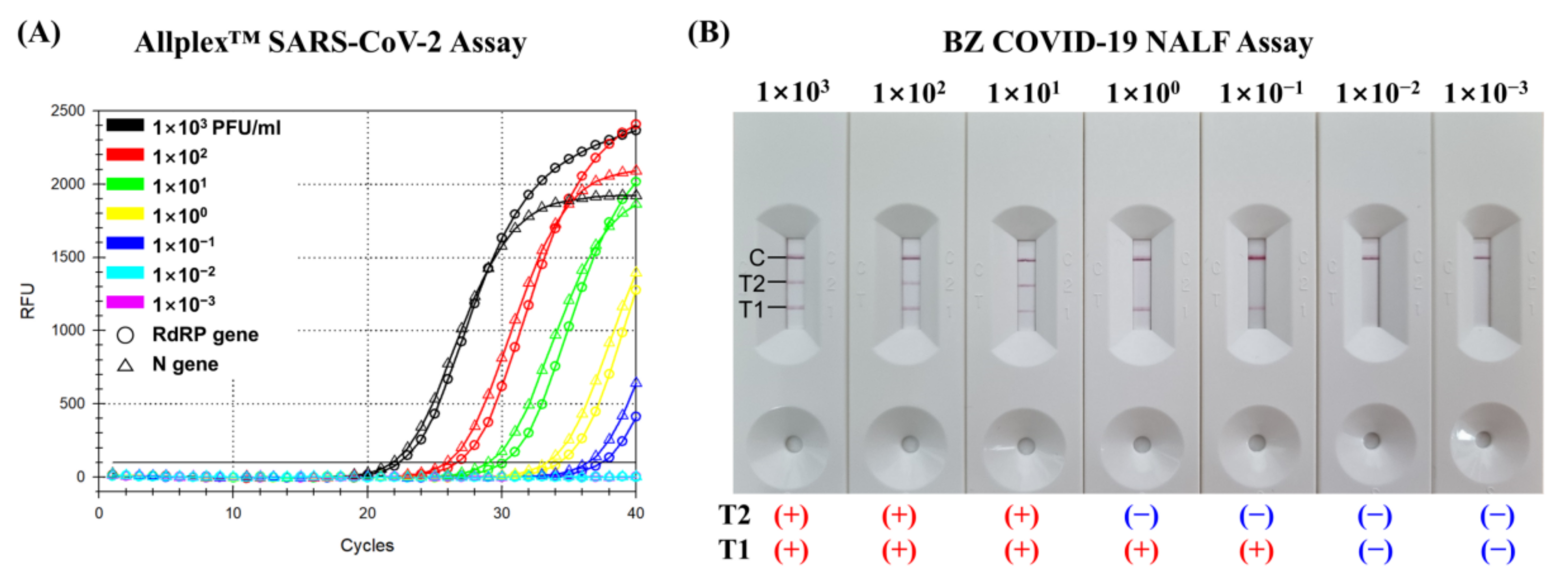
2.4. Limit of Detection
2.5. Statistical Analysis
3. Results
3.1. Limit of Detection of the BZ COVID-19 NALF Assay
3.2. Clinical Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hui, D.S.; I Azhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The Continuing 2019-NCoV Epidemic Threat of Novel Coronaviruses to Global Health—The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamadian, M.; Chiti, H.; Shoghli, A.; Biglari, S.; Parsamanesh, N.; Esmaeilzadeh, A. COVID-19: Virology, Biology and Novel Laboratory Diagnosis. J. Gene Med. 2021, 23, e3303. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 10 January 2022).
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The Reproductive Number of COVID-19 Is Higher Compared to SARS Coronavirus. J. Travel Med. 2020, 27, taaa021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, S.S.; Attrish, D.; Ghosh, S.; Choudhury, P.P.; Uversky, V.N.; Aljabali, A.A.A.; Lundstrom, K.; Uhal, B.D.; Rezaei, N.; Seyran, M.; et al. Notable Sequence Homology of the ORF10 Protein Introspects the Architecture of SARS-CoV-2. Int. J. Biol. Macromol. 2021, 181, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-W.; Wu, X.-X.; Jiang, X.-G.; Xu, K.-J.; Ying, L.-J.; Ma, C.-L.; Li, S.-B.; Wang, H.-Y.; Zhang, S.; Gao, H.-N.; et al. Clinical Findings in a Group of Patients Infected with the 2019 Novel Coronavirus (SARS-CoV-2) Outside of Wuhan, China: Retrospective Case Series. BMJ 2020, 368, m606. [Google Scholar] [CrossRef] [Green Version]
- Cirulli, E.T.; Schiabor Barrett, K.M.; Riffle, S.; Bolze, A.; Neveux, I.; Dabe, S.; Grzymski, J.J.; Lu, J.T.; Washington, N.L. Long-Term COVID-19 Symptoms in a Large Unselected Population. medRxiv 2020. [Google Scholar] [CrossRef]
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 Variant: A New Chapter in the COVID-19 Pandemic. Lancet 2021, 398, 2126–2128. [Google Scholar] [CrossRef]
- Taleghani, N.; Taghipour, F. Diagnosis of COVID-19 for Controlling the Pandemic: A Review of the State-of-the-Art. Biosens. Bioelectron. 2021, 174, 112830. [Google Scholar] [CrossRef]
- Pecoraro, V.; Negro, A.; Pirotti, T.; Trenti, T. Estimate False-negative RT-PCR Rates for SARS-CoV-2. A Systematic Review and Meta-analysis. Eur. J. Clin. Investig. 2022, 52, e13706. [Google Scholar] [CrossRef]
- Kilic, T.; Weissleder, R.; Lee, H. Molecular and Immunological Diagnostic Tests of COVID-19: Current Status and Challenges. iScience 2020, 23, 101406. [Google Scholar] [CrossRef]
- Nyaruaba, R.; Mwaliko, C.; Hong, W.; Amoth, P.; Wei, H. SARS-CoV-2/COVID-19 Laboratory Biosafety Practices and Current Molecular Diagnostic Tools. J. Biosaf. Biosecur. 2021, 3, 131–140. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Muenchhoff, M.; Mairhofer, H.; Nitschko, H.; Grzimek-Koschewa, N.; Hoffmann, D.; Berger, A.; Rabenau, H.; Widera, M.; Ackermann, N.; Konrad, R.; et al. Multicentre Comparison of Quantitative PCR-Based Assays to Detect SARS-CoV-2, Germany, March 2020. Eurosurveillance 2020, 25, 2001057. [Google Scholar] [CrossRef]
- Kahn, M.; Schuierer, L.; Bartenschlager, C.; Zellmer, S.; Frey, R.; Freitag, M.; Dhillon, C.; Heier, M.; Ebigbo, A.; Denzel, C.; et al. Performance of Antigen Testing for Diagnosis of COVID-19: A Direct Comparison of a Lateral Flow Device to Nucleic Acid Amplification Based Tests. BMC Infect. Dis. 2021, 21, 798. [Google Scholar] [CrossRef]
- Häuser, F.; Sprinzl, M.F.; Dreis, K.J.; Renzaho, A.; Youhanen, S.; Kremer, W.M.; Podlech, J.; Galle, P.R.; Lackner, K.J.; Rossmann, H.; et al. Evaluation of a Laboratory-Based High-Throughput SARS-CoV-2 Antigen Assay for Non-COVID-19 Patient Screening at Hospital Admission. Med. Microbiol. Immunol. 2021, 210, 165–171. [Google Scholar] [CrossRef]
- Park, Y.; Hong, K.H.; Lee, S.-K.; Hyun, J.; Oh, E.-J.; Lee, J.; Lee, H.; Song, S.H.; Kee, S.-J.; Kwon, G.C.; et al. Performance Comparison of Five SARS-CoV-2 Antibody Assays for Seroprevalence Studies. Ann. Lab. Med. 2022, 42, 71–78. [Google Scholar] [CrossRef]
- Kamphee, H.; Chaiprasert, A.; Prammananan, T.; Wiriyachaiporn, N.; Kanchanatavee, A.; Dharakul, T. Rapid Molecular Detection of Multidrug-Resistant Tuberculosis by PCR-Nucleic Acid Lateral Flow Immunoassay. PLoS ONE 2015, 10, e0137791. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Wang, K.; Zheng, W.; Cheng, Y.; Li, T.; Cao, B.; Jin, Q.; Cui, D. Rapid Developments in Lateral Flow Immunoassay for Nucleic Acid Detection. Analyst 2021, 146, 1514–1528. [Google Scholar] [CrossRef]
- Anupama, K.P.; Chakraborty, A.; Karunasagar, I.; Karunasagar, I.; Maiti, B. Loop-Mediated Isothermal Amplification Assay as a Point-of-Care Diagnostic Tool for Vibrio Parahaemolyticus: Recent Developments and Improvements. Expert Rev. Mol. Diagn. 2019, 19, 229–239. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Yoon, J.; Kim, A.; Lim, M.; Nam, J.; Yanagihara, R.; Ryu, S.-W.; Jung, B.K.; Ryoo, N.-H.; et al. Development of a Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for on-Site Diagnosis of SARS CoV-2. PLoS ONE 2021, 16, e0248042. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, Y.P.; Choy, K.W.; Doerig, C.; Lim, C.X. SARS-CoV-2 Testing Strategies in the Diagnosis and Management of COVID-19 Patients in Low-Income Countries: A Scoping Review. Mol. Diagn. Ther. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cai, K.; Zhang, R.; He, X.; Shen, X.; Liu, J.; Xu, J.; Qiu, F.; Lei, W.; Wang, J.; et al. Novel One-Step Single-Tube Nested Quantitative Real-Time PCR Assay for Highly Sensitive Detection of SARS-CoV-2. Anal. Chem. 2020, 92, 9399–9404. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Wu, X.; Wan, Z.; Li, Y.; Zuo, L.; Qin, J.; Jin, X.; Zhang, C. Development of a Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Virol. Sin. 2020, 35, 344–347. [Google Scholar] [CrossRef] [Green Version]
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef]
- Hu, X.; Deng, Q.; Li, J.; Chen, J.; Wang, Z.; Zhang, X.; Fang, Z.; Li, H.; Zhao, Y.; Yu, P.; et al. Development and Clinical Application of a Rapid and Sensitive Loop-Mediated Isothermal Amplification Test for SARS-CoV-2 Infection. mSphere 2020, 5, e00808-20. [Google Scholar] [CrossRef]
- Rödel, J.; Egerer, R.; Suleyman, A.; Sommer-Schmid, B.; Baier, M.; Henke, A.; Edel, B.; Löffler, B. Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J. Clin. Virol. 2020, 132, 104616. [Google Scholar] [CrossRef]
- Kitajima, H.; Tamura, Y.; Yoshida, H.; Kinoshita, H.; Katsuta, H.; Matsui, C.; Matsushita, A.; Arai, T.; Hashimoto, S.; Iuchi, A.; et al. Clinical COVID-19 diagnostic methods: Comparison of reverse transcription loop-mediated isothermal amplification (RT-LAMP) and quantitative RT-PCR (qRT-PCR). J. Clin. Virol. 2021, 139, 104813. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Y.; Ding, L.; Huang, X.; Xiong, Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. Trends Anal. Chem. 2021, 145, 116452. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Sakai-Tagawa, Y.; Koga, M.; Akasaka, O.; Nakachi, I.; Koh, H.; Maeda, K.; Adachi, E.; Saito, M.; Nagai, H.; et al. Comparison of Rapid Antigen Tests for COVID-19. Viruses 2020, 12, 1420. [Google Scholar] [CrossRef]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.-P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for COVID-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Heskin, J.; Pallett, S.J.C.; Al-Hindawi, A.; Davies, G.W.; Rayment, M.; Mughal, N.; Randell, P.; Jones, R.; Moore, L.S.P. Evaluating the Performance Characteristics of Five Lateral Flow Assays for the Detection of the SARS-CoV-2 Nucleocapsid Antigen. Sci. Rep. 2022, 12, 8811. [Google Scholar] [CrossRef]
- Irsara, C.; Egger, A.E.; Prokop, W.; Nairz, M.; Loacker, L.; Sahanic, S.; Pizzini, A.; Sonnweber, T.; Mayer, W.; Schennach, H.; et al. Evaluation of Four Commercial, Fully Automated SARS-CoV-2 Antibody Tests Suggests a Revision of the Siemens SARS-CoV-2 IgG Assay. Clin. Chem. Lab. Med. (CCLM) 2021, 59, 1143–1154. [Google Scholar] [CrossRef]

| SARS-Cov-2 | Allplex™ SARS-CoV-2 Assay | BZ COVID-19 NALF Assay | ||
|---|---|---|---|---|
| Detected | Not Detected | Detected | Not Detected | |
| Positive samples | 180 | 2 | 173 | 9 |
| Negative samples | 0 | 207 | 2 | 205 |
| Clinical Samples | Threshold Cycle (Ct) | Allplex™ SARS-CoV-2 Assay | BZ COVID-19 NALF Assay |
|---|---|---|---|
| Positive Samples (n = 182) | 10.00–19.99 | 53 Detected | 53 Detected |
| 20.00–29.99 | 85 Detected | 84 Detected/1 Not detected | |
| 30.00–39.99 | 42 Detected | 36 Detected/6 Not detected | |
| No amplification | 2 Not detected | 2 Not Detected | |
| Negative Samples (n = 207) | No amplification | 207 Not detected | 2 Detected/205 Not detected |
| Sample | Allplex™ SARS-CoV-2 Assay | BZ COVID-19 NALF Assay |
|---|---|---|
| 1 | Detected (Ct = 29.81) | Not detected |
| 2 | Detected (Ct = 36.57) | Not detected |
| 3 | Detected (Ct = 37.79) | Not detected |
| 4 | Detected (Ct = 37.69) | Not detected |
| 5 | Detected (Ct = 36.19) | Not detected |
| 6 | Detected (Ct = 36.65) | Not detected |
| 7 | Detected (Ct = 39.27) | Not detected |
| 8 | Not detected | Detected |
| 9 | Not detected | Detected |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, W.S.; Jee, H.; Lee, J.M.; Lim, C.S.; Kim, J. Performance Evaluation of a BZ COVID-19 NALF Assay for Rapid Diagnosis of SARS-CoV-2. Diagnostics 2023, 13, 1118. https://doi.org/10.3390/diagnostics13061118
Jang WS, Jee H, Lee JM, Lim CS, Kim J. Performance Evaluation of a BZ COVID-19 NALF Assay for Rapid Diagnosis of SARS-CoV-2. Diagnostics. 2023; 13(6):1118. https://doi.org/10.3390/diagnostics13061118
Chicago/Turabian StyleJang, Woong Sik, Hyunseul Jee, Joon Min Lee, Chae Seung Lim, and Jeeyong Kim. 2023. "Performance Evaluation of a BZ COVID-19 NALF Assay for Rapid Diagnosis of SARS-CoV-2" Diagnostics 13, no. 6: 1118. https://doi.org/10.3390/diagnostics13061118
APA StyleJang, W. S., Jee, H., Lee, J. M., Lim, C. S., & Kim, J. (2023). Performance Evaluation of a BZ COVID-19 NALF Assay for Rapid Diagnosis of SARS-CoV-2. Diagnostics, 13(6), 1118. https://doi.org/10.3390/diagnostics13061118







